BAL Proteomic Signature of Lung Adenocarcinoma in IPF Patients and Its Transposition in Serum Samples for Less Invasive Diagnostic Procedures
Abstract
1. Introduction
2. Results
2.1. Population
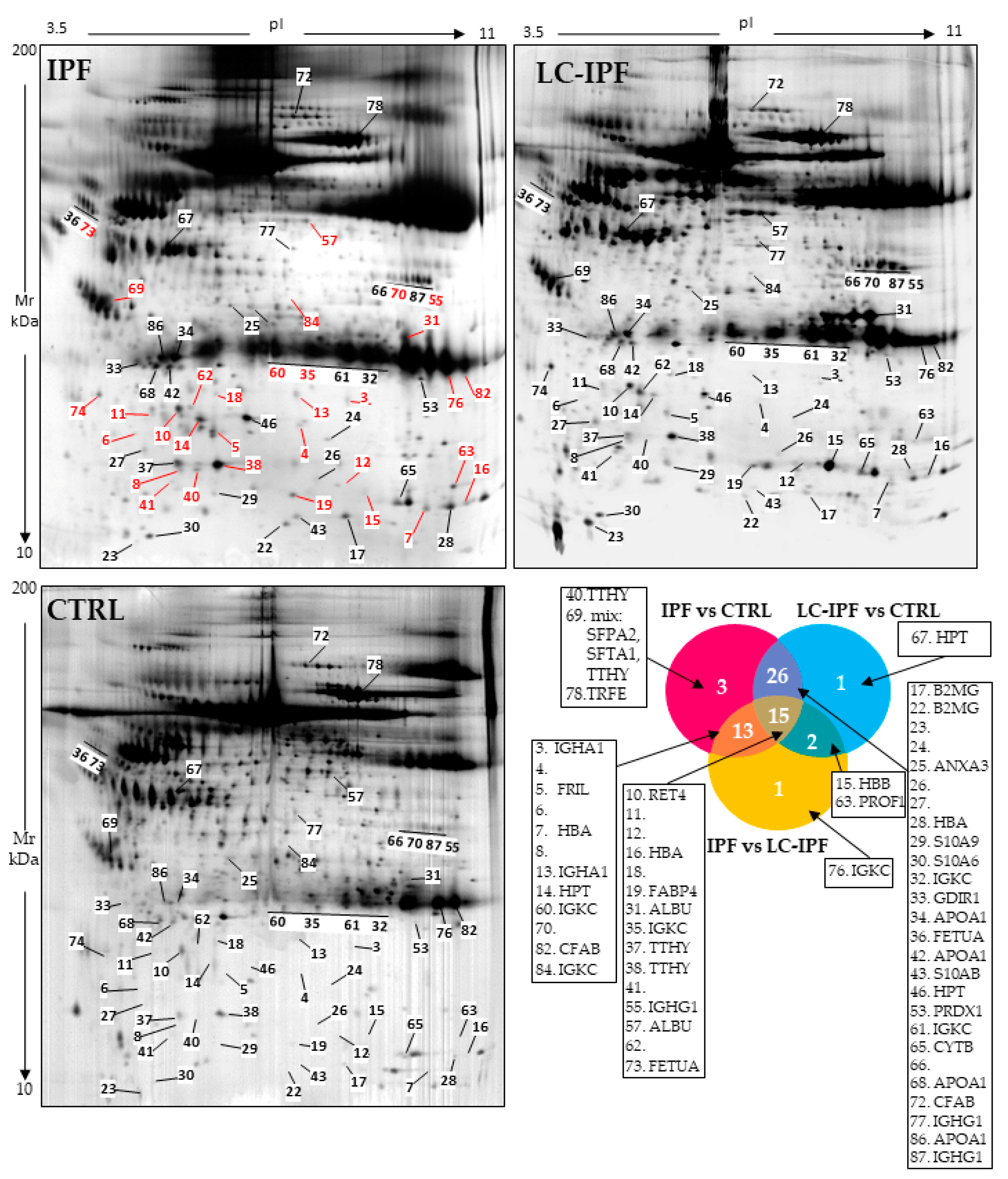
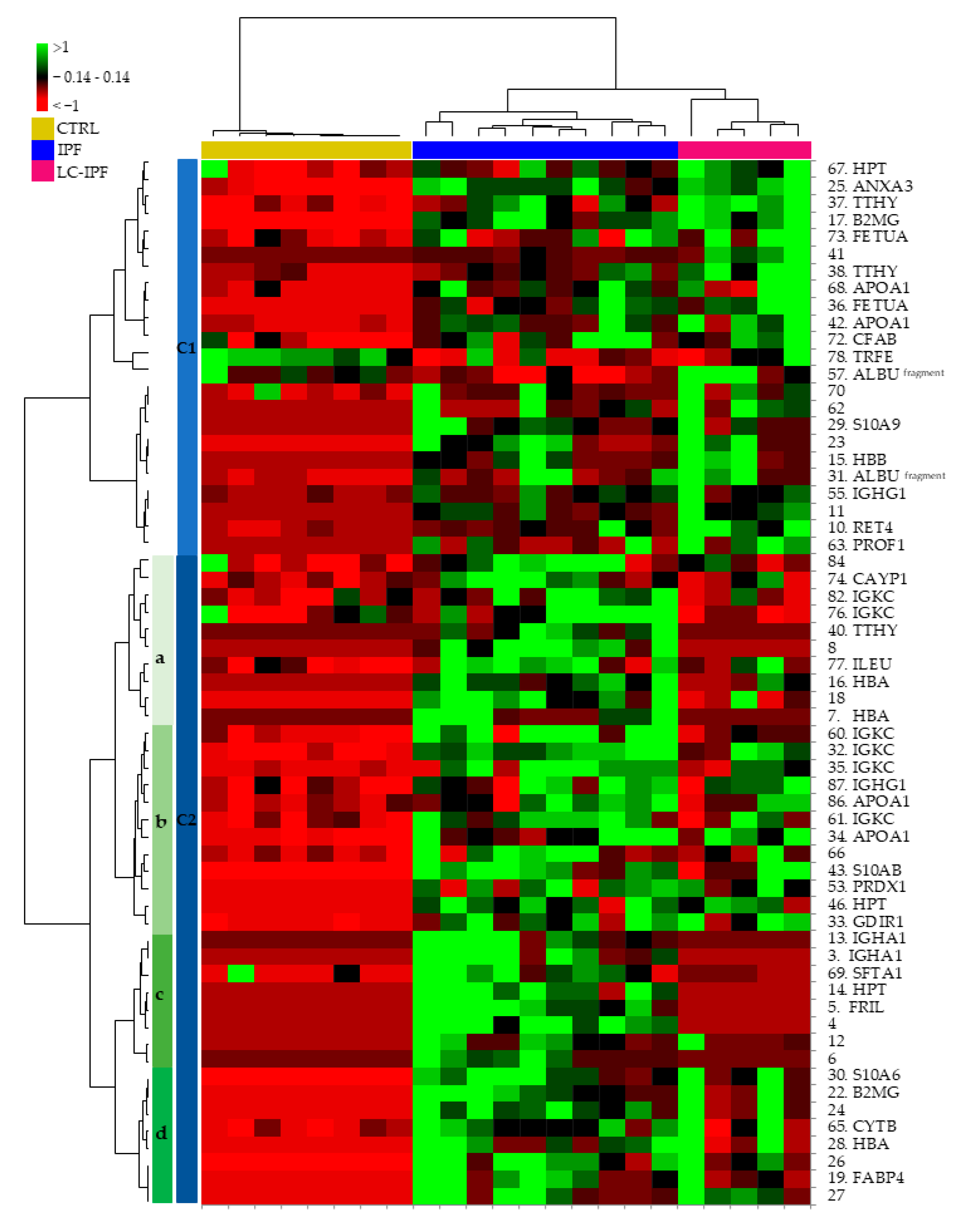
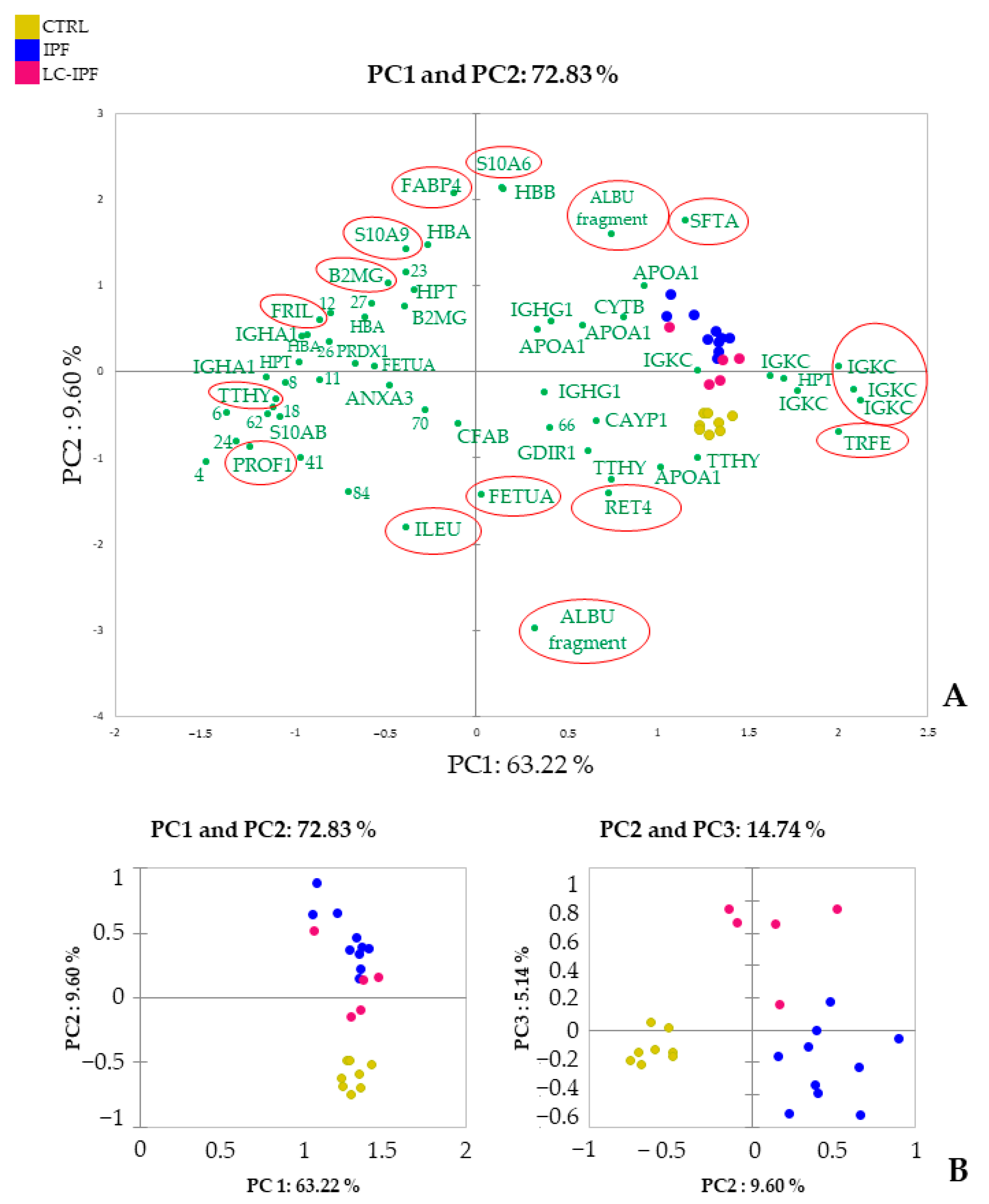
2.2. Proteomic and Multivariate Analyses Revealed a Differential BALF Protein Pattern between IPF and LC-IPF
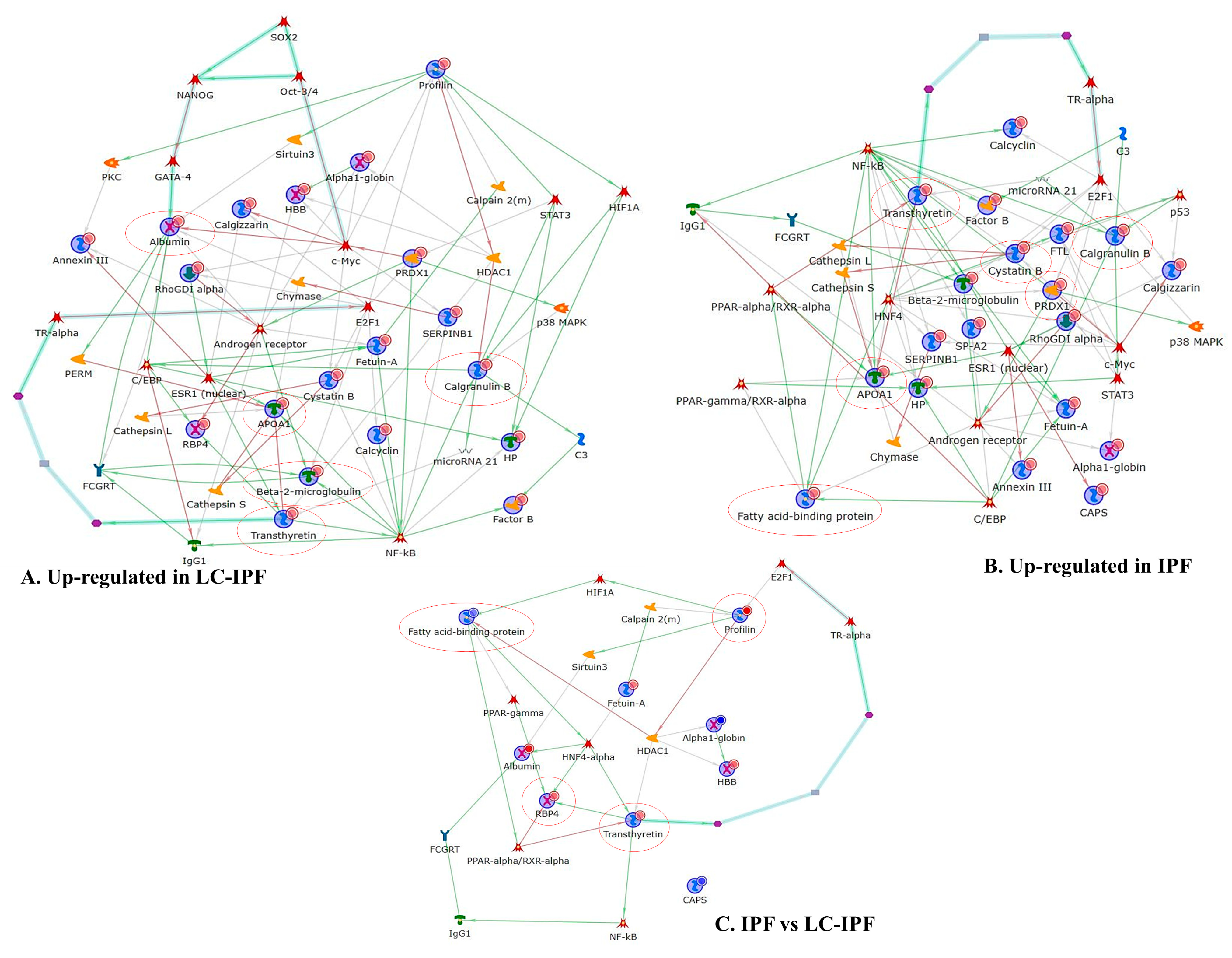
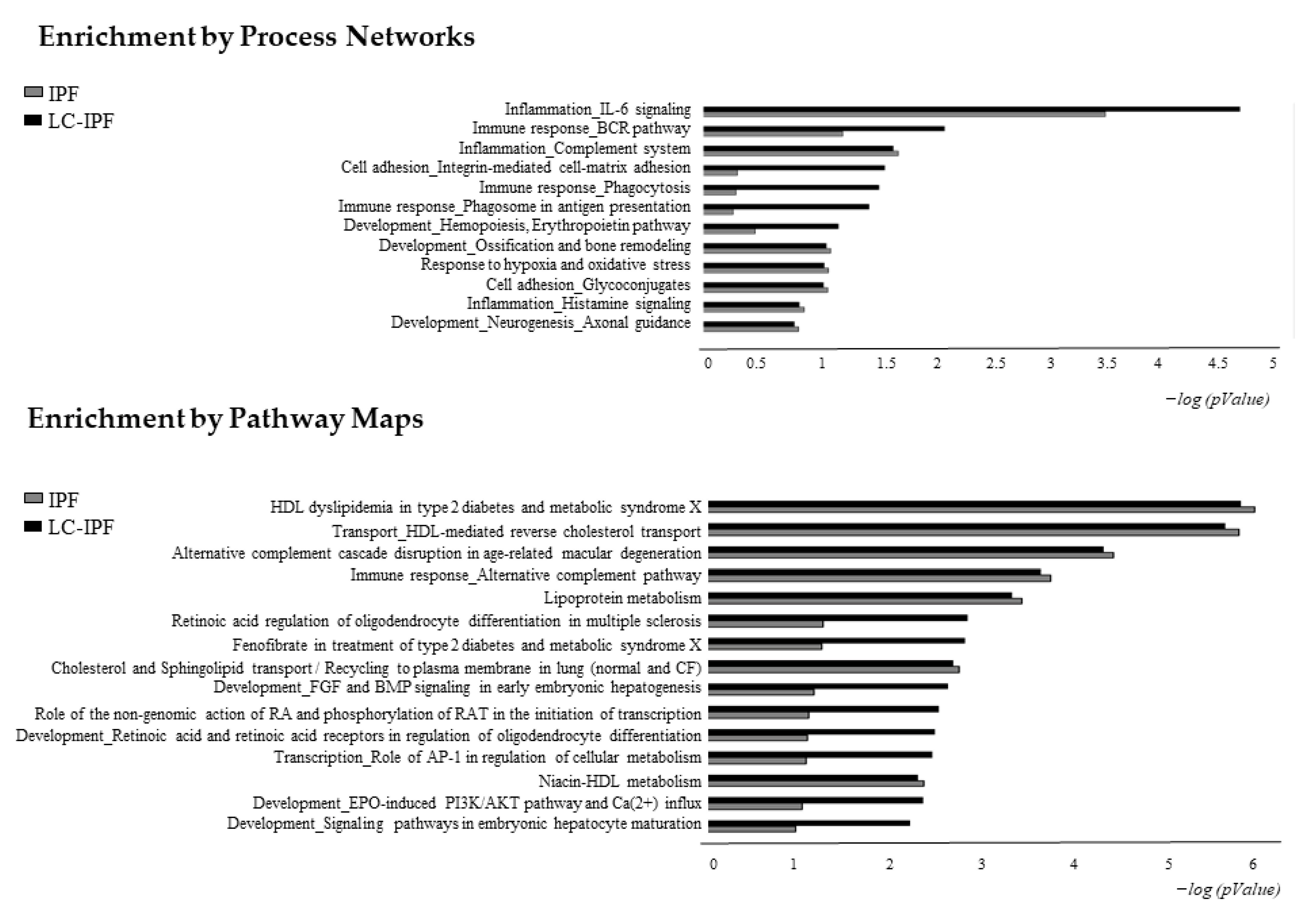
2.3. Enrichment Analysis Discovered Dynamics of Inflammation, Lipid Metabolism, and Cell Adhesion Biomarkers in IPF and LC-IPF
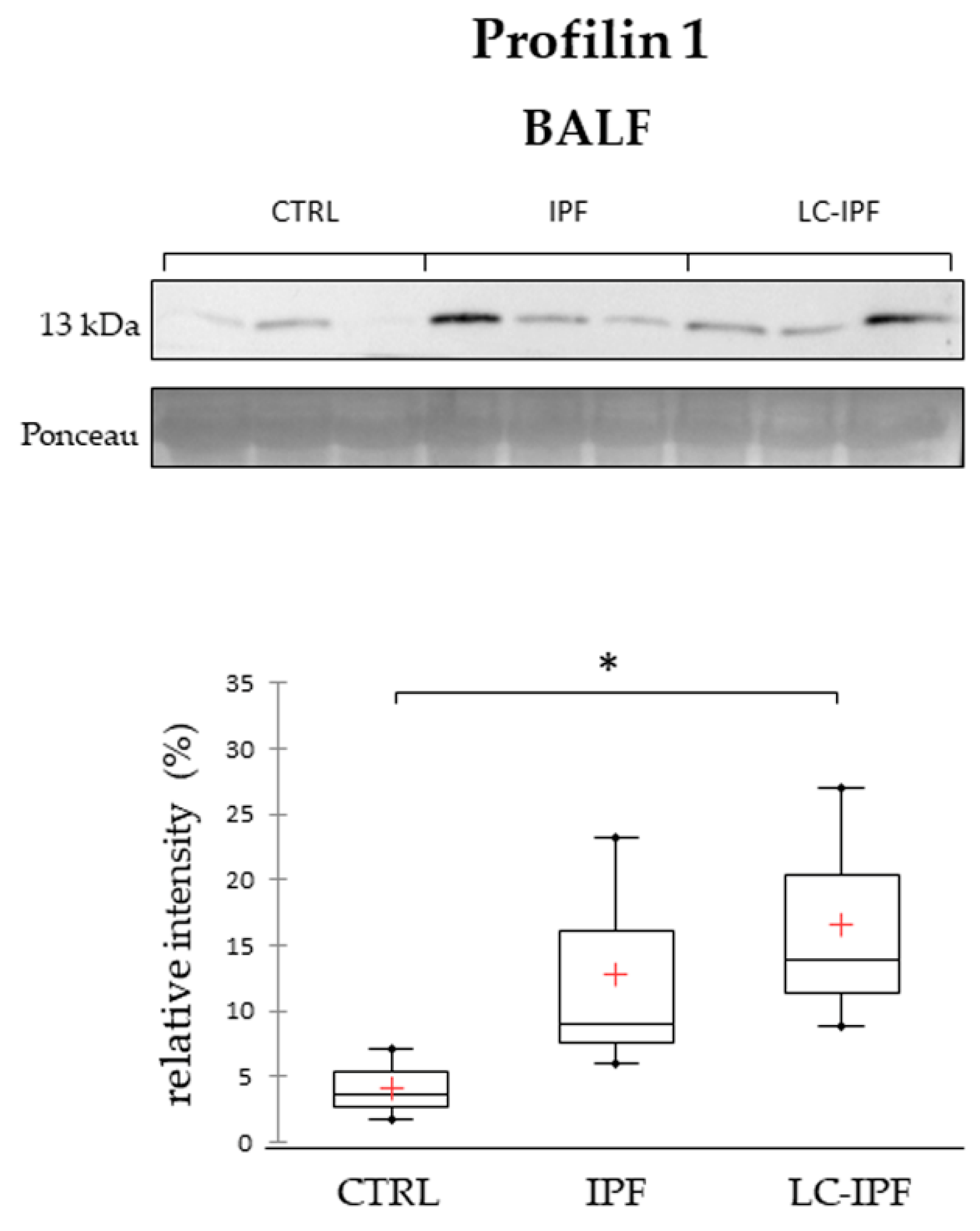
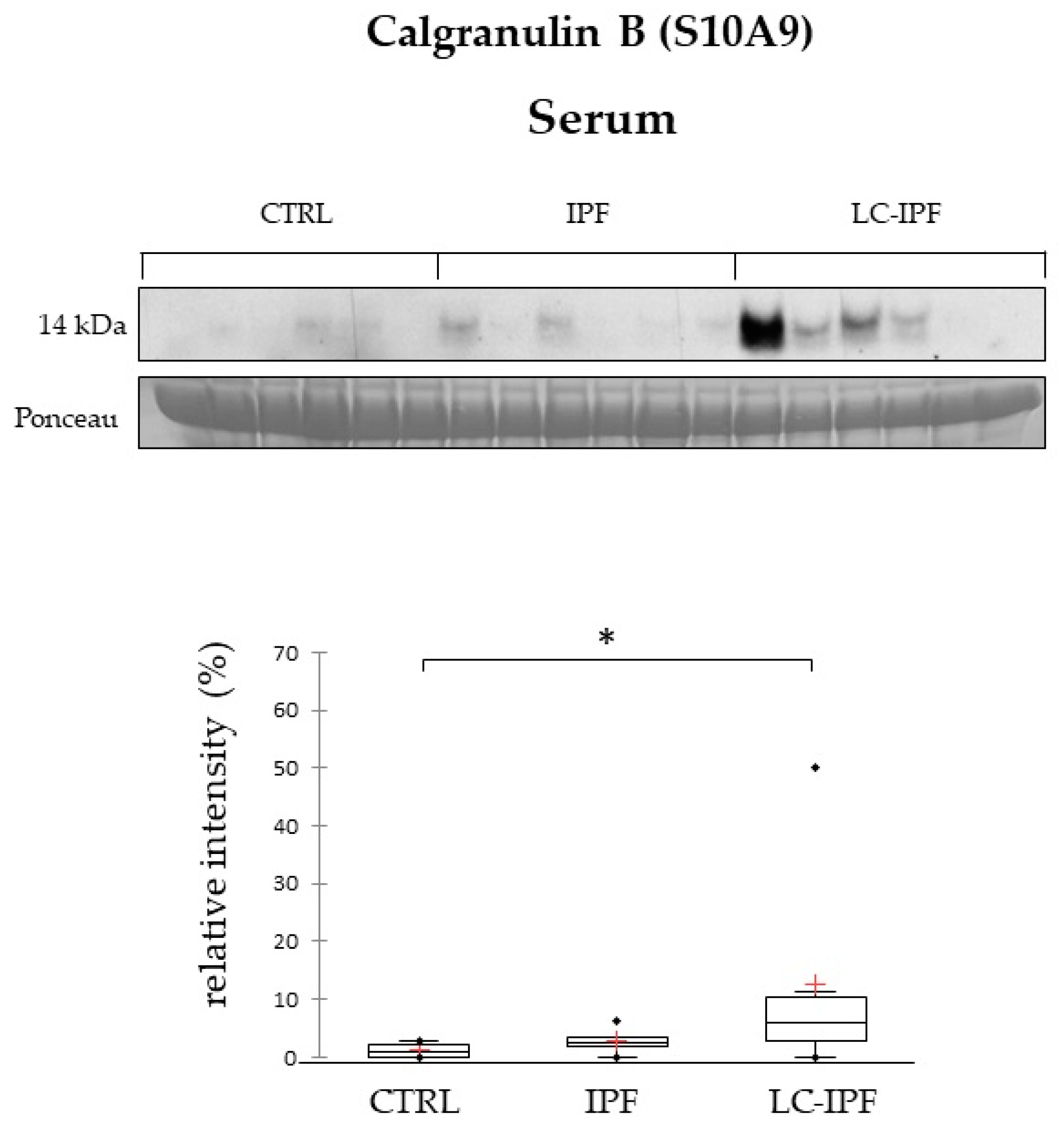
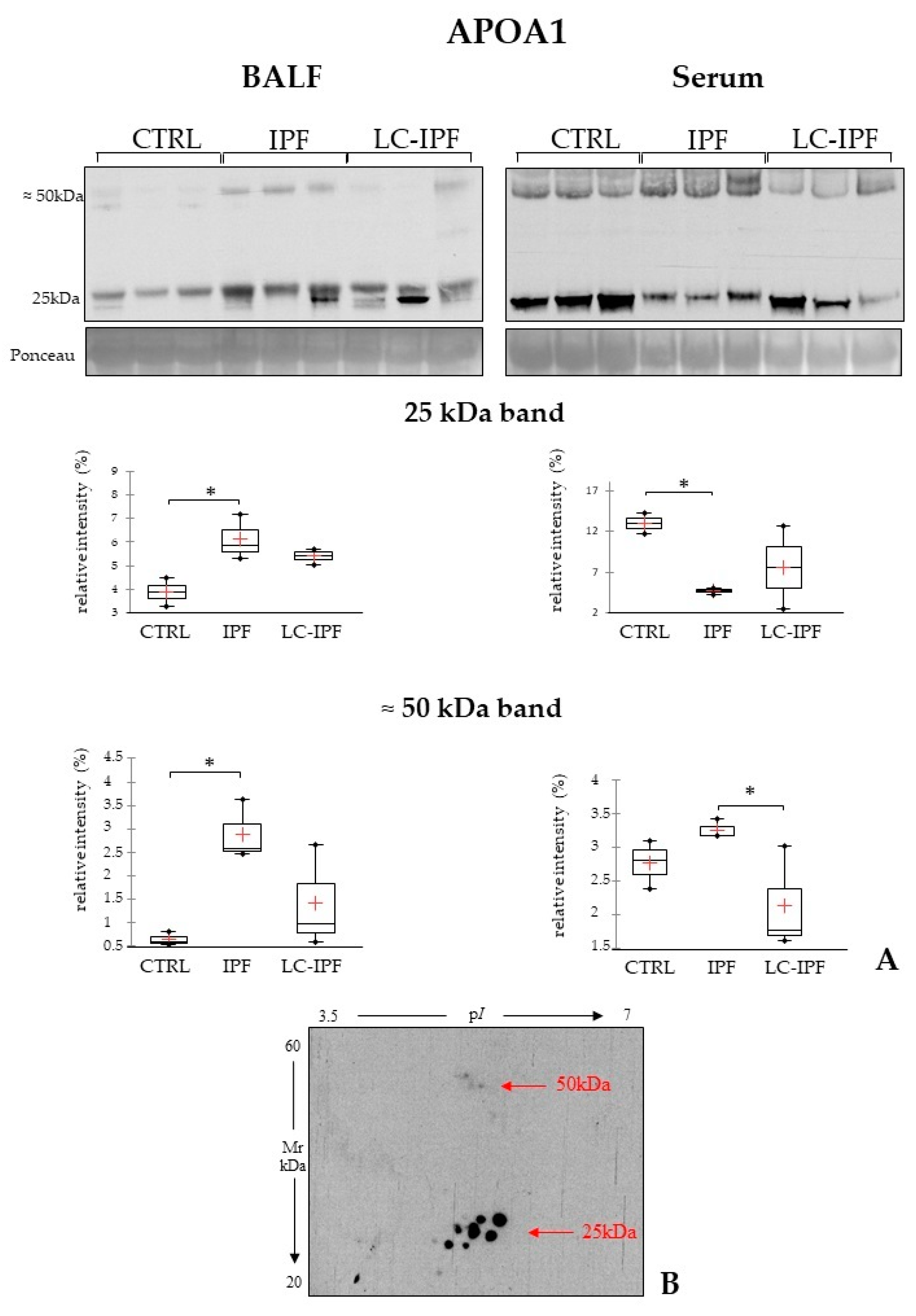
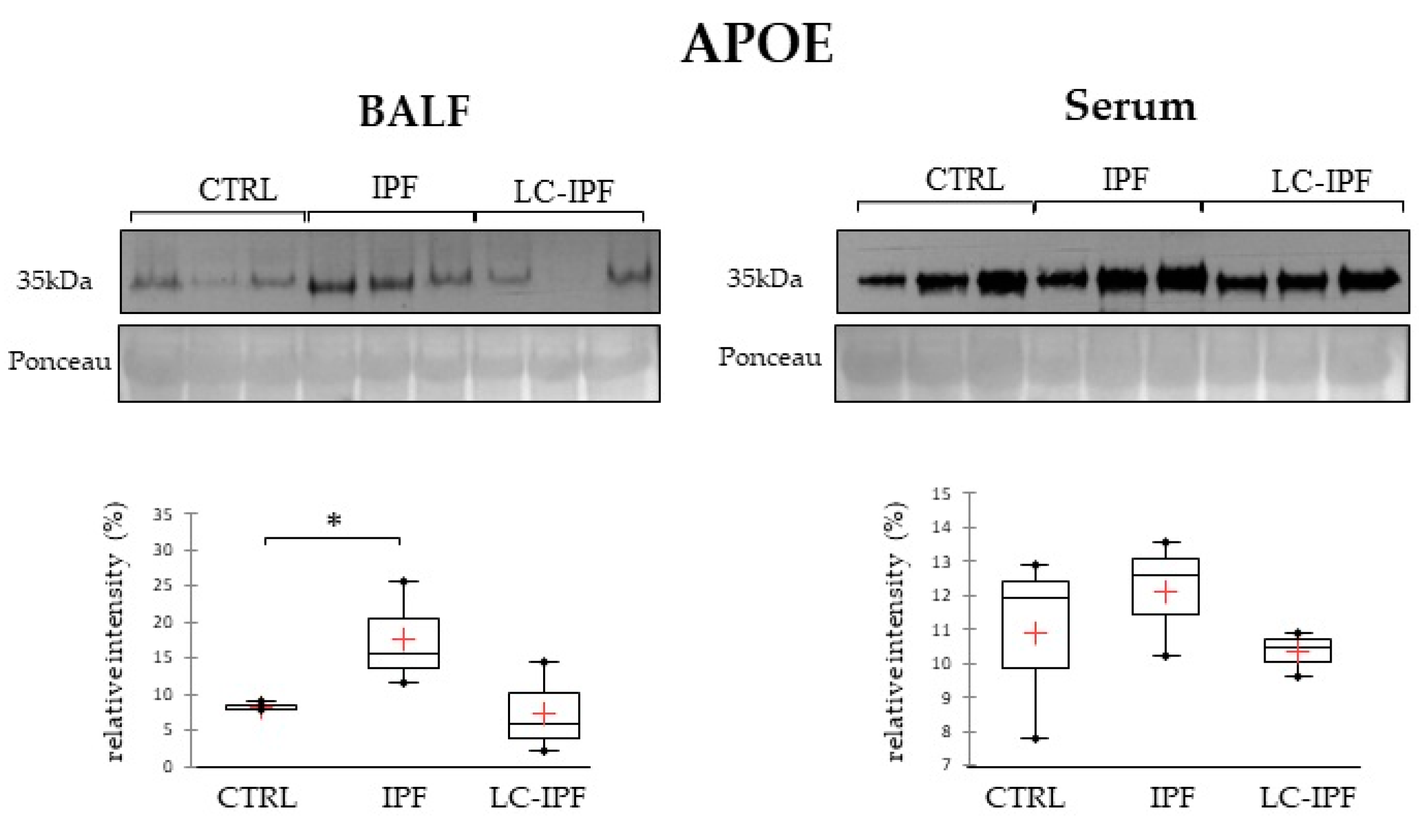
2.4. Western Blot Analyses in BALF and Serum Samples of APOAI, APOE, PROF1 and S10A9, Pivotal in IPF and/or LC-IPF
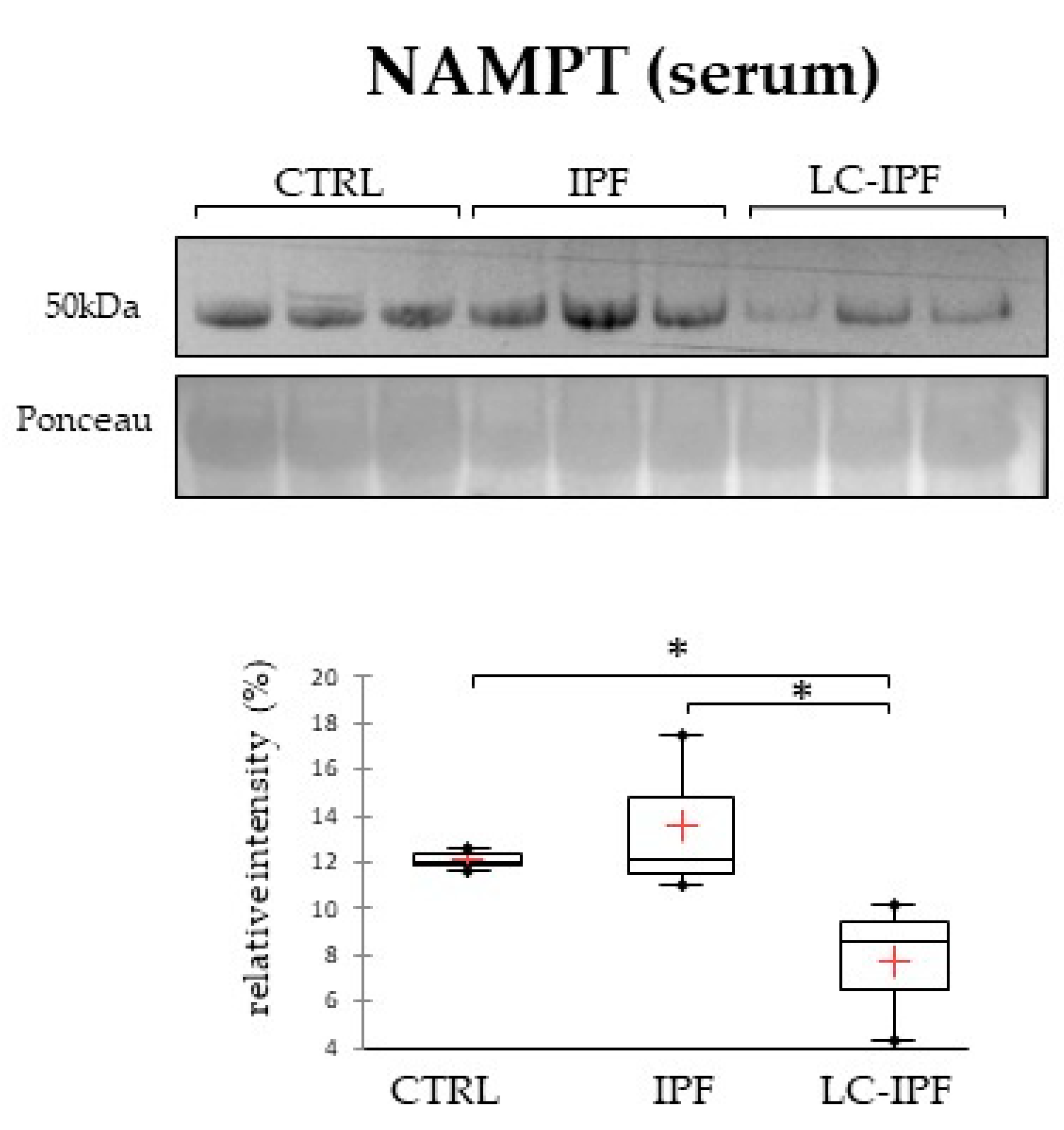
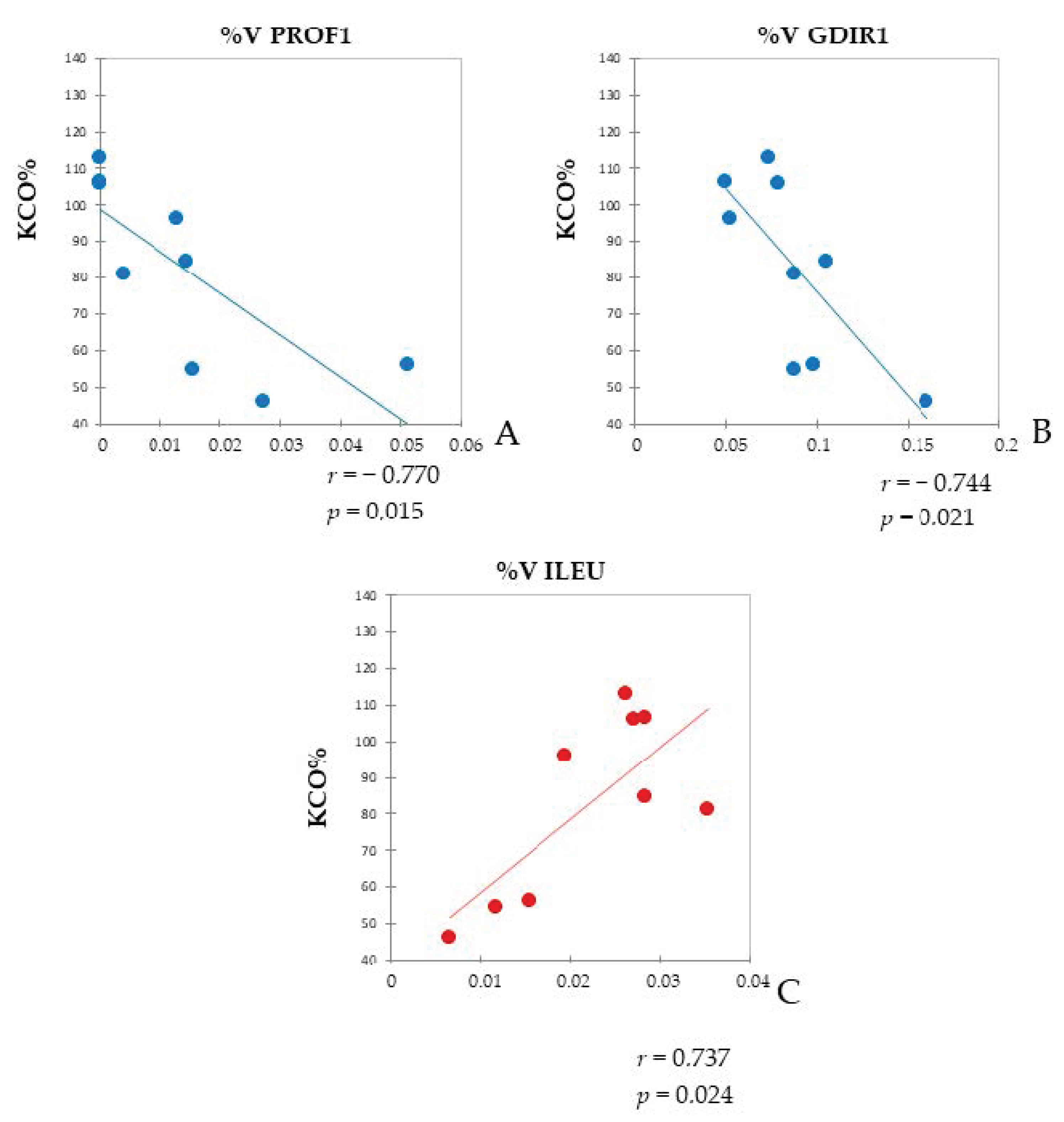
