Kinesin Family Member C1 (KIFC1/HSET) Underlies Aggressive Disease in Androgen Receptor-Low and Basal-Like Triple-Negative Breast Cancers
Abstract
:1. Introduction
2. Results
2.1. AR-Low Tumors Display Aggressive Clinical Features Compared to AR-High Tumors in TNBC
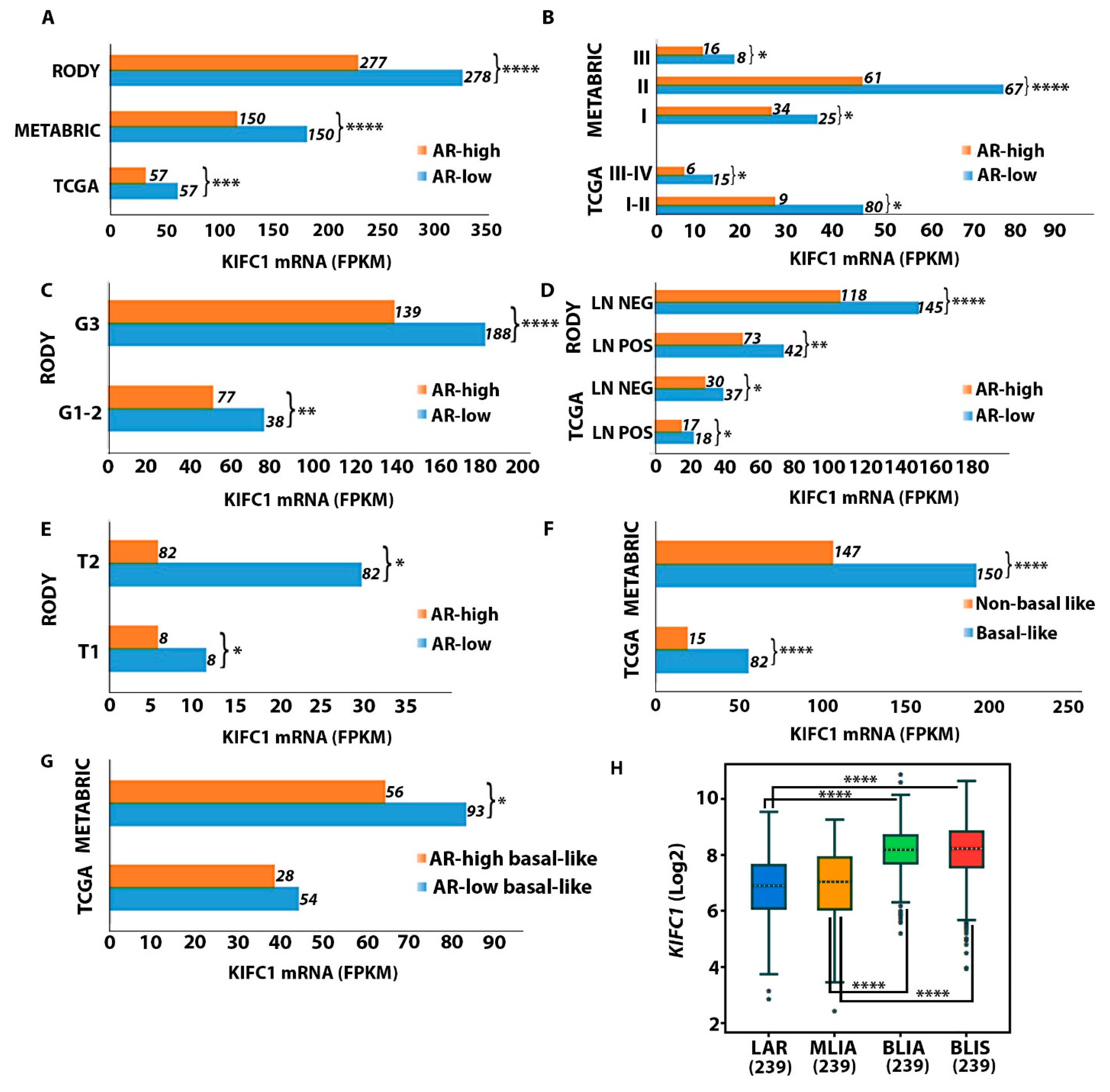
2.2. KIFC1 RNA Levels Are Upregulated in AR-Low vs. AR-High TNBC Tumors
2.3. KIFC1 RNA Is Upregulated in Black/AA and Premenopausal AR-Low Tumors
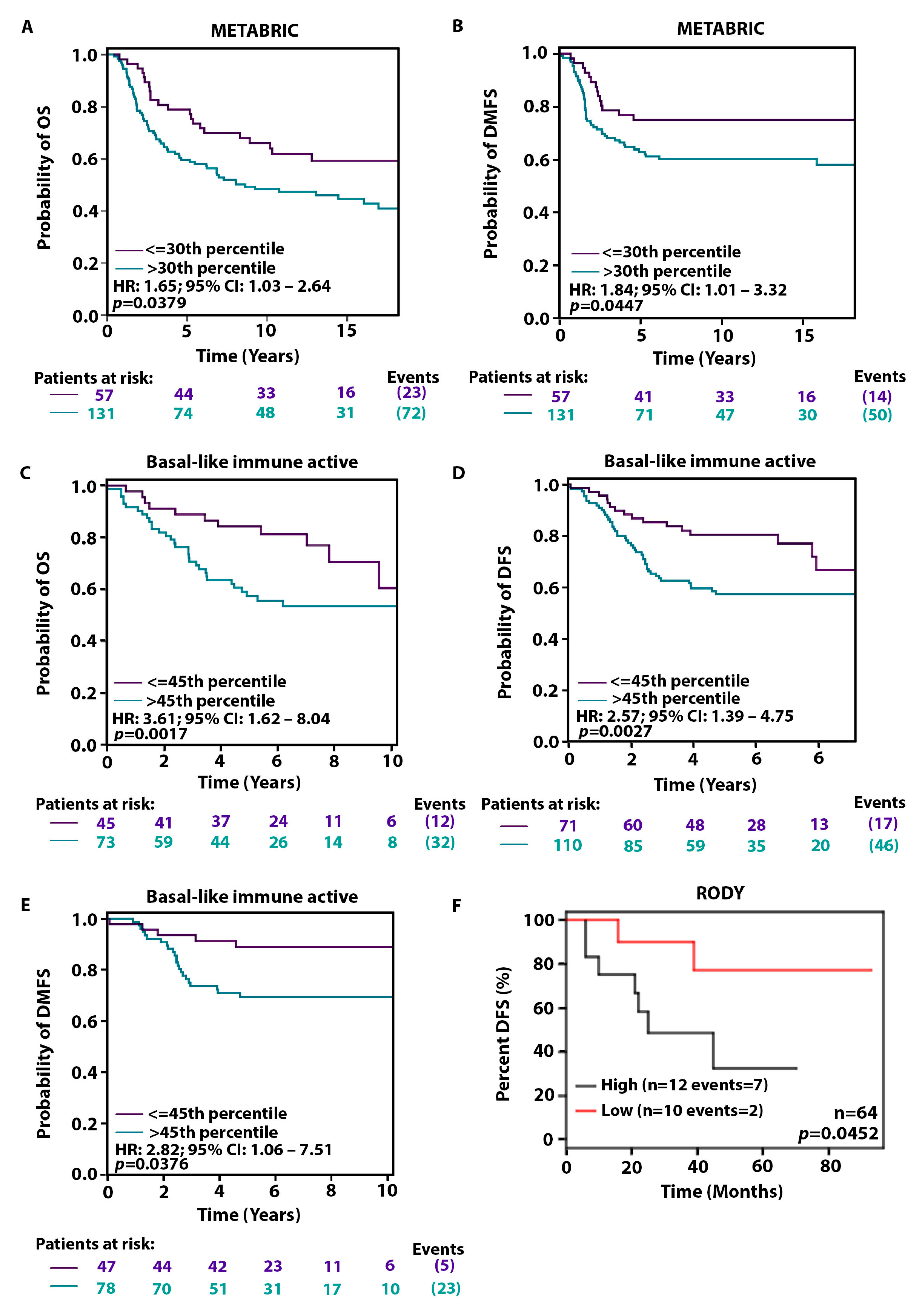
2.4. High KIFC1 Levels Are Associated with a Poorer Prognosis among AR-Low Patients
2.5. Genes Driving Centrosome Amplification Are Upregulated in AR-Low TNBCs Compared to AR-High TNBCs
3. Discussion
4. Materials and Methods
4.1. Study Populations
4.2. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Giaquinto, A.N.; Sung, H.; Miller, K.D.; Kramer, J.L.; Newman, L.A.; Minihan, A.; Jemal, A.; Siegel, R.L. Breast Cancer Statistics, 2022. CA Cancer J. Clin. 2022, 72, 524–541. [Google Scholar] [CrossRef]
- Giaquinto, A.N.; Miller, K.D.; Tossas, K.Y.; Winn, R.A.; Jemal, A.; Siegel, R.L. Cancer statistics for African American/Black People 2022. CA Cancer J. Clin. 2022, 72, 202–229. [Google Scholar] [CrossRef]
- Gennaro, P.; Gabriele, G.; Salini, C.; Chisci, G.; Cascino, F.; Xu, J.F.; Ungari, C. Our supramicrosurgical experience of lymphaticovenular anastomosis in lymphoedema patients to prevent cellulitis. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 674–679. [Google Scholar]
- Gennaro, P.; Chisci, G.; Mazzei, F.; Gabriele, G. Magnetic resonance lymphangiography: How to prove it? J. Magn. Reson. Imaging 2016, 44, 509–510. [Google Scholar] [CrossRef] [PubMed]
- Hara, H.; Mihara, M. The relationship between the degree of subcutaneous fluid accumulation and the lymphatic diameter. J. Plast. Reconstr. Aesthet. Surg. 2023, 82, 163–169. [Google Scholar] [CrossRef]
- Hara, H.; Yoshida, M.; Ikehata, N.; Tachibana, S.; Hamanaka, N.; Nakakawaji, K.; Mihara, M. Compression Pressure Variability in Upper Limb Multilayer Bandaging Applied by Lymphedema Therapists. Lymphat. Res. Biol. 2021, 19, 378–382. [Google Scholar] [CrossRef] [PubMed]
