Seasonal Azithromycin Use in Paediatric Protracted Bacterial Bronchitis Does Not Promote Antimicrobial Resistance but Does Modulate the Nasopharyngeal Microbiome
Abstract
1. Introduction
2. Results
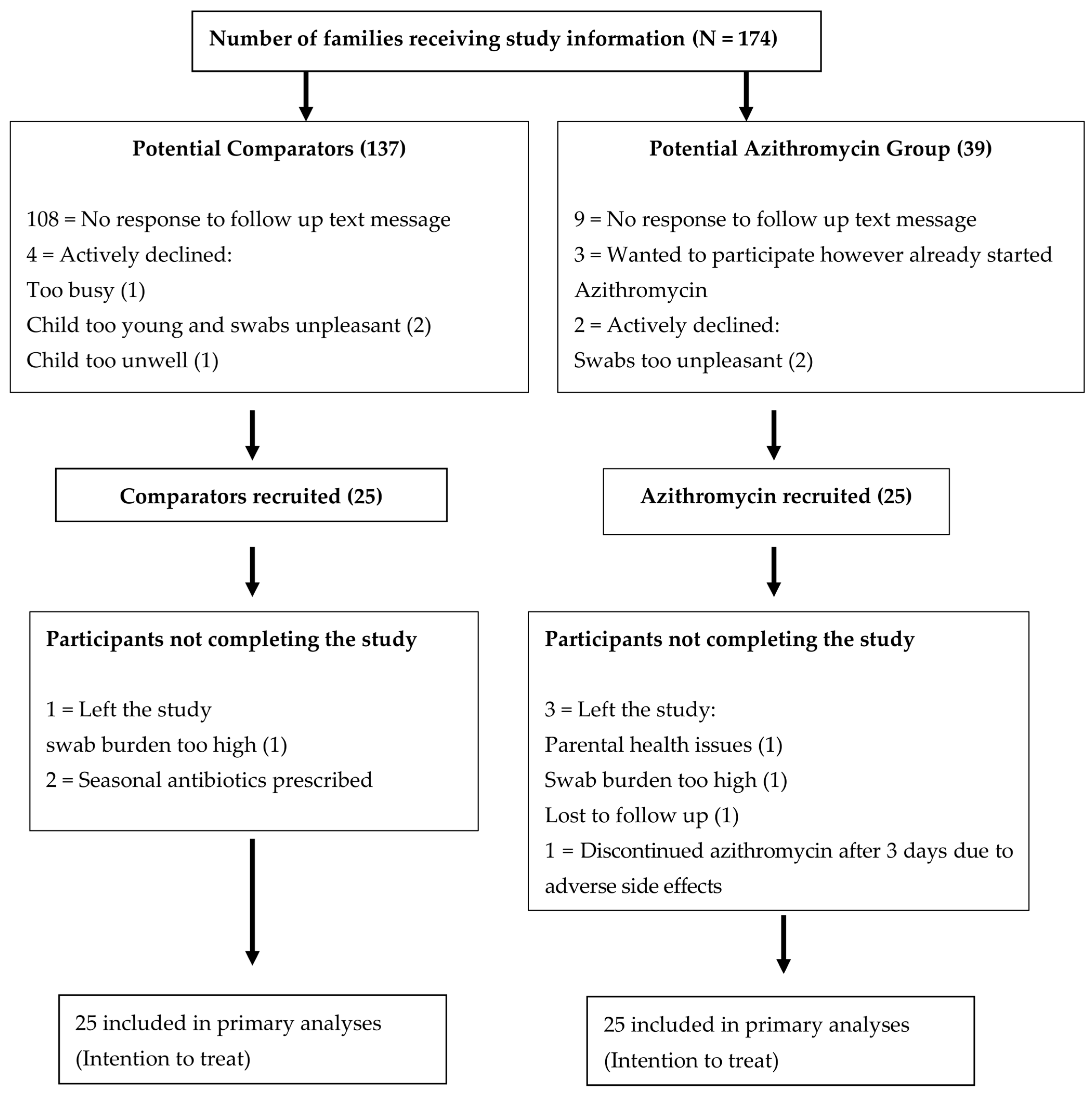
2.1. Antibiotic Exposure
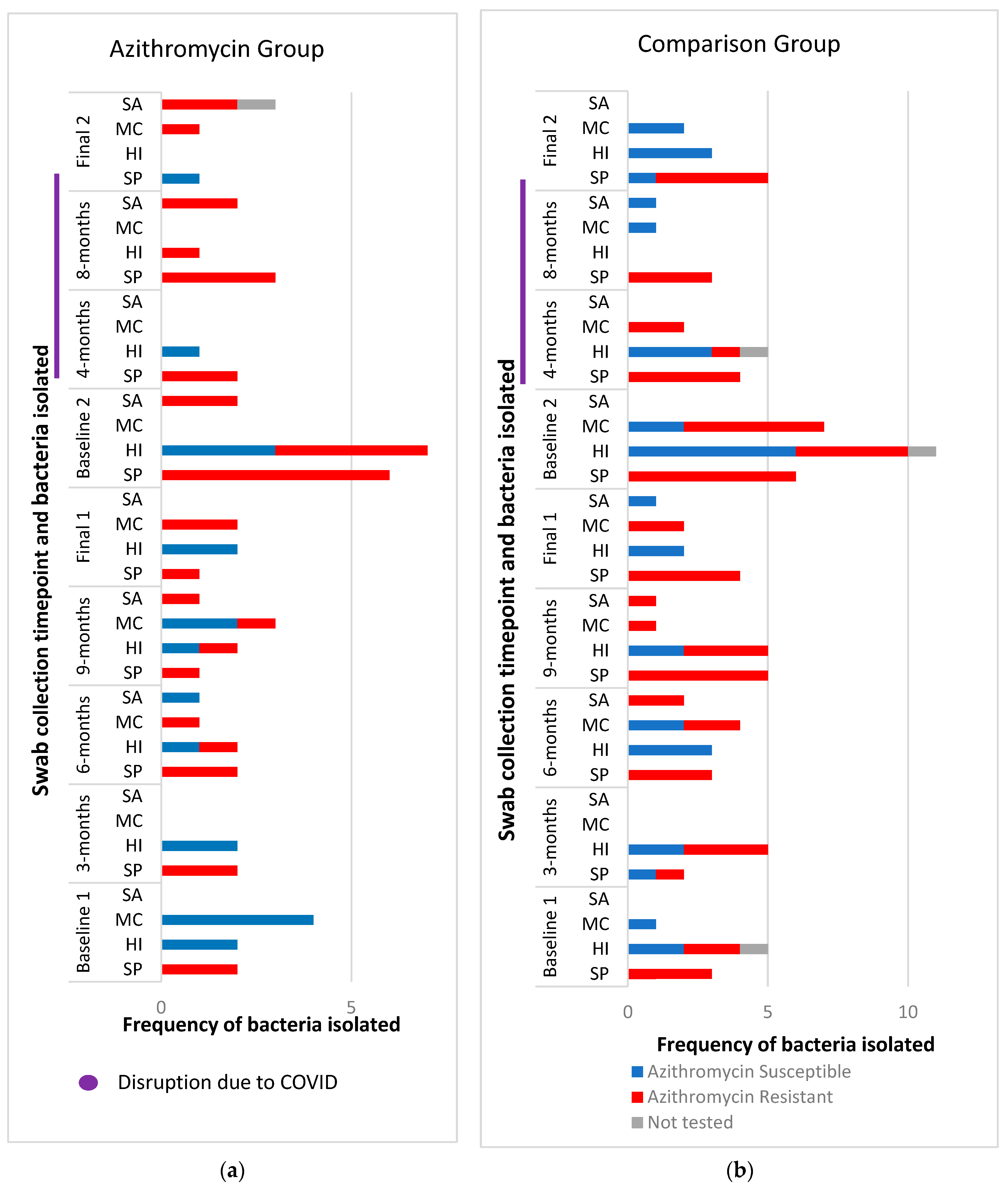
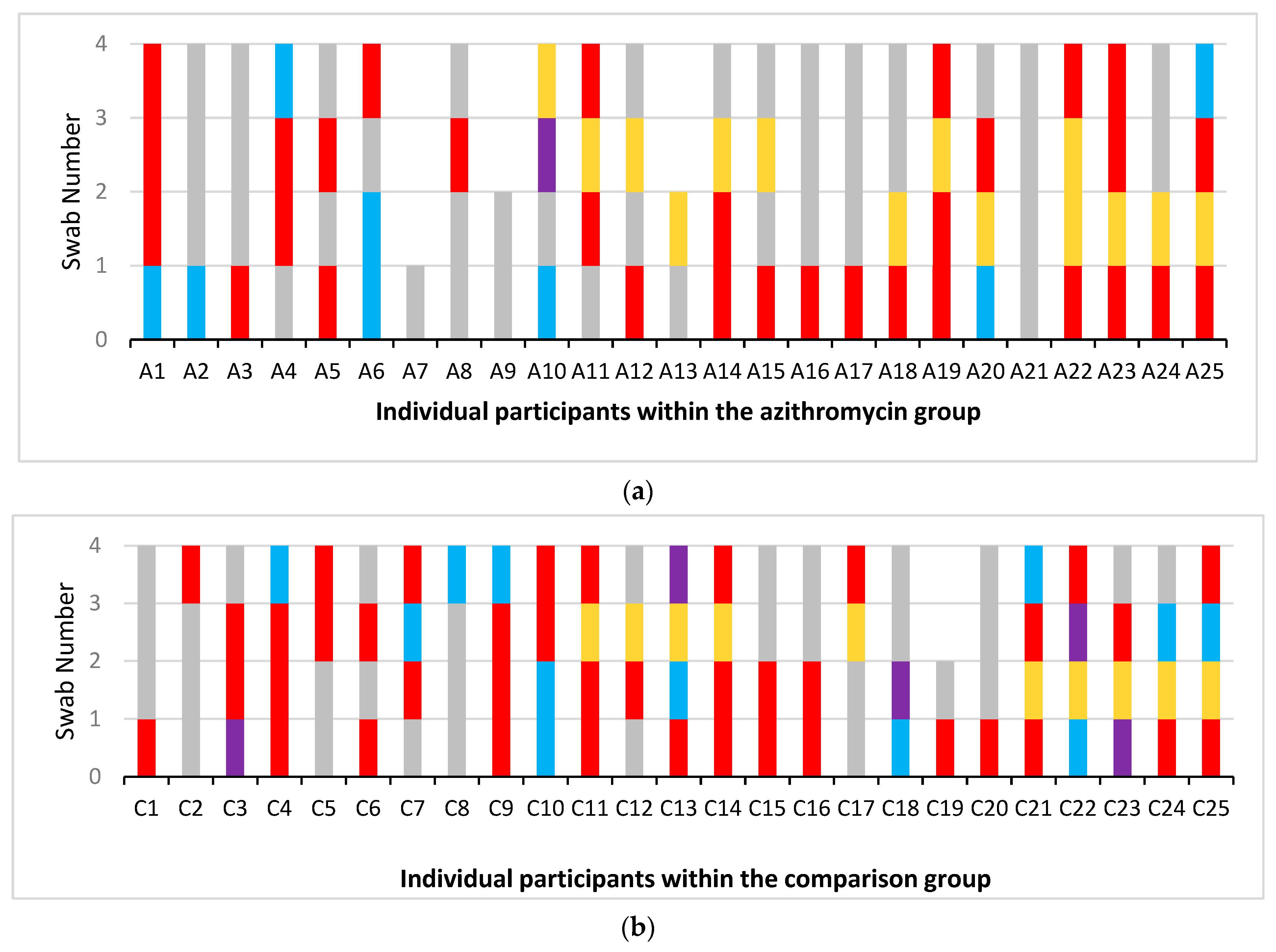
2.2. Nasopharyngeal Bacterial Culture Results
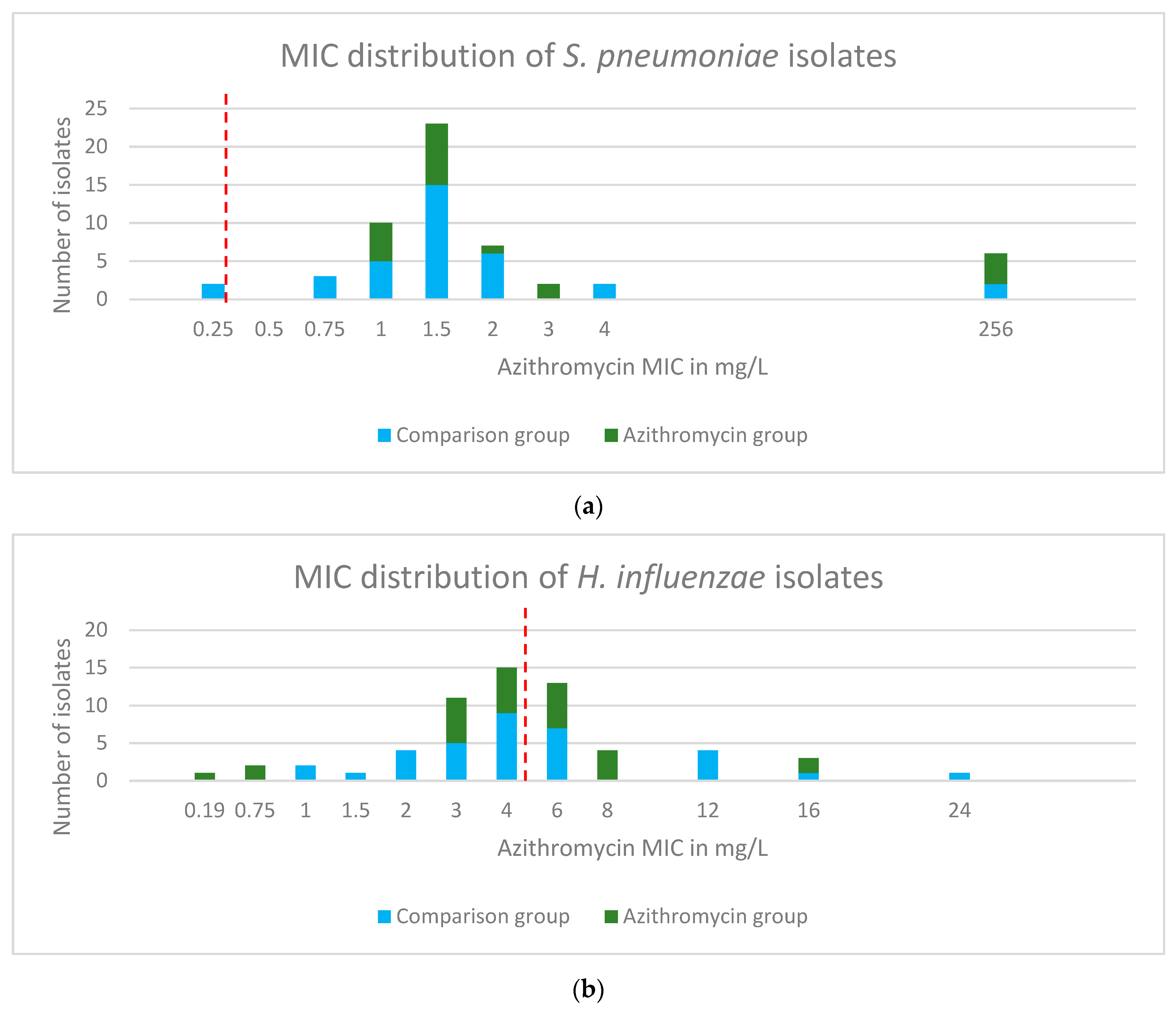
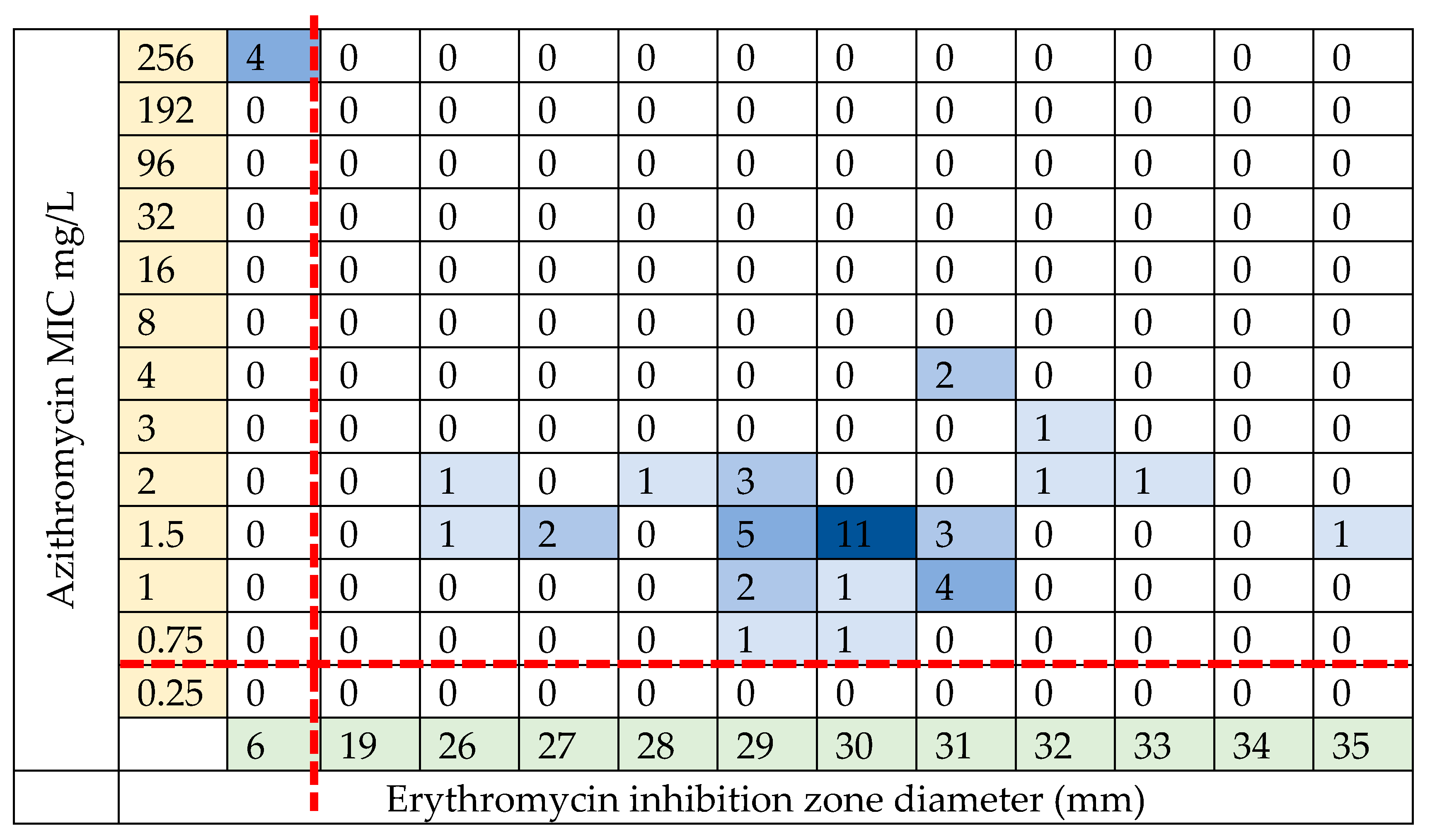
2.3. Phenotypic Antibiotic Resistance Testing
2.4. Genotypic Resistance of S. pneumoniae Isolates
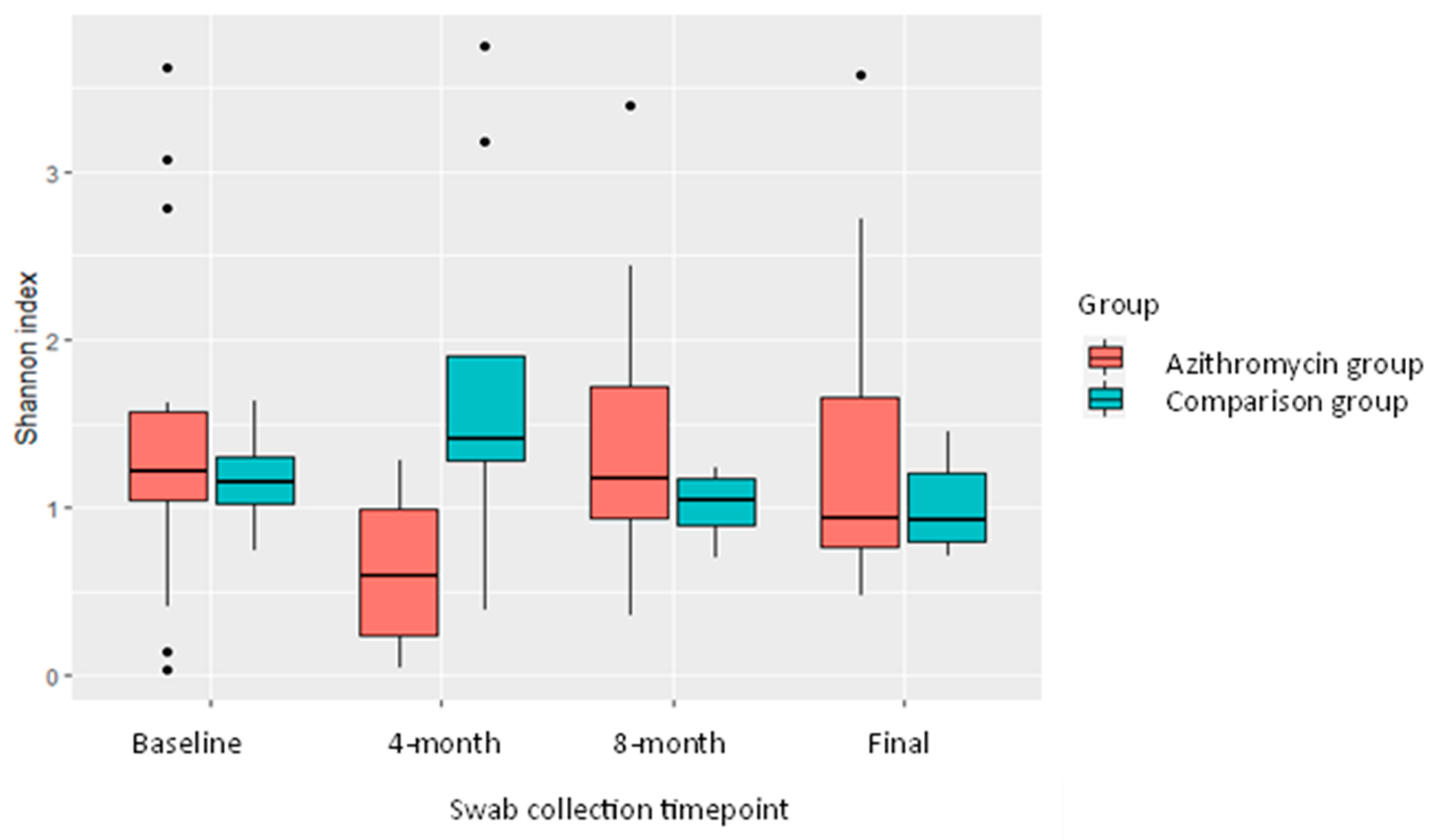
2.5. Nasopharyngeal Microbiome
3. Discussion
4. Materials and Methods
4.1. Study Design and Recruitment
4.2. Sample and Data Collection
4.3. Bacterial Culture and Phenotypic/Genomic Susceptibility Testing
4.4. The 16s Analysis
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mallet, M.C.; Mozun, R.; Ardura-Garcia, C.; Latzin, P.; Moeller, A.; Kuehni, C.E. Prevalence and risk factors of chronic cough in Swiss schoolchildren. Eur. Respir. J. 2020, 56 (Suppl. S64), 3506. [Google Scholar] [CrossRef]
- Chang, A.B.; Robertson, C.F.; Van Asperen, P.P.; Glasgow, N.J.; Mellis, C.M.; Masters, I.B.; Teoh, L.; Tjhung, I.; Morris, P.S.; Petsky, H.L.; et al. A multicenter study on chronic cough in children: Burden and etiologies based on a standardized management pathway. Chest 2012, 142, 943–950. [Google Scholar] [CrossRef] [PubMed]