2.5. Pearson’s Correlation of Differential Proteins Highlighted a Relationship between Inflammation and Lipid Metabolism and Transport in IPF and LC-IPF and the Crossroad Role of NAMPT
3. Discussion
4. Materials and Methods
4.1. Population, Bronchoscopy, and Samples Collection
4.2. BALF Preparation for Two-Dimensional Electrophoresis
4.3. Two-Dimensional Electrophoresis
4.4. Statistical Analysis
4.5. MALDI-TOF Identification by PMF
4.6. MetaCore Enrichment Analysis
4.7. Western Blot Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| %V | percentage relative volume |
| 1DEWB | 1-Dimensional Western Blot |
| 2-DE | Two-Dimensional Electrophoresis |
| 2DEWB | Two-Dimensional Western Blot |
| AACT | Alpha-1-antichymotrypsin |
| ACN | acetonitrile |
| ADC | Adenocarcinoma |
| ALBU | Albumin |
| ANXA3 | Annexin A3 |
| APOA1 | Apolipoprotein A1 |
| APOE | Apolipoprotein E |
| ATS/ERS | American Thoracic Society/European Respiratory Society |
| B2MG | Beta-2-Microglobulin |
| BAL | Bronchoalveolar Lavage |
| BALF | Fluidic part of BAL |
| BCR | B Cell Antigen Receptor |
| BMP | Bone Morphogenetic Protein |