- Yersal, O.; Barutca, S. Biological subtypes of breast cancer: Prognostic and therapeutic implications. World J. Clin. Oncol. 2014, 5, 412–424. [Google Scholar] [CrossRef]
- Yin, L.; Duan, J.J.; Bian, X.W.; Yu, S.C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 61. [Google Scholar] [CrossRef] [PubMed]
- Dent, R.; Trudeau, M.; Pritchard, K.I.; Hanna, W.M.; Kahn, H.K.; Sawka, C.A.; Lickley, L.A.; Rawlinson, E.; Sun, P.; Narod, S.A. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin. Cancer Res. 2007, 13, 4429–4434. [Google Scholar] [CrossRef] [PubMed]
- Lin, N.U.; Claus, E.; Sohl, J.; Razzak, A.R.; Arnaout, A.; Winer, E.P. Sites of distant recurrence and clinical outcomes in patients with metastatic triple-negative breast cancer: High incidence of central nervous system metastases. Cancer 2008, 113, 2638–2645. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, B.D.; Pietenpol, J.A. Identification and use of biomarkers in treatment strategies for triple-negative breast cancer subtypes. J. Pathol. 2014, 232, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.J.; Naidu, S.; Topham, A.K.; Guiles, F.; Xu, Y.; McCue, P.; Schwartz, G.F.; Park, P.K.; Rosenberg, A.L.; Brill, K.; et al. Differences in breast carcinoma characteristics in newly diagnosed African-American and Caucasian patients: A single-institution compilation compared with the National Cancer Institute’s Surveillance, Epidemiology, and End Results database. Cancer 2007, 110, 876–884. [Google Scholar] [CrossRef]
- Lindner, R.; Sullivan, C.; Offor, O.; Lezon-Geyda, K.; Halligan, K.; Fischbach, N.; Shah, M.; Bossuyt, V.; Schulz, V.; Tuck, D.P.; et al. Molecular phenotypes in triple negative breast cancer from African American patients suggest targets for therapy. PLoS ONE 2013, 8, e71915. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.; Tripathi, S.; Hughley, R.; He, Q.; Bae, S.; Karanam, B.; Martini, R.; Newman, L.; Colomb, W.; Grizzle, W.; et al. AR negative triple negative or “quadruple negative” breast cancers in African American women have an enriched basal and immune signature. PLoS ONE 2018, 13, e0196909. [Google Scholar] [CrossRef] [PubMed]