- Marchant, J.M.; Masters, I.B.; Taylor, S.M.; Cox, N.C.; Seymour, G.J.; Chang, A.B. Evaluation and outcome of young children with chronic cough. Chest 2006, 129, 1132–1141. [Google Scholar] [CrossRef] [PubMed]
- Bergamini, M.; Kantar, A.; Cutrera, R.; Interest Group, I.P.C. Analysis of the Literature on Chronic Cough in Children. Open Respir. Med. J. 2017, 11, 1–9. [Google Scholar] [CrossRef]
- Kantar, A.; Chang, A.B.; Shields, M.D.; Marchant, J.M.; Grimwood, K.; Grigg, J.; Priftis, K.N.; Cutrera, R.; Midulla, F.; Brand, P.L.P.; et al. ERS statement on protracted bacterial bronchitis in children. Eur. Respir. J. 2017, 50, 1602139. [Google Scholar] [CrossRef]
- Ruffles, T.J.C.; Marchant, J.M.; Masters, I.B.; Yerkovich, S.T.; Wurzel, D.F.; Gibson, P.G.; Busch, G.; Baines, K.J.; Simpson, J.L.; Smith-Vaughan, H.C.; et al. Outcomes of protracted bacterial bronchitis in children: A 5-year prospective cohort study. Respirology 2021, 26, 241–248. [Google Scholar] [CrossRef]
- Marchant, J.M.; Newcombe, P.A.; Juniper, E.F.; Sheffield, J.K.; Stathis, S.L.; Chang, A.B. What is the burden of chronic cough for families? Chest 2008, 134, 303–309. [Google Scholar] [CrossRef]
- Zimmermann, P.; Ziesenitz, V.C.; Curtis, N.; Ritz, N. The Immunomodulatory Effects of Macrolides-A Systematic Review of the Underlying Mechanisms. Front. Immunol. 2018, 9, 302. [Google Scholar] [CrossRef]