| C/EBP | CCAAT-Enhancer-Binding Proteins |
| CAYP1 | Calcyphosin |
| CFAB | Complement Factor B |
| CHAPS | 3-((3-cholamidopropyl) dimethylammonio)-1-propanesulfonate |
| CHCA | α-cyano-4-hydroxycinnamic acid |
| CTRL | Healthy Controls |
| CYTB | Cystatin |
| DASs | Differentially Abundant Spots |
| DLCO | Diffusing Capacity Of The Lungs For Carbon Monoxide |
| DTE | 1,4-Dithioerythritol |
| E2F1 | E2F Transcription Factor 1 |
| ESR1 | Estrogen Receptor 1 |
| FABP4 | Fatty Acid Binding Protein 4 |
| FEV1 | Forced Expiratory Volume in the 1st second |
| FETUA | Alpha-2-HS-glycoprotein |
| FOXA3 | Forkhead Box A3 |
| FRIL | Ferritin Light Chain |
| FVC | Forced Vital Capacity |
| GDIR1 | rho-specific guanine nucleotide dissociation inhibitor α |
| HRCT | High-resolution computed tomography of the chest |
| HIF-1A | Hypoxia-Inducible Factor 1-Alpha |
| HBA | Hemoglobin subunit alpha |
| HBB | Hemoglobin subunit beta |
| HDL | High Density Lipoproteins |
| HNF-4α | Hepatocyte Nuclear Factor 4 Alpha |
| HP | Haptoglobin |