- Jinna, N.; Jovanovic-Talisman, T.; LaBarge, M.; Natarajan, R.; Kittles, R.; Sistrunk, C.; Rida, P.; Seewaldt, V.L. Racial Disparity in Quadruple Negative Breast Cancer: Aggressive Biology and Potential Therapeutic Targeting and Prevention. Cancers 2022, 14, 4484. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Xu, S.; Zhang, Q.; Zhao, W. The expression and clinical significance of the androgen receptor and E-cadherin in triple-negative breast cancer. Med. Oncol. 2012, 29, 526–533. [Google Scholar] [CrossRef]
- He, J.; Peng, R.; Yuan, Z.; Wang, S.; Peng, J.; Lin, G.; Jiang, X.; Qin, T. Prognostic value of androgen receptor expression in operable triple-negative breast cancer: A retrospective analysis based on a tissue microarray. Med. Oncol. 2012, 29, 406–410. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.L.; Macarthur, S.; Ross-Innes, C.S.; Tilley, W.D.; Neal, D.E.; Mills, I.G.; Carroll, J.S. Androgen receptor driven transcription in molecular apocrine breast cancer is mediated by FoxA1. EMBO J. 2011, 30, 3019–3027. [Google Scholar] [CrossRef]
- Ogawa, Y.; Hai, E.; Matsumoto, K.; Ikeda, K.; Tokunaga, S.; Nagahara, H.; Sakurai, K.; Inoue, T.; Nishiguchi, Y. Androgen receptor expression in breast cancer: Relationship with clinicopathological factors and biomarkers. Int. J. Clin. Oncol. 2008, 13, 431–435. [Google Scholar] [CrossRef]
- Sutton, L.M.; Cao, D.; Sarode, V.; Molberg, K.H.; Torgbe, K.; Haley, B.; Peng, Y. Decreased androgen receptor expression is associated with distant metastases in patients with androgen receptor-expressing triple-negative breast carcinoma. Am. J. Clin. Pathol. 2012, 138, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Rakha, E.A.; El-Sayed, M.E.; Green, A.R.; Lee, A.H.; Robertson, J.F.; Ellis, I.O. Prognostic markers in triple-negative breast cancer. Cancer 2007, 109, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Qu, Q.; Mao, Y.; Fei, X.C.; Shen, K.W. The impact of androgen receptor expression on breast cancer survival: A retrospective study and meta-analysis. PLoS ONE 2013, 8, e82650. [Google Scholar] [CrossRef]