- Southern, K.W.; Barker, P.M.; Solis-Moya, A.; Patel, L. Macrolide antibiotics for cystic fibrosis. Cochrane Database Syst. Rev. 2012, 11, CD002203. [Google Scholar] [CrossRef]
- Stick, S.M.; Foti, A.; Ware, R.S.; Tiddens, H.; Clements, B.S.; Armstrong, D.S.; Selvadurai, H.; Tai, A.; Cooper, P.J.; Byrnes, C.A.; et al. The effect of azithromycin on structural lung disease in infants with cystic fibrosis (COMBAT CF): A phase 3, randomised, double-blind, placebo-controlled clinical trial. Lancet Respir. Med. 2022, 10, 776–784. [Google Scholar] [CrossRef]
- Peter, J.; Mogayzel, J.; Naureckas, E.T.; Robinson, K.A.; Mueller, G.; Hadjiliadis, D.; Hoag, J.B.; Lubsch, L.; Hazle, L.; Sabadosa, K.; et al. Cystic Fibrosis Pulmonary Guidelines. Am. J. Respir. Crit. Care Med. 2013, 187, 680–689. [Google Scholar] [CrossRef]
- Hill, A.T.; Sullivan, A.L.; Chalmers, J.D.; De Soyza, A.; Elborn, S.J.; Floto, A.R.; Grillo, L.; Gruffydd-Jones, K.; Harvey, A.; Haworth, C.S.; et al. British Thoracic Society Guideline for bronchiectasis in adults. Thorax 2019, 74 (Suppl. S1), 1–69. [Google Scholar] [CrossRef]
- Kobbernagel, H.E.; Buchvald, F.F.; Haarman, E.G.; Casaulta, C.; Collins, S.A.; Hogg, C.; Kuehni, C.E.; Lucas, J.S.; Moser, C.E.; Quittner, A.L.; et al. Efficacy and safety of azithromycin maintenance therapy in primary ciliary dyskinesia (BESTCILIA): A multicentre, double-blind, randomised, placebo-controlled phase 3 trial. Lancet Respir. Med. 2020, 8, 493–505. [Google Scholar] [CrossRef] [PubMed]
