Impact of Ivermectin on the Gut Microbial Ecosystem
Abstract
:1. Introduction
2. Results and Discussion
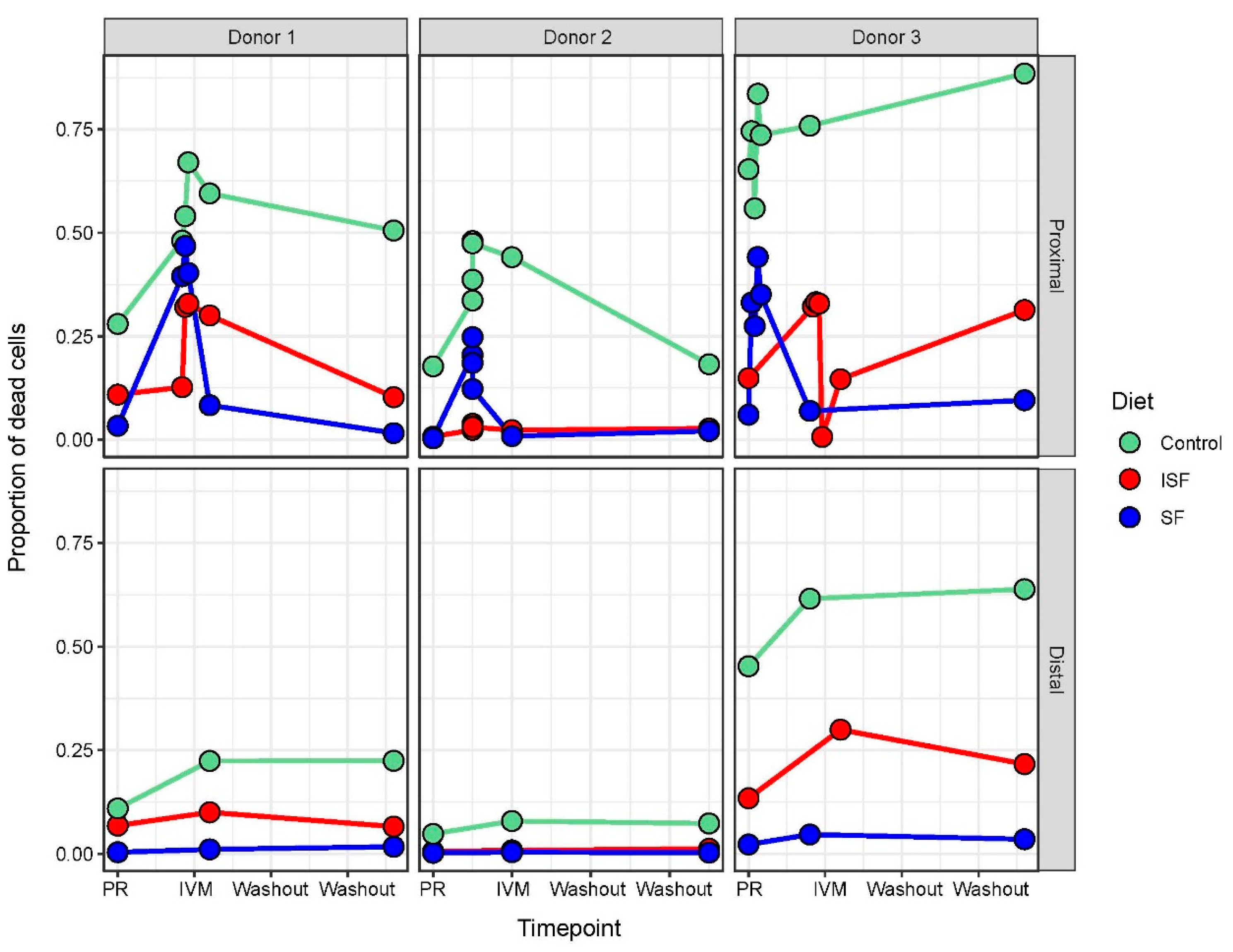
2.1. IVM Administration Increased Cell Mortality in the Proximal Colon Region
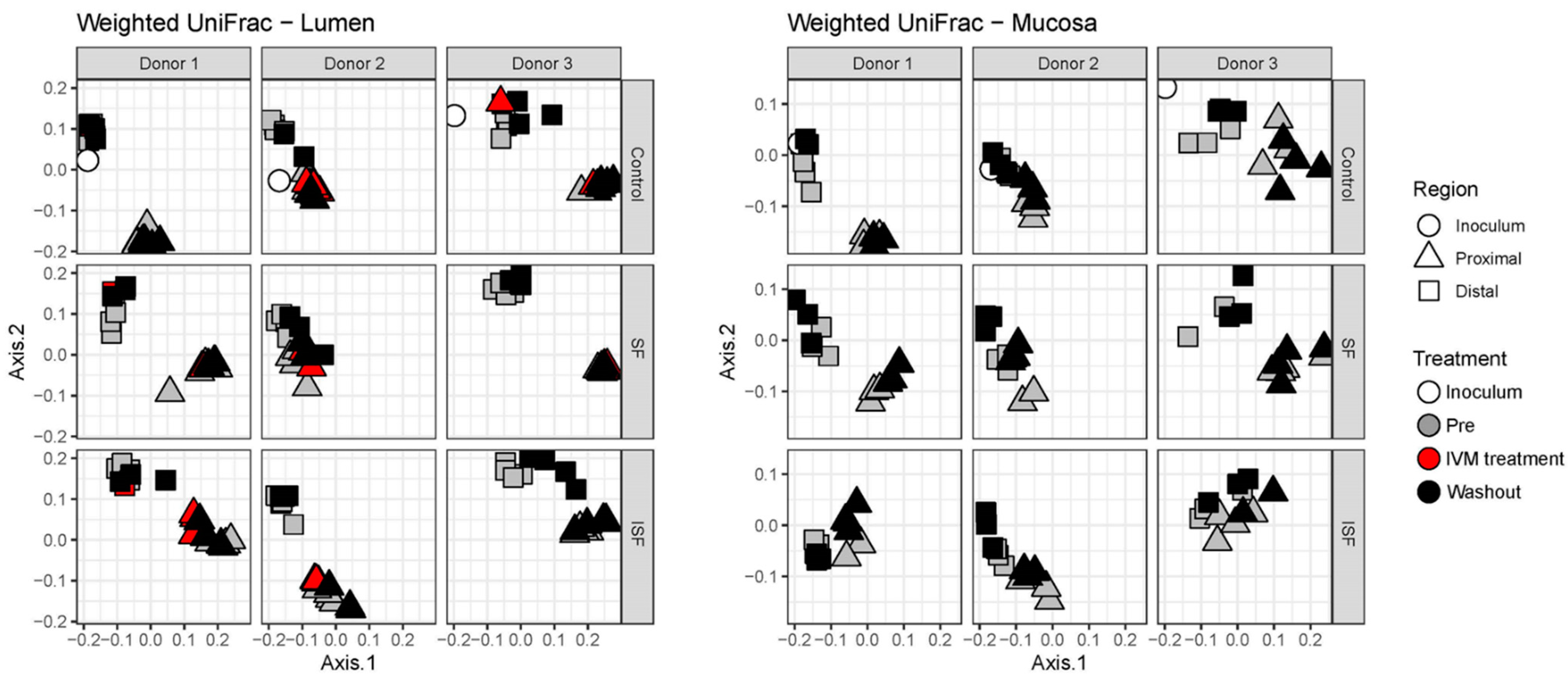
2.2. Effect of IVM on the Gut Microbiota Structure in Terms of Alpha and Beta Diversity
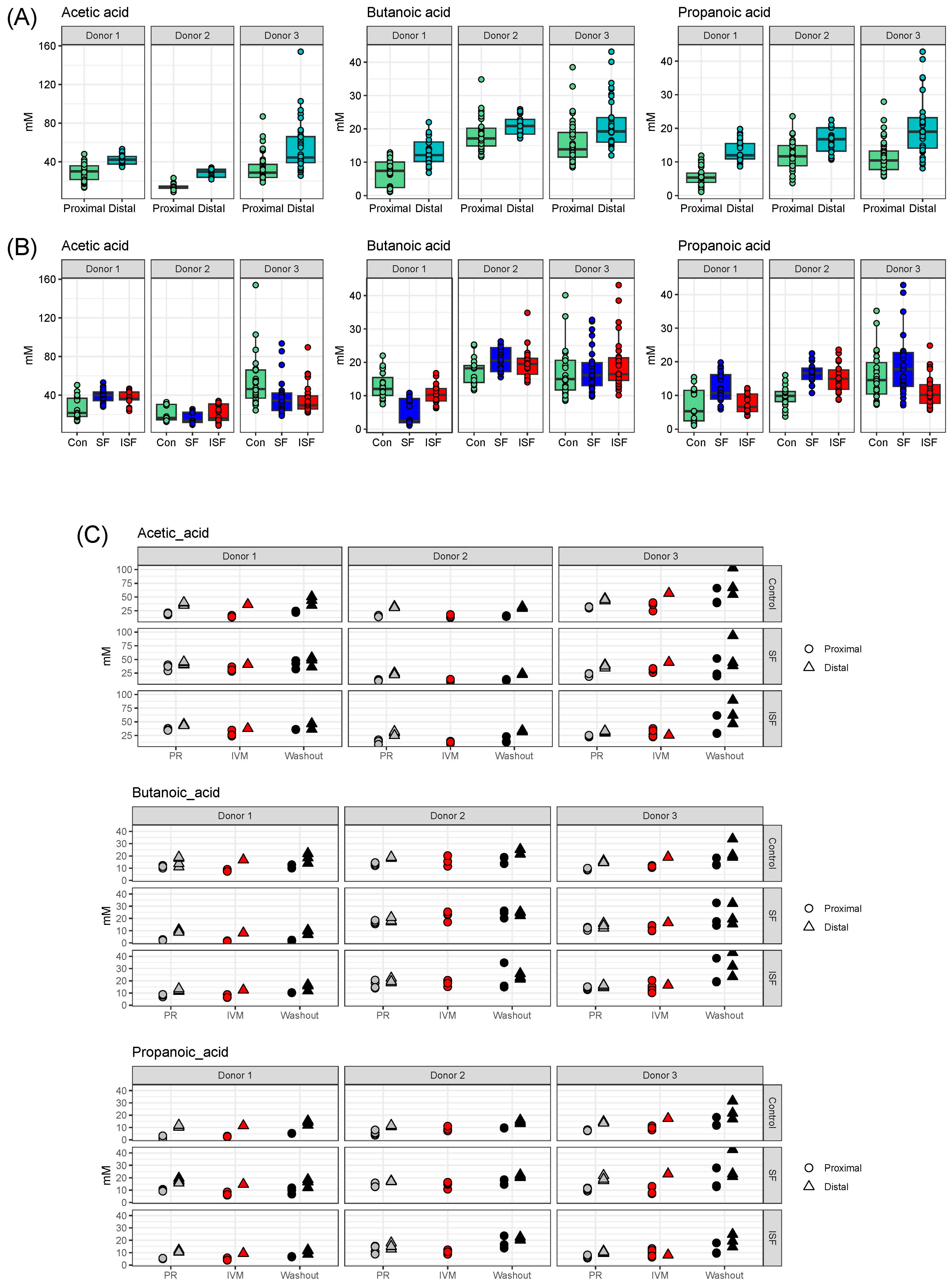
2.3. Effect of IVM on the Functionality of the Gut Microbiota
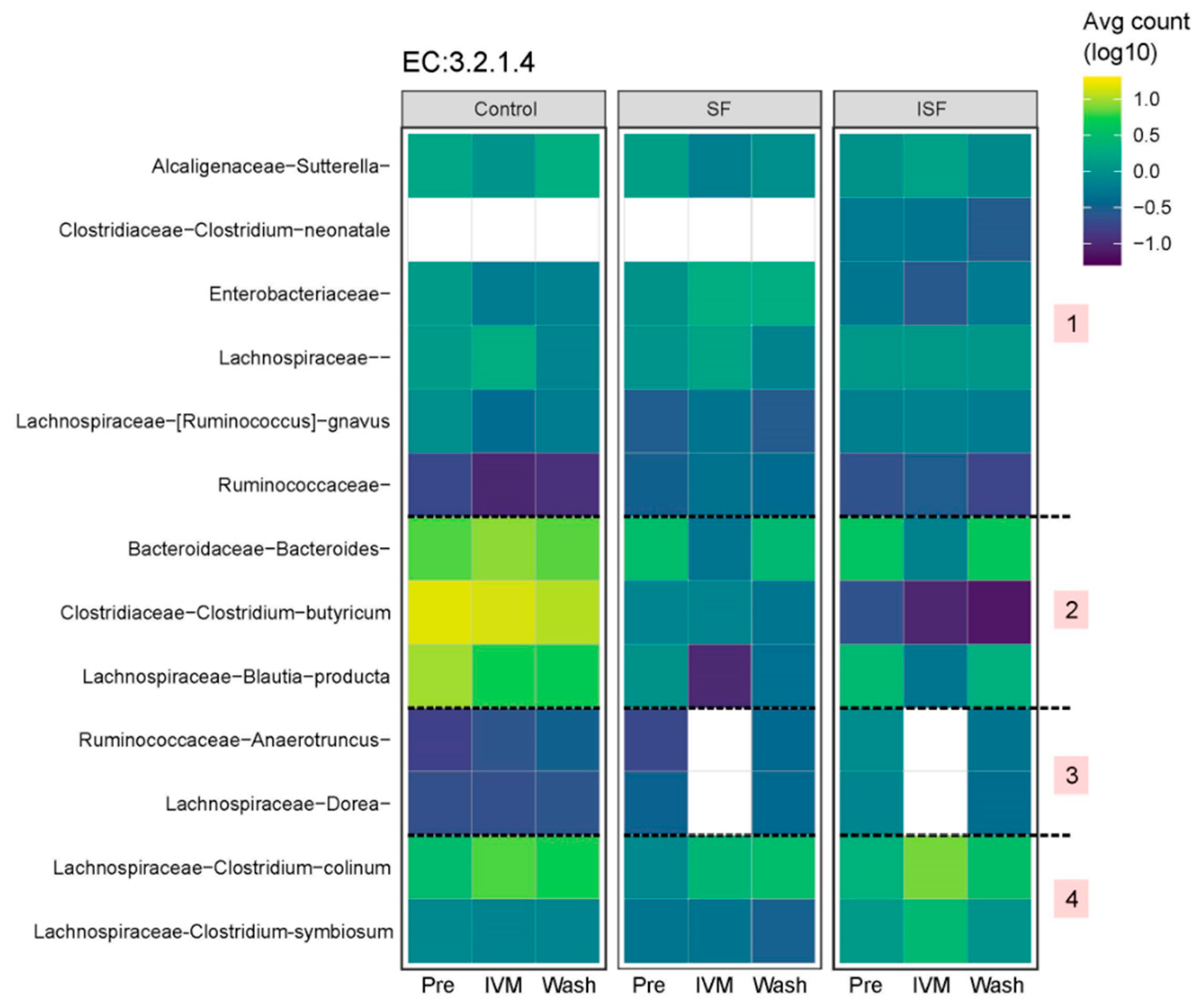
2.4. Predicting Functional Pathways
3. Materials and Methods
3.1. Materials
3.2. Triple SHIME® In Vitro Experiment
3.3. 16S rRNA Sequencing
3.4. Flow Cytometry
3.5. Bioinformatics and Statistical Analysis
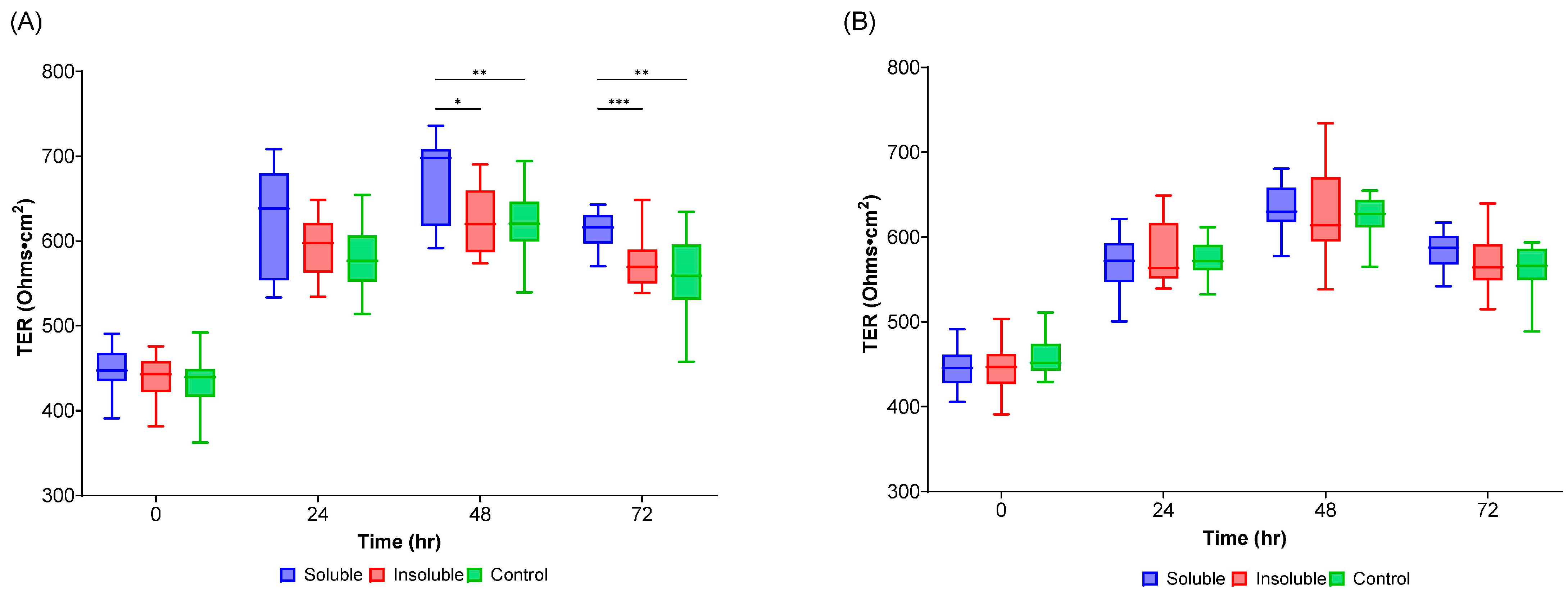
3.6. Transepithelial Electrical Resistance (TEER) Measurement
3.7. Short-Chain Fatty Acid (SCFA) Determination
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Crump, A.; Omura, S. Ivermectin, ‘wonder drug’ from Japan: The human use perspective. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2011, 87, 13–28. [Google Scholar] [CrossRef] [PubMed]
- Roman, Y.M.; Burela, P.A.; Pasupuleti, V.; Piscoya, A.; Vidal, J.E.; Hernandez, A.V. Ivermectin for the Treatment of Coronavirus Disease 2019: A Systematic Review and Meta-analysis of Randomized Controlled Trials. Clin. Infect. Dis. 2022, 74, 1022–1029. [Google Scholar] [CrossRef]
- Martin, R.J.; Robertson, A.P.; Choudhary, S. Ivermectin: An Anthelmintic, an Insecticide, and Much More. Trends Parasitol. 2021, 37, 48–64. [Google Scholar] [CrossRef]
- Sharpton, T.J.; Combrink, L.; Arnold, H.K.; Gaulke, C.A.; Kent, M. Harnessing the gut microbiome in the fight against anthelminthic drug resistance. Curr. Opin. Microbiol. 2020, 53, 26–34. [Google Scholar] [CrossRef]