- Gasparini, P.; Fassan, M.; Cascione, L.; Guler, G.; Balci, S.; Irkkan, C.; Paisie, C.; Lovat, F.; Morrison, C.; Zhang, J.; et al. Androgen receptor status is a prognostic marker in non-basal triple negative breast cancers and determines novel therapeutic options. PLoS ONE 2014, 9, e88525. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, S.; Klimov, S.; Mittal, K.; Krishnamurti, U.; Li, X.B.; Oprea-Ilies, G.; Wetherilt, C.S.; Riaz, A.; Aleskandarany, M.A.; Green, A.R.; et al. Prognostic Role of Androgen Receptor in Triple Negative Breast Cancer: A Multi-Institutional Study. Cancers 2019, 11, 995. [Google Scholar] [CrossRef]
- Kleylein-Sohn, J.; Pollinger, B.; Ohmer, M.; Hofmann, F.; Nigg, E.A.; Hemmings, B.A.; Wartmann, M. Acentrosomal spindle organization renders cancer cells dependent on the kinesin HSET. J. Cell Sci. 2012, 125, 5391–5402. [Google Scholar] [CrossRef]
- Kwon, M.; Godinho, S.A.; Chandhok, N.S.; Ganem, N.J.; Azioune, A.; Thery, M.; Pellman, D. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes. Dev. 2008, 22, 2189–2203. [Google Scholar] [CrossRef] [PubMed]
- Ogden, A.; Garlapati, C.; Li, X.B.; Turaga, R.C.; Oprea-Ilies, G.; Wright, N.; Bhattarai, S.; Mittal, K.; Wetherilt, C.S.; Krishnamurti, U.; et al. Multi-institutional study of nuclear KIFC1 as a biomarker of poor prognosis in African American women with triple-negative breast cancer. Sci. Rep. 2017, 7, 42289. [Google Scholar] [CrossRef] [PubMed]
- Pannu, V.; Rida, P.C.; Ogden, A.; Turaga, R.C.; Donthamsetty, S.; Bowen, N.J.; Rudd, K.; Gupta, M.V.; Reid, M.D.; Cantuaria, G.; et al. HSET overexpression fuels tumor progression via centrosome clustering-independent mechanisms in breast cancer patients. Oncotarget 2015, 6, 6076–6091. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lu, W.; Chen, D.; Boohaker, R.J.; Zhai, L.; Padmalayam, I.; Wennerberg, K.; Xu, B.; Zhang, W. KIFC1 is a novel potential therapeutic target for breast cancer. Cancer Biol. Ther. 2015, 16, 1316–1322. [Google Scholar] [CrossRef]
- Patel, N.; Weekes, D.; Drosopoulos, K.; Gazinska, P.; Noel, E.; Rashid, M.; Mirza, H.; Quist, J.; Braso-Maristany, F.; Mathew, S.; et al. Integrated genomics and functional validation identifies malignant cell specific dependencies in triple negative breast cancer. Nat. Commun. 2018, 9, 1044. [Google Scholar] [CrossRef]