- Albert, R.K.; Connett, J.; Bailey, W.C.; Casaburi, R.; Cooper, J.A.D., Jr.; Criner, G.J.; Curtis, J.L.; Dransfield, M.T.; Han, M.K.; Lazarus, S.C.; et al. Azithromycin for Prevention of Exacerbations of COPD. N. Engl. J. Med. 2011, 365, 689–698. [Google Scholar] [CrossRef] [PubMed]
- Saiman, L.; Anstead, M.; Mayer-Hamblett, N.; Lands, L.C.; Kloster, M.; Hocevar-Trnka, J.; Goss, C.H.; Rose, L.M.; Burns, J.L.; Marshall, B.C.; et al. Effect of azithromycin on pulmonary function in patients with cystic fibrosis uninfected with Pseudomonas aeruginosa: A randomized controlled trial. JAMA 2010, 303, 1707–1715. [Google Scholar] [CrossRef] [PubMed]
- Serisier, D.J.; Martin, M.L.; McGuckin, M.A.; Lourie, R.; Chen, A.C.; Brain, B.; Biga, S.; Schlebusch, S.; Dash, P.; Bowler, S.D. Effect of long-term, low-dose erythromycin on pulmonary exacerbations among patients with non-cystic fibrosis bronchiectasis: The BLESS randomized controlled trial. JAMA 2013, 309, 1260–1267. [Google Scholar] [CrossRef] [PubMed]
- Valery, P.C.; Morris, P.S.; Byrnes, C.A.; Grimwood, K.; Torzillo, P.J.; Bauert, P.A.; Masters, I.B.; Diaz, A.; McCallum, G.B.; Mobberley, C.; et al. Long-term azithromycin for Indigenous children with non-cystic-fibrosis bronchiectasis or chronic suppurative lung disease (Bronchiectasis Intervention Study): A multicentre, double-blind, randomised controlled trial. Lancet Respir. Med. 2013, 1, 610–620. [Google Scholar] [CrossRef]