| HRCT | High-resolution computed tomography |
| IGHA1 | Immunoglobulin Heavy Constant Alpha 1 |
| IGHG1 | Immunoglobulin Heavy Constant Gamma |
| IGKC | Immunoglobulin Kappa Constant |
| IL-6 | Interleukin-6 |
| ILDs | Interstitial Lung Diseases |
| ILEU | Leukocyte Elastase Inhibitor |
| IPF | Idiopathic Pulmonary Fibrosis |
| LC | Lung Cancer |
| LC-IPF | Lung Cancer associated with Idiopathic Pulmonary Fibrosis |
| MALDI | Matrix Assisted Laser Desorption/Ionization |
| MW | Molecular Weight |
| NAMPT | Nicotinamide Phosphoribosyltransferase |
| NF-Kb | Nuclear Factor Kappa-Light-Chain-Enhancer of Activated B Cells |
| NSCLC | Non-Small Cell Lung Cancer |
| PC1 | First Principal Component |
| PC2 | Second Principal Component |
| PC3 | Third Principal Component |
| PCA | Principal Component Analysis |
| PCs | Principal Components |
| PFT | Pulmonary Function Test |
| pI | Isoelectric Point |
| PMF | Peptide Mass Fingerprinting |
| PRDX1 | Peroxiredoxin 1 |
| PROF1 | Profilin 1 |
| RBP4 | Binding Protein 4 |
| RET4 | Retinol Binding Protein 4 |