- Wu, J.; Mikule, K.; Wang, W.; Su, N.; Petteruti, P.; Gharahdaghi, F.; Code, E.; Zhu, X.; Jacques, K.; Lai, Z.; et al. Discovery and mechanistic study of a small molecule inhibitor for motor protein KIFC1. ACS Chem. Biol. 2013, 8, 2201–2208. [Google Scholar] [CrossRef] [PubMed]
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef]
- Acosta-Lopez, S.; Diaz-Bethencourt, D.; Concepcion-Massip, T.; Martin-Fernandez de Basoa, M.C.; Plata-Bello, A.; Gonzalez-Rodriguez, A.; Perez-Hernandez, F.; Plata-Bello, J. The androgen receptor expression and its activity have different relationships with prognosis in hepatocellular carcinoma. Sci. Rep. 2020, 10, 22046. [Google Scholar] [CrossRef] [PubMed]
- Govindan, S.; Siraganahalli Eswaraiah, M.; Basavaraj, C.; Adinarayan, M.; Sankaran, S.; Bakre, M. Androgen Receptor mRNA levels determine the prognosis in triple-negative breast cancer patients. BMC Cancer 2020, 20, 745. [Google Scholar] [CrossRef] [PubMed]
- Quintyne, N.J.; Reing, J.E.; Hoffelder, D.R.; Gollin, S.M.; Saunders, W.S. Spindle multipolarity is prevented by centrosomal clustering. Science 2005, 307, 127–129. [Google Scholar] [CrossRef] [PubMed]
- Ogden, A.; Rida, P.C.; Aneja, R. Prognostic value of CA20, a score based on centrosome amplification-associated genes, in breast tumors. Sci. Rep. 2017, 7, 262. [Google Scholar] [CrossRef] [PubMed]
- Riaz, N.; Idress, R.; Habib, S.; Lalani, E.N. Lack of Androgen Receptor Expression Selects for Basal-Like Phenotype and Is a Predictor of Poor Clinical Outcome in Non-Metastatic Triple Negative Breast Cancer. Front. Oncol. 2020, 10, 1083. [Google Scholar] [CrossRef]
- Astvatsaturyan, K.; Yue, Y.; Walts, A.E.; Bose, S. Androgen receptor positive triple negative breast cancer: Clinicopathologic, prognostic, and predictive features. PLoS ONE 2018, 13, e0197827. [Google Scholar] [CrossRef]
- Rampurwala, M.; Wisinski, K.B.; O’Regan, R. Role of the androgen receptor in triple-negative breast cancer. Clin. Adv. Hematol. Oncol. 2016, 14, 186–193. [Google Scholar]
- Teoh, P.Y.; Tan, G.C.; Mahsin, H.; Wong, Y.P. Androgen receptor expression in triple negative breast carcinoma and its association with the clinicopathological parameters. Malays. J. Pathol. 2019, 41, 125–132. [Google Scholar]