- Foy, B.D.; Alout, H.; Seaman, J.A.; Rao, S.; Magalhaes, T.; Wade, M.; Parikh, S.; Soma, D.D.; Sagna, A.B.; Fournet, F.; et al. Efficacy and risk of harms of repeat ivermectin mass drug administrations for control of malaria (RIMDAMAL): A cluster-randomised trial. Lancet 2019, 393, 1517–1526. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.R.; Ricci, F.M.; Ottesen, E.A. Ivermectin: Effectiveness in lymphatic filariasis. Parasitology 2000, 12, S133–S146. [Google Scholar] [CrossRef]
- De Souza, D.K.; Otchere, J.; Ahorlu, C.S.; Adu-Amankwah, S.; Larbi, I.A.; Dumashie, E.; McCarthy, F.A.; King, S.A.; Otoo, S.; Osabutey, D.; et al. Low Microfilaremia Levels in Three Districts in Coastal Ghana with at Least 16 Years of Mass Drug Administration and Persistent Transmission of Lymphatic Filariasis. Trop. Med. Infect. Dis. 2018, 3, 105. [Google Scholar] [CrossRef] [PubMed]
- Byakika-Kibwika, P.; Kamya, M.R.; Nankabirwa, J. Ivermectin for mass drug administration against malaria. Lancet Infect. Dis. 2022, 22, 433–435. [Google Scholar] [CrossRef]
- Llinas-Caballero, K.; Caraballo, L. Helminths and Bacterial Microbiota: The Interactions of Two of Humans’ “Old Friends”. Int. J. Mol. Sci. 2022, 23, 13358. [Google Scholar] [CrossRef]
- Rashid, M.; Rashid, M.I.; Akbar, H.; Ahmad, L.; Hassan, M.A.; Ashraf, K.; Saeed, K.; Gharbi, M. A systematic review on modelling approaches for economic losses studies caused by parasites and their associated diseases in cattle. Parasitology 2019, 146, 129–141. [Google Scholar] [CrossRef]
- Ola-Fadunsin, S.D.; Ganiyu, I.A.; Rabiu, M.; Hussain, K.; Sanda, I.M.; Baba, A.Y.; Furo, N.A.; Balogun, R.B. Helminth infections of great concern among cattle in Nigeria: Insight to its prevalence, species diversity, patterns of infections and risk factors. Vet. World 2020, 13, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.S.; Kubo, Y. Ivermectin and its target molecules: Shared and unique modulation mechanisms of ion channels and receptors by ivermectin. J. Physiol. 2018, 596, 1833–1845. [Google Scholar] [CrossRef]