| S10A9 | Calgranulin B |
| S10AB | Protein S100-A11 |
| SFTA | Pulmonary Surfactant-Associated Protein A |
| SDS | Sodium Dodecyl Sulfate |
| SP-A | Surfactant A2 |
| SPDEF | SAM Pointed Domain-Containing Ets Transcription Factor |
| STAT3 | Signal Transducer and Activator Of Transcription 3 |
| TLC | Total Lung Capacity |
| TFA | Trifluoroacetic Acid |
| TLR4 | Toll-like receptor 4 |
| ToF | Time Of Flight |
| TR-alpha | Thyroid Hormone Receptor Alpha |
| TRFE | Serotransferrin |
| TTHY | Transthyretin |
| WB | Western Blot |
References
- Liu, Y.; Zhu, M.; Geng, J.; Ban, C.; Zhang, S.; Chen, W.; Ren, Y.; He, X.; Chen, W.; Dai, H. Incidence and Radiologic-Pathological Features of Lung Cancer in Idiopathic Pulmonary Fibrosis. Clin. Respir. J. 2018, 12, 1700–1705. [Google Scholar] [CrossRef] [PubMed]
- Landi, C.; Bargagli, E.; Carleo, A.; Bianchi, L.; Gagliardi, A.; Prasse, A.; Perari, M.G.; Refini, R.M.; Bini, L.; Rottoli, P. A System Biology Study of BALF from Patients Affected by Idiopathic Pulmonary Fibrosis (IPF) and Healthy Controls. Proteom. Clin. Appl. 2014, 8, 932–950. [Google Scholar] [CrossRef] [PubMed]