- Sunar, V.; Dogan, H.T.; Sarici, F.; Ates, O.; Akin, S.; Baspinar, B.; Aksoy, S.; Altundag, K. Association between androgen receptor status and prognosis in triple negative breast cancer. J. BUON 2018, 23, 1325–1330. [Google Scholar] [PubMed]
- Haruna, M.; Daramola, A.O.; Awolola, N.A.; Badr, N.M.; Banjo, A.A.F.; Shaaban, A. Clinicopathological features and androgen receptor expression in triple negative breast cancer at Lagos, Nigeria. Ecancermedicalscience 2022, 16, 1452. [Google Scholar] [CrossRef] [PubMed]
- Li, T.F.; Zeng, H.J.; Shan, Z.; Ye, R.Y.; Cheang, T.Y.; Zhang, Y.J.; Lu, S.H.; Zhang, Q.; Shao, N.; Lin, Y. Overexpression of kinesin superfamily members as prognostic biomarkers of breast cancer. Cancer Cell Int. 2020, 20, 123. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhang, Y.; Lang, Z.; Huang, J.; Zou, Z. Prognostic and clinicopathological significance of kinesin family member C1 in various cancers: A meta-analysis. Medicine 2019, 98, e17346. [Google Scholar] [CrossRef]
- Fleisher, B.; Werkman, C.; Jacobs, B.; Varkey, J.; Taha, K.; Ait-Oudhia, S. KIFC1, A Reliable Prognostic Biomarker in Rb-positive Triple-negative Breast Cancer Patients Treated With Doxorubicin in Combination With Abemaciclib. Cancer Diagn. Progn. 2022, 2, 525–532. [Google Scholar] [CrossRef] [PubMed]

| AR Higher 50% (N = 58) 1 | AR Lower 50% (N = 57) | Total (N = 115) | p-Value 2 | |
|---|---|---|---|---|
| KIFC1 | 22.9 (1.04–86.62) | 30.02 (10.1–72.1) | 26.49 (1.04–86.62) | 0.0029 |
| AR | 2.22 (0.62–56.7) | 0.28 (0.03–0.58) | 0.62 (0.03–56.71) | Not applicable |
| Age At Diagnosis (years) | 55 (29–90) | 53 (38–82) | 54 (29–90) | 0.871 |
| Race | 0.690 | |||
| Asian | 4 (7) | 4 (7) | 8 (7) | |
| Black or African American | 17 (29) | 15 (26) | 32 (28) | |
| White | 32 (55) | 36 (63) | 68 (59) | |
| NA | 5 (9) | 2 (4) | 7 (6) | |
| TNM_T_stage | 1.000 | |||
| M0 | 49 (84) | 49 (86) | 98 (85) | |
| M1 | 1 (2) | 1 (2) | 2 (2) | |
| MX | 8 (14) | 7 (12) | 15 (13) | |
| TNM_N_stage | 0.764 | |||
| N0 | 36 (62) | 38 (67) | 74 (64) | |
| N1 | 12 (21) | 13 (23) | 25 (22) | |
| N2 | 7 (12) | 5 (9) | 12 (10) | |
| N3 | 3 (5) | 1 (2) | 4 (3) | |
| Overall_stage | 0.366 | |||
| Missing | 3 (0) | 4 (0) | 7 (6.1) | |