- Loghry, H.J.; Yuan, W.; Zamanian, M.; Wheeler, N.J.; Day, T.A.; Kimber, M.J. Ivermectin inhibits extracellular vesicle secretion from parasitic nematodes. J. Extracell. Vesicles 2020, 10, e12036. [Google Scholar] [CrossRef] [PubMed]
- Drurey, C.; Maizels, R.M. Helminth extracellular vesicles: Interactions with the host immune system. Mol. Immunol. 2021, 137, 124–133. [Google Scholar] [CrossRef]
- Zhang, X.; Song, Y.; Ci, X.; An, N.; Ju, Y.; Li, H.; Wang, X.; Han, C.; Cui, J.; Deng, X. Ivermectin inhibits LPS-induced production of inflammatory cytokines and improves LPS-induced survival in mice. Inflamm. Res. 2008, 57, 524–529. [Google Scholar] [CrossRef]
- Wagstaff, K.M.; Sivakumaran, H.; Heaton, S.M.; Harrich, D.; Jans, D.A. Ivermectin is a specific inhibitor of importin alpha/beta-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 2012, 443, 851–856. [Google Scholar] [CrossRef]
- Zaidi, A.K.; Dehgani-Mobaraki, P. The mechanisms of action of ivermectin against SARS-CoV-2-an extensive review. J. Antibiot. 2022, 75, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Nabi-Afjadi, M.; Mohebi, F.; Zalpoor, H.; Aziziyan, F.; Akbari, A.; Moradi-Sardareh, H.; Bahreini, E.; Moeini, A.M.; Effatpanah, H. A cellular and molecular biology-based update for ivermectin against COVID-19: Is it effective or non-effective? Inflammopharmacology 2023, 31, 21–35. [Google Scholar] [CrossRef]
- Tang, M.; Hu, X.; Wang, Y.; Yao, X.; Zhang, W.; Yu, C.; Cheng, F.; Li, J.; Fang, Q. Ivermectin, a potential anticancer drug derived from an antiparasitic drug. Pharmacol. Res. 2021, 163, 105207. [Google Scholar] [CrossRef]
- Sharmeen, S.; Skrtic, M.; Sukhai, M.A.; Hurren, R.; Gronda, M.; Wang, X.; Fonseca, S.B.; Sun, H.; Wood, T.E.; Ward, R.; et al. The antiparasitic agent ivermectin induces chloride-dependent membrane hyperpolarization and cell death in leukemia cells. Blood 2010, 116, 3593–3603. [Google Scholar] [CrossRef]