- Malhotra-Kumar, S.; Lammens, C.; Coenen, S.; Van Herck, K.; Goossens, H. Effect of azithromycin and clarithromycin therapy on pharyngeal carriage of macrolide-resistant streptococci in healthy volunteers: A randomised, double-blind, placebo-controlled study. Lancet 2007, 369, 482–490. [Google Scholar] [CrossRef]
- Oldenburg, C.E.; Sié, A.; Coulibaly, B.; Ouermi, L.; Dah, C.; Tapsoba, C.; Bärnighausen, T.; Ray, K.J.; Zhong, L.; Cummings, S.; et al. Effect of Commonly Used Pediatric Antibiotics on Gut Microbial Diversity in Preschool Children in Burkina Faso: A Randomized Clinical Trial. Open Forum Infect. Dis. 2018, 5, ofy289. [Google Scholar] [CrossRef]
- Cox, T.O.; Lundgren, P.; Nath, K.; Thaiss, C.A. Metabolic control by the microbiome. Genome Med. 2022, 14, 80. [Google Scholar] [CrossRef]
- Man, W.H.; de Steenhuijsen Piters, W.A.; Bogaert, D. The microbiota of the respiratory tract: Gatekeeper to respiratory health. Nat. Rev. Microbiol. 2017, 15, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Bosch, A.A.T.M.; Piters, W.A.A.d.S.; Houten, M.A.v.; Chu, M.L.J.N.; Biesbroek, G.; Kool, J.; Pernet, P.; Groot, P.-K.C.M.d.; Eijkemans, M.J.C.; Keijser, B.J.F.; et al. Maturation of the Infant Respiratory Microbiota, Environmental Drivers, and Health Consequences. A Prospective Cohort Study. Am. J. Respir. Crit. Care Med. 2017, 196, 1582–1590. [Google Scholar] [CrossRef] [PubMed]