| Stage I | 12 (22) | 7 (13) | 19 (17) | |
| Stage II | 32 (58) | 39 (74) | 71 (62) | |
| Stage III | 10 (18) | 6 (11) | 16 (14) | |
| Stage IV | 1 (2) | 1 (2) | 2 (2) | |
| Tumor subtype | <0.001 | |||
| Basal | 30 (52) | 54 (95) | 84 (73) | |
| Other | 28 (48) | 3 (5) | 31 (27) | |
| Lymph-Node-Positive Number Group | 0.685 | |||
| 0 | 31 (63) | 37 (67) | 68 (59) | |
| ≥1 | 18 (37) | 18 (33) | 36 (31) | |
| Missing | 9 (0) | 2 (0) | 11 (10) | |
| Lymph-Node-Positive Number | 0 (0–14) | 0 (0–10) | 0 (0–14) | 0.509 |
| Menopause Status | 0.061 | |||
| Perimenopausal | 1 (2) | 4 (7) | 5 (4) | |
| Postmenopausal | 36 (62) | 33 (58) | 69 (60) | |
| Premenopausal | 19 (33) | 11 (19) | 30 (26) | |
| Indeterminate | 0 (0) | 2 (4) | 2 (2) | |
| NA | 2 (4) | 7 (12) | 9 (8) |
| AR Higher 50% (N = 149) 1 | AR Lower 50% (N = 150) | Total (N = 299) | p-Value 2 | |
|---|---|---|---|---|
| KIFC1 | 7.30 (5.53–9.14) | 7.94 (6–10) | 7.69 (5.53–9.68) | <0.001 |
| AR | 6.62 (5.88–10.16) | 5.67 (5.22–5.87) | 5.87 (5.22–10.16) | Not applicable |
| Age at Diagnosis (years) | 57 (27–96) | 54 (28–85) | 56 (27–96) | 0.032 |
| Menopausal time point | 0.127 | |||
| Post | 102 (69) | 90 (60) | 192 (64) | |
| Pre | 47 (32) | 60 (40) | 107 (36) | |
| PAM50 Subtype | <0.001 | |||
| Basal | 56 (38) | 95 (63) | 151 (51) | |
| Others | 93 (62) | 55 (37) | 148 (50) | |
| The Nottingham Prognostic Index (NPI) | 4 (2–6) | 4 (1–6) | 4 (1–6) | 0.820 |
| Grade | 0.026 | |||
| Grade 1 or 2 | 26 (18) | 13 (9) | 39 (13) | |
| Grade 3 | 122 (82) | 135 (91) | 257 (86) | |
| Missing | 1 (0) | 2 (0) | 3 (1) | |
| Tumor size (mm) | 22 (1–182) | 25 (0–84) | 25 (0–182) | 0.552 |
| Tumor Stage | 0.098 | |||
| 1 | 35 (24) | 27 (18) | 62 (21) | |
| 2 | 62 (42) | 68 (45) | 130 (43) | |
| 3 | 17 (11) | 8 (5) | 25 (8) | |
| null | 35 (23) | 47 (31) | 82 (27) | |
| Chemotherapy | 0.453 | |||
| No | 74 (50) | 68 (45) | 142 (47) | |
| Yes | 75 (50) | 82 (55) | 157 (53) |
| Variables | AR Lower 50%, N = 278 1 | AR Higher 50%, N = 278 1 | Overall, N = 556 1 | p-Value 2 |
|---|---|---|---|---|
| AR | −0.00285 (−0.01445, −0.00155) | 0.00015 (−0.00155, 0.00837) | −0.00155 (−0.01445, 0.00837) | Not applicable |