- Juarez, M.; Schcolnik-Cabrera, A.; Duenas-Gonzalez, A. The multitargeted drug ivermectin: From an antiparasitic agent to a repositioned cancer drug. Am. J. Cancer Res. 2018, 8, 317–331. [Google Scholar]
- Chen, L.; Bi, S.; Wei, Q.; Zhao, Z.; Wang, C.; Xie, S. Ivermectin suppresses tumour growth and metastasis through degradation of PAK1 in oesophageal squamous cell carcinoma. J. Cell. Mol. Med. 2020, 24, 5387–5401. [Google Scholar] [CrossRef] [PubMed]
- Hussain, R. Effect of Ivermectin on Pulmonary Aspergillosis in Racing Pigeons. Diyala J. Pure Sci. 2016, 12, 83–93. [Google Scholar]
- Cardwell, L.A.; Alinia, H.; Moradi Tuchayi, S.; Feldman, S.R. New developments in the treatment of rosacea—Role of once-daily ivermectin cream. Clin. Cosmet. Investig. Dermatol. 2016, 9, 71–77. [Google Scholar] [CrossRef]
- Hu, X.; Xu, Y.; Liu, G.; Hu, D.; Wang, Y.; Zhang, W.; Zheng, Y. The impact of anthelmintic treatment on gut bacterial and fungal communities in diagnosed parasite-free sika deer Cervus nippon. Appl. Microbiol. Biotechnol. 2020, 104, 9239–9250. [Google Scholar] [CrossRef] [PubMed]
- Miro-Canturri, A.; Ayerbe-Algaba, R.; Smani, Y. Drug Repurposing for the Treatment of Bacterial and Fungal Infections. Front. Microbiol. 2019, 10, 41. [Google Scholar] [CrossRef]
- Shirazi, F.M.; Mirzaei, R.; Nakhaee, S.; Nejatian, A.; Ghafari, S.; Mehrpour, O. Repurposing the drug, ivermectin, in COVID-19: Toxicological points of view. Eur. J. Med. Res. 2022, 27, 21. [Google Scholar] [CrossRef] [PubMed]
- Schneeberger, P.H.H.; Gueuning, M.; Welsche, S.; Hurlimann, E.; Dommann, J.; Haberli, C.; Frey, J.E.; Sayasone, S.; Keiser, J. Different gut microbial communities correlate with efficacy of albendazole-ivermectin against soil-transmitted helminthiases. Nat. Commun. 2022, 13, 1063. [Google Scholar] [CrossRef]