- Toivonen, L.; Karppinen, S.; Schuez-Havupalo, L.; Waris, M.; He, Q.; Hoffman, K.L.; Petrosino, J.F.; Dumas, O.; Camargo, C.A.; Hasegawa, K.; et al. Longitudinal Changes in Early Nasal Microbiota and the Risk of Childhood Asthma. Pediatrics 2020, 146, e20200421. [Google Scholar] [CrossRef] [PubMed]
- Toivonen, L.; Schuez-Havupalo, L.; Karppinen, S.; Waris, M.; Hoffman, K.L.; Camargo, C.A.; Hasegawa, K.; Peltola, V. Antibiotic Treatments During Infancy, Changes in Nasal Microbiota, and Asthma Development: Population-based Cohort Study. Clin. Infect. Dis. 2021, 72, 1546–1554. [Google Scholar] [CrossRef]
- Van der Gast, C.J.; Cuthbertson, L.; Rogers, G.B.; Pope, C.; Marsh, R.L.; Redding, G.J.; Bruce, K.D.; Chang, A.B.; Hoffman, L.R. Three clinically distinct chronic pediatric airway infections share a common core microbiota. Ann. Am. Thorac. Soc. 2014, 11, 1039–1048. [Google Scholar] [CrossRef]
- Cuthbertson, L.; Craven, V.; Bingle, L.; Cookson, W.O.C.M.; Everard, M.L.; Moffatt, M.F. The impact of persistent bacterial bronchitis on the pulmonary microbiome of children. PLoS ONE 2017, 12, e0190075. [Google Scholar] [CrossRef]
- Kansra, S. Diagnosis and Management of Children with Protracted Bacterial Bronchitis. Diagnosis and Management of Children with Protracted Bacterial Bronchitis. Sheffield Children's Hospital Medical Guideline. 2018. Available online: https://www.sheffieldchildrens.nhs.uk/clinical-guidelines-handbook/ (accessed on 1 January 2021).