| KIFC1 | 0.003 (−0.009, 0.009) | 0.001 (−0.009, 0.007) | 0.002 (−0.009, 0.009) | <0.001 |
| KIFC1 Quartile | <0.001 | |||
| KIFC1 Q1 | 36 (13) | 103 (37) | 139 (25) | |
| KIFC1 Q2 | 66 (24) | 73 (26) | 139 (25) | |
| KIFC1 Q3 | 82 (29) | 57 (21) | 139 (25) | |
| KIFC1 Q4 | 94 (34) | 45 (16) | 139 (25) | |
| Age at diagnosis (years) | 50 (28, 82) | 53 (29, 88) | 51 (28, 88) | 0.003 |
| Missing | 53 | 77 | 130 | |
| Lymph node status | <0.001 | |||
| Negative | 146 (77) | 118 (61) | 264 (69) | |
| Positive | 43 (23) | 74 (39) | 117 (31) | |
| Missing | 89 | 86 | 175 | |
| Tumor size | >0.999 | |||
| Up to 1 cm | 8 (25) | 8 (25) | 16 (25) | |
| >1 cm | 24 (75) | 24 (75) | 48 (75) | |
| Missing | 246 | 246 | 492 | |
| Histological grade | <0.001 | |||
| Grade 1 or 2 | 39 (17) | 78 (36) | 117 (26) | |
| Grade 3 | 189 (83) | 139 (64) | 328 (74) | |
| Missing | 50 | 61 | 111 | |
| Chemotherapy treatment | 0.476 | |||
| No | 3 (9.1) | 6 (18) | 9 (13) | |
| Yes | 30 (91) | 28 (82) | 58 (87) | |
| Missing | 245 | 244 | 489 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jinna, N.; Yuan, Y.-C.; Rida, P. Kinesin Family Member C1 (KIFC1/HSET) Underlies Aggressive Disease in Androgen Receptor-Low and Basal-Like Triple-Negative Breast Cancers. Int. J. Mol. Sci. 2023, 24, 16072. https://doi.org/10.3390/ijms242216072
Jinna N, Yuan Y-C, Rida P. Kinesin Family Member C1 (KIFC1/HSET) Underlies Aggressive Disease in Androgen Receptor-Low and Basal-Like Triple-Negative Breast Cancers. International Journal of Molecular Sciences. 2023; 24(22):16072. https://doi.org/10.3390/ijms242216072
Chicago/Turabian StyleJinna, Nikita, Yate-Ching Yuan, and Padmashree Rida. 2023. "Kinesin Family Member C1 (KIFC1/HSET) Underlies Aggressive Disease in Androgen Receptor-Low and Basal-Like Triple-Negative Breast Cancers" International Journal of Molecular Sciences 24, no. 22: 16072. https://doi.org/10.3390/ijms242216072
APA StyleJinna, N., Yuan, Y.-C., & Rida, P. (2023). Kinesin Family Member C1 (KIFC1/HSET) Underlies Aggressive Disease in Androgen Receptor-Low and Basal-Like Triple-Negative Breast Cancers. International Journal of Molecular Sciences, 24(22), 16072. https://doi.org/10.3390/ijms242216072