- Landi, C.; Bargagli, E.; Bianchi, L.; Gagliardi, A.; Carleo, A.; Bennett, D.; Perari, M.G.; Armini, A.; Prasse, A.; Rottoli, P.; et al. Towards a Functional Proteomics Approach to the Comprehension of Idiopathic Pulmonary Fibrosis, Sarcoidosis, Systemic Sclerosis and Pulmonary Langerhans Cell Histiocytosis. J. Proteom. 2013, 83, 60–75. [Google Scholar] [CrossRef] [PubMed]
- Raghu, G.; Remy-Jardin, M.; Myers, J.L.; Richeldi, L.; Ryerson, C.J.; Lederer, D.J.; Behr, J.; Cottin, V.; Danoff, S.K.; Morell, F.; et al. Diagnosis of Idiopathic Pulmonary Fibrosis. An Official ATS/ERS/JRS/ALAT Clinical Practice Guideline. Am. J. Respir. Crit. Care Med. 2018, 198, e44–e68. [Google Scholar] [CrossRef] [PubMed]
- Kewalramani, N.; Machahua, C.; Poletti, V.; Cadranel, J.; Wells, A.U.; Funke-Chambour, M. Lung Cancer in Patients with Fibrosing Interstitial Lung Diseases: An Overview of Current Knowledge and Challenges. ERJ Open Res. 2022, 8, 00115–02022. [Google Scholar] [CrossRef] [PubMed]