- EUCAST. Clinical Breakpoints—Breakpoints and Guidance. 2022. Available online: https://www.eucast.org/clinical_breakpoints/ (accessed on 5 July 2022).
- Stevens, E.J.; Morse, D.J.; Bonini, D.; Duggan, S.; Brignoli, T.; Recker, M.; Lees, J.A.; Croucher, N.J.; Bentley, S.; Wilson, D.J.; et al. Targeted control of pneumolysin production by a mobile genetic element in Streptococcus pneumoniae. Microb. Genom. 2022, 8, 000784. [Google Scholar] [CrossRef]
- Abotsi, R.E.; Nicol, M.P.; McHugh, G.; Simms, V.; Rehman, A.M.; Barthus, C.; Ngwira, L.G.; Kwambana-Adams, B.; Heyderman, R.S.; Odland, J.; et al. The impact of long-term azithromycin on antibiotic resistance in HIV-associated chronic lung disease. ERJ Open Res. 2022, 8, 00491–02021. [Google Scholar] [CrossRef]
- Samson, C.; Tamalet, A.; Thien, H.V.; Taytard, J.; Perisson, C.; Nathan, N.; Clement, A.; Boelle, P.Y.; Corvol, H. Long-term effects of azithromycin in patients with cystic fibrosis. Respir. Med. 2016, 117, 1–6. [Google Scholar] [CrossRef]
- ECDC. Antimicrobial Resistance in the EU/EEA (EARS-Net)—Annual Epidemiological Report for 2019. Antimicrobial Resistance Surveillance in Europe 2020. Available online: https://www.ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2019 (accessed on 1 January 2021).
- Jorgensen, J.H.; Howell, A.W.; Maher, L.A. Quantitative antimicrobial susceptibility testing of Haemophilus influenzae and Streptococcus pneumoniae by using the E-test. J. Clin. Microbiol. 1991, 29, 109–114. [Google Scholar] [CrossRef]
- Johnson, J.; Bouchillon, S.; Pontani, D. The effect of carbon dioxide on susceptibility testing of azithromycin, clarithromycin and roxithromycin against clinical isolates of Streptococcus pneumoniae and Streptococcus pyogenes by broth microdilution and the Etest: Artemis Project-first-phase study. Clin. Microbiol. Infect. 1999, 5, 327–330. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mosleh, M.N.; Gharibi, M.; Alikhani, M.Y.; Saidijam, M.; Vakhshiteh, F. Antimicrobial susceptibility and analysis of macrolide resistance genes in Streptococcus pneumoniae isolated in Hamadan. Iran. J. Basic. Med. Sci. 2014, 17, 595–599. [Google Scholar] [PubMed]
- Jia, X.; Ren, H.; Nie, X.; Li, Y.; Li, J.; Qin, T. Antibiotic Resistance and Azithromycin Resistance Mechanism of Legionella pneumophila Serogroup 1 in China. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef] [PubMed]
- Pittman, J.E.; Wylie, K.M.; Akers, K.; Storch, G.A.; Hatch, J.; Quante, J.; Frayman, K.B.; Clarke, N.; Davis, M.; Stick, S.M.; et al. Association of Antibiotics, Airway Microbiome, and Inflammation in Infants with Cystic Fibrosis. Ann. Am. Thorac. Soc. 2017, 14, 1548–1555. [Google Scholar] [CrossRef]
- EUCAST. Media Preparation for EUCAST Disk Diffusion Testing and for Determination of MIC Values by the Borth Microdilution Method. 2022. Available online: https://www.eucast.org/ast_of_bacteria/media_preparation (accessed on 5 July 2022).
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Seemann, T. Abricate. Available online: https://github.com/tseemann/abricate (accessed on 1 January 2021).
- Kapatai, G.; Sheppard, C.L.; Al-Shahib, A.; Litt, D.J.; Underwood, A.P.; Harrison, T.G.; Fry, N.K. Whole genome sequencing of Streptococcus pneumoniae: Development, evaluation and verification of targets for serogroup and serotype prediction using an automated pipeline. PeerJ 2016, 4, e2477. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Brynildsrud, O.; Bohlin, J.; Scheffer, L.; Eldholm, V. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol. 2016, 17, 238. [Google Scholar] [CrossRef]
- Man, W.H.; van Houten, M.A.; Merelle, M.E.; Vlieger, A.M.; Chu, M.; Jansen, N.J.G.; Sanders, E.A.M.; Bogaert, D. Bacterial and viral respiratory tract microbiota and host characteristics in children with lower respiratory tract infections: A matched case-control study. Lancet Respir. Med. 2019, 7, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Davis, N.M.; Proctor, D.M.; Holmes, S.P.; Relman, D.A.; Callahan, B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. bioRxiv 2018, 6, 226. [Google Scholar] [CrossRef] [PubMed]
- Salter, S.J.; Cox, M.J.; Turek, E.M.; Calus, S.T.; Cookson, W.O.; Moffatt, M.F.; Turner, P.; Parkhill, J.; Loman, N.J.; Walker, A.W. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014, 12, 87. [Google Scholar] [CrossRef]
- Subramanian, S.; Huq, S.; Yatsunenko, T.; Haque, R.; Mahfuz, M.; Alam, M.A.; Benezra, A.; DeStefano, J.; Meier, M.F.; Muegge, B.D.; et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 2014, 510, 417–421. [Google Scholar] [CrossRef]
- Von Elm, E.; Altman, D.G.; Egger, M.; Pocock, S.J.; Gøtzsche, P.C.; Vandenbroucke, J.P. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: Guidelines for reporting observational studies. J. Clin. Epidemiol. 2008, 61, 344–349. [Google Scholar] [CrossRef]