- Schneeberger, P.H.H.; Coulibaly, J.T.; Gueuning, M.; Moser, W.; Coburn, B.; Frey, J.E.; Keiser, J. Off-target effects of tribendimidine, tribendimidine plus ivermectin, tribendimidine plus oxantel-pamoate, and albendazole plus oxantel-pamoate on the human gut microbiota. Int. J. Parasitol. Drugs Drug Resist. 2018, 8, 372–378. [Google Scholar] [CrossRef]
- Barra, N.G.; Anhe, F.F.; Cavallari, J.F.; Singh, A.M.; Chan, D.Y.; Schertzer, J.D. Micronutrients impact the gut microbiota and blood glucose. J. Endocrinol. 2021, 250, R1–R21. [Google Scholar] [CrossRef]
- Dicks, L.M.T.; Deane, S.M.; Grobbelaar, M.J. Could the COVID-19-Driven Increased Use of Ivermectin Lead to Incidents of Imbalanced Gut Microbiota and Dysbiosis? Probiotics Antimicrob. Proteins 2022, 14, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Van de Wiele, T.; Van den Abbeele, P.; Ossieur, W.; Possemiers, S.; Marzorati, M. The Simulator of the Human Intestinal Microbial Ecosystem (SHIME((R))). In The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models; Verhoeckx, K., Cotter, P., Lopez-Exposito, I., Kleiveland, C., Lea, T., Mackie, A., Requena, T., Swiatecka, D., Wichers, H., Eds.; Spinger: Cham, Switzerland, 2015; pp. 305–317. [Google Scholar]
- Liu, L.; Firrman, J.; Tanes, C.; Bittinger, K.; Thomas-Gahring, A.; Wu, G.D.; Van den Abbeele, P.; Tomasula, P.M. Establishing a mucosal gut microbial community in vitro using an artificial simulator. PLoS ONE 2018, 13, e0197692. [Google Scholar] [CrossRef] [PubMed]
- Piras, C.; Gugliandolo, E.; Castagna, F.; Palma, E.; Britti, D. Ivermectin (IVM) Possible Side Activities and Implications in Antimicrobial Resistance and Animal Welfare: The Authors’ Perspective. Vet. Sci. 2022, 9, 24. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, S.; Chaudhry, U.; Raza, A.; Ghosh, D.; Zhao, X. In vitro activity of ivermectin against Staphylococcus aureus clinical isolates. Antimicrob. Resist. Infect. Control 2018, 7, 27. [Google Scholar] [CrossRef] [PubMed]