- Raghu, G.; Amatto, V.C.; Behr, J.; Stowasser, S. Comorbidities in Idiopathic Pulmonary Fibrosis Patients: A Systematic Literature Review. Eur. Respir. J. 2015, 46, 1113–1130. [Google Scholar] [CrossRef]
- Kato, E.; Takayanagi, N.; Takaku, Y.; Kagiyama, N.; Kanauchi, T.; Ishiguro, T.; Sugita, Y. Incidence and Predictive Factors of Lung Cancer in Patients with Idiopathic Pulmonary Fibrosis. ERJ Open Res. 2018, 4, 00111–02016. [Google Scholar] [CrossRef]
- Spagnolo, P.; Tonelli, R.; Cocconcelli, E.; Stefani, A.; Richeldi, L. Idiopathic Pulmonary Fibrosis: Diagnostic Pitfalls and Therapeutic Challenges. Multidiscip. Respir. Med. 2012, 7, 42. [Google Scholar] [CrossRef]
- Wynn, T.A. Integrating Mechanisms of Pulmonary Fibrosis. J. Exp. Med. 2011, 208, 1339–1350. [Google Scholar] [CrossRef]
- d’Alessandro, M.; Bergantini, L.; Torricelli, E.; Cameli, P.; Lavorini, F.; Pieroni, M.; Refini, R.M.; Sestini, P.; Bargagli, E. Systematic Review and Metanalysis of Oncomarkers in IPF Patients and Serial Changes of Oncomarkers in a Prospective Italian Real-Life Case Series. Cancers 2021, 13, 539. [Google Scholar] [CrossRef]