- Paulson, J.N.; Stine, O.C.; Bravo, H.C.; Pop, M. Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 2013, 10, 1200–1202. [Google Scholar] [CrossRef]


| Demographic | Comparison Group N = 25 (%) | Azithromycin Group N = 25 (%) | p Value for t-Test or Fisher’s Exact Test * |
|---|---|---|---|
| Mean age in months | 39.24 (SD 14.2) | 50.24 (SD 24.9) | p = 0.063 |
| Female | 12 (48) | 11 (44) | p = ns |
| Vaginal delivery | 21 (84) | 15 (60) | p = 0.113 |
| Breast fed | 13 (52) | 12 (48) | p = ns |
| Microbiological diagnosis of PBB | 13 (52) | 20 (80) | p = 0.071 |
| Premature < 37 weeks | 2 (8) | 4 (16) | p = 0.667 |
| Ventilated for prematurity | 1 (4) | 2 (8) | p = ns |
| Parent smoker | 6 (24) | 5 (20) | p = 0.71 |
| Fully immunised (UK vaccination schedule) | 25 (100) | 25 (100) | p = ns |
| Penicillin allergy | 1 (4) | 0 (0) | p = ns |
| Exposure | Comparison Group Median (25th–75th) | Azithromycin Group Median (25th–75th) | Statistics * |
|---|---|---|---|
| Acute courses of antibiotics before study enrolment | 7 (4–10) | 10 (6–13) | IRR, p = 0.037 |
| Antibiotics > 2 weeks duration before study enrolment | 1 (1–2) | 2 (2–3) | IRR, p = 0.070 |
| Time from last course of antibiotics (weeks) | 11 (2–25) | 5 (4–21) | t-test p = 0.953 |
| Previous macrolide use before study enrolment | 15/25 (56%) | 13/25 (52%) | Fisher’s p = 0.776 |
| (a) | |||||||
| Bacterial Isolate | Antibiotic Resistance Tested Using Disc Diffusion Unless Otherwise Stated | ||||||
| Azithromycin * | Erythromycin | Tetracycline | Oxacillin | ||||
| S. pneumoniae N = 46 | 46 | 5 | 3 | 5 | |||
| (b) | |||||||
| Bacterial isolate | Antibiotic resistance tested using disc diffusion unless otherwise stated | ||||||
| Azithromycin * | Ampicillin | Cefuroxime | Chloramphenicol | Co-amoxiclav | |||
| H. influenzae N = 54 | 19 | 12 | 1 | 2 | 7 | ||
| Swab Round | Number in AZM * Group Actively Taking AZM | Mean Shannon Index AZM Group | Mean Shannon Index Comparison Group | t-Test p Value |
|---|---|---|---|---|
| Baseline | 0 | 1.44 | 1.17 | 0.38 |
| 4 month | 15 | 0.63 | 1.71 | 0.02 |
| 8 month | 2 | 1.48 | 1.01 | 0.22 |
| Final | 4 | 1.33 | 0.97 | 0.22 |
| Azithromycin Group | Comparison Group | |
|---|---|---|
| Inclusion criteria | Age:18 months to 10 years | |
| At least 1 episode of PBB over the preceding 18 months | ||
| Not had seasonal azithromycin prophylaxis for at least 6 months | Never been exposed to seasonal or prophylactic antibiotics | |
| Due to start seasonal azithromycin over the winter months—standard clinic care (local guideline [27]) | Less than 3 exacerbations over the preceding 12 months. Not anticipated to require seasonal azithromycin over the study period | |
| Exclusion criteria | Ongoing investigations for cystic fibrosis or other underlying lung disease | |
| Known diagnosis of cystic fibrosis, bronchiectasis, primary immune deficiency | ||
| Those prescribed 3 monthly courses of intravenous antibiotics | ||
| Previous prescription of non-azithromycin prophylaxis | ||
| Bleeding disorder or anticoagulation | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hardman, S.J.; Shackley, F.M.; Ugonna, K.; Darton, T.C.; Rigby, A.S.; Bogaert, D.; Binkowska, J.M.; Condliffe, A.M. Seasonal Azithromycin Use in Paediatric Protracted Bacterial Bronchitis Does Not Promote Antimicrobial Resistance but Does Modulate the Nasopharyngeal Microbiome. Int. J. Mol. Sci. 2023, 24, 16053. https://doi.org/10.3390/ijms242216053
Hardman SJ, Shackley FM, Ugonna K, Darton TC, Rigby AS, Bogaert D, Binkowska JM, Condliffe AM. Seasonal Azithromycin Use in Paediatric Protracted Bacterial Bronchitis Does Not Promote Antimicrobial Resistance but Does Modulate the Nasopharyngeal Microbiome. International Journal of Molecular Sciences. 2023; 24(22):16053. https://doi.org/10.3390/ijms242216053
Chicago/Turabian StyleHardman, Simon J., Fiona M. Shackley, Kelechi Ugonna, Thomas C. Darton, Alan S. Rigby, Debby Bogaert, Justyna M. Binkowska, and Alison M. Condliffe. 2023. "Seasonal Azithromycin Use in Paediatric Protracted Bacterial Bronchitis Does Not Promote Antimicrobial Resistance but Does Modulate the Nasopharyngeal Microbiome" International Journal of Molecular Sciences 24, no. 22: 16053. https://doi.org/10.3390/ijms242216053
APA StyleHardman, S. J., Shackley, F. M., Ugonna, K., Darton, T. C., Rigby, A. S., Bogaert, D., Binkowska, J. M., & Condliffe, A. M. (2023). Seasonal Azithromycin Use in Paediatric Protracted Bacterial Bronchitis Does Not Promote Antimicrobial Resistance but Does Modulate the Nasopharyngeal Microbiome. International Journal of Molecular Sciences, 24(22), 16053. https://doi.org/10.3390/ijms242216053