- Penumutchu, S.; Korry, B.J.; Hewlett, K.; Belenky, P. Fiber supplementation protects from antibiotic-induced gut microbiome dysbiosis by modulating gut redox potential. Nat. Commun. 2023, 14, 5161. [Google Scholar] [CrossRef]
- Muller, M.; Hermes, G.D.A.; Canfora, E.E.; Smidt, H.; Masclee, A.A.M.; Zoetendal, E.G.; Blaak, E.E. Distal colonic transit is linked to gut microbiota diversity and microbial fermentation in humans with slow colonic transit. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 318, G361–G369. [Google Scholar] [CrossRef]
- Martinez-Guryn, K.; Leone, V.; Chang, E.B. Regional Diversity of the Gastrointestinal Microbiome. Cell Host Microbe 2019, 26, 314–324. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Flint, H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef]
- Pedro, F.-J.; Daniel, M.C.; Van Douwe, S.; Jose, M.-M. Cross-feeding interactions between human gut commensals belonging to the Bacteroides and Bifidobacterium genera when grown on dietary glycans. Microbiome Res. Rep. 2022, 1, 12. [Google Scholar]
- Mazhar, M.; Zhu, Y.; Qin, L. The Interplay of Dietary Fibers and Intestinal Microbiota Affects Type 2 Diabetes by Generating Short-Chain Fatty Acids. Foods 2023, 12, 1023. [Google Scholar] [CrossRef]
- Usta-Gorgun, B.; Yilmaz-Ersan, L. Short-chain fatty acid production by the Bifidobacterium species in the presence of salep. Electron. J. Biotechnol. 2020, 47, 29–35. [Google Scholar] [CrossRef]
- Fusco, W.; Lorenzo, M.B.; Cintoni, M.; Porcari, S.; Rinninella, E.; Kaitsas, F.; Lener, E.; Mele, M.C.; Gasbarrini, A.; Collado, M.C.; et al. Short-Chain Fatty-Acid-Producing Bacteria: Key Components of the Human Gut Microbiota. Nutrients 2023, 15, 2211. [Google Scholar] [CrossRef] [PubMed]
- Christopherson, M.R.; Dawson, J.A.; Stevenson, D.M.; Cunningham, A.C.; Bramhacharya, S.; Weimer, P.J.; Kendziorski, C.; Suen, G. Unique aspects of fiber degradation by the ruminal ethanologen Ruminococcus albus 7 revealed by physiological and transcriptomic analysis. BMC Genom. 2014, 15, 1066. [Google Scholar] [CrossRef] [PubMed]
- Leschine, S.B. Cellulose degradation in anaerobic environments. Annu. Rev. Microbiol. 1995, 49, 399–426. [Google Scholar] [CrossRef]
- Dilnessa, T.; Getaneh, A.; Hailu, W.; Moges, F.; Gelaw, B. Prevalence and antimicrobial resistance pattern of Clostridium difficile among hospitalized diarrheal patients: A systematic review and meta-analysis. PLoS ONE 2022, 17, e0262597. [Google Scholar] [CrossRef] [PubMed]
- Tomas-Pejo, E.; Gonzalez-Fernandez, C.; Greses, S.; Kennes, C.; Otero-Logilde, N.; Veiga, M.C.; Bolzonella, D.; Muller, B.; Passoth, V. Production of short-chain fatty acids (SCFAs) as chemicals or substrates for microbes to obtain biochemicals. Biotechnol. Biofuels Bioprod. 2023, 16, 96. [Google Scholar] [CrossRef]
- Van den Abbeele, P.; Roos, S.; Eeckhaut, V.; MacKenzie, D.A.; Derde, M.; Verstraete, W.; Marzorati, M.; Possemiers, S.; Vanhoecke, B.; Van Immerseel, F.; et al. Incorporating a mucosal environment in a dynamic gut model results in a more representative colonization by lactobacilli. Microb. Biotechnol. 2012, 5, 106–115. [Google Scholar] [CrossRef]
- Firrman, J.; Liu, L.; Van den Abbeele, P.; Tanes, C.; Bittinger, K.; Tomasula, P. Applying Advanced In Vitro Culturing Technology to Study the Human Gut Microbiota. J. Vis. Exp. 2019, 15, e59054. [Google Scholar]
- Mahalak, K.K.; Firrman, J.; Lee, J.J.; Bittinger, K.; Nunez, A.; Mattei, L.M.; Zhang, H.; Fett, B.; Bobokalonov, J.; Arango-Argoty, G.; et al. Triclosan has a robust, yet reversible impact on human gut microbial composition in vitro. PLoS ONE 2020, 15, e0234046. [Google Scholar] [CrossRef]
- Mayo Clinic. Ivermectin (Oral Route). Available online: https://www.mayoclinic.org/drugs-supplements/ivermectin-oral-route/proper-use/drg-20064397 (accessed on 30 October 2023).
- Laffont, C.; Toutain, P.L.; Alvinerie, M.; Bousquet-Mélou, A. Intestinal secretion is a major route for parent ivermectin elimination in the rat. Drug Metab. Dispos. 2002, 30, 626–630. [Google Scholar] [CrossRef]
- Cesar, T.; Salgaco, M.K.; Mesa, V.; Sartoratto, A.; Sivieri, K. Exploring the Association between Citrus Nutraceutical Eriocitrin and Metformin for Improving Pre-Diabetes in a Dynamic Microbiome Model. Pharmaceuticals 2023, 16, 650. [Google Scholar] [CrossRef]
- Mahalak, K.K.; Firrman, J.; Bobokalonov, J.; Narrowe, A.B.; Bittinger, K.; Daniel, S.; Tanes, C.; Mattei, L.M.; Zeng, W.B.; Soares, J.W.; et al. Persistence of the Probiotic Lacticaseibacillus rhamnosus Strain GG (LGG) in an In Vitro Model of the Gut Microbiome. Int. J. Mol. Sci. 2022, 23, 12973. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; D’Agostino McGowan, L.; François, R.; Garrett Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Mallick, H.; Rahnavard, A.; McIver, L.J.; Ma, S.; Zhang, Y.; Nguyen, L.H.; Tickle, T.L.; Weingart, G.; Ren, B.; Schwager, E.H.; et al. Multivariable association discovery in population-scale meta-omics studies. PLOS Comput. Biol. 2021, 17, e1009442. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, B.; Kolli, A.R.; Esch, M.B.; Abaci, H.E.; Shuler, M.L.; Hickman, J.J. TEER measurement techniques for in vitro barrier model systems. J. Lab. Autom. 2015, 20, 107–126. [Google Scholar] [CrossRef] [PubMed]
- Mahalak, K.K.; Bobokalonov, J.; Firrman, J.; Williams, R.; Evans, B.; Fanelli, B.; Soares, J.W.; Kobori, M.; Liu, L. Analysis of the Ability of Capsaicin to Modulate the Human Gut Microbiota In Vitro. Nutrients 2022, 14, 1283. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Mahalak, K.K.; Bobokalonov, J.T.; Narrowe, A.B.; Firrman, J.; Lemons, J.M.S.; Bittinger, K.; Hu, W.; Jones, S.M.; Moustafa, A.M. Impact of Ivermectin on the Gut Microbial Ecosystem. Int. J. Mol. Sci. 2023, 24, 16125. https://doi.org/10.3390/ijms242216125
Liu L, Mahalak KK, Bobokalonov JT, Narrowe AB, Firrman J, Lemons JMS, Bittinger K, Hu W, Jones SM, Moustafa AM. Impact of Ivermectin on the Gut Microbial Ecosystem. International Journal of Molecular Sciences. 2023; 24(22):16125. https://doi.org/10.3390/ijms242216125
Chicago/Turabian StyleLiu, LinShu, Karley K. Mahalak, Jamshed T. Bobokalonov, Adrienne B. Narrowe, Jenni Firrman, Johanna M. S. Lemons, Kyle Bittinger, Weiming Hu, Steven M. Jones, and Ahmed M. Moustafa. 2023. "Impact of Ivermectin on the Gut Microbial Ecosystem" International Journal of Molecular Sciences 24, no. 22: 16125. https://doi.org/10.3390/ijms242216125
APA StyleLiu, L., Mahalak, K. K., Bobokalonov, J. T., Narrowe, A. B., Firrman, J., Lemons, J. M. S., Bittinger, K., Hu, W., Jones, S. M., & Moustafa, A. M. (2023). Impact of Ivermectin on the Gut Microbial Ecosystem. International Journal of Molecular Sciences, 24(22), 16125. https://doi.org/10.3390/ijms242216125