- Ballester, B.; Milara, J.; Cortijo, J. Idiopathic Pulmonary Fibrosis and Lung Cancer: Mechanisms and Molecular Targets. Int. J. Mol. Sci. 2019, 20, E593. [Google Scholar] [CrossRef] [PubMed]
- Aubry, M.-C.; Myers, J.L.; Douglas, W.W.; Tazelaar, H.D.; Washington Stephens, T.L.; Hartman, T.E.; Deschamps, C.; Pankratz, V.S. Primary Pulmonary Carcinoma in Patients with Idiopathic Pulmonary Fibrosis. Mayo Clin. Proc. 2002, 77, 763–770. [Google Scholar] [CrossRef] [PubMed]
- Kreuter, M.; Ehlers-Tenenbaum, S.; Schaaf, M.; Oltmanns, U.; Palmowski, K.; Hoffmann, H.; Schnabel, P.A.; Heußel, C.-P.; Puderbach, M.; Herth, F.J.F.; et al. Treatment and Outcome of Lung Cancer in Idiopathic Interstitial Pneumonias. Sarcoidosis Vasc. Diffuse Lung Dis 2015, 31, 266–274. [Google Scholar] [PubMed]
- Guo, M.; Tomoshige, K.; Meister, M.; Muley, T.; Fukazawa, T.; Tsuchiya, T.; Karns, R.; Warth, A.; Fink-Baldauf, I.M.; Nagayasu, T.; et al. Gene Signature Driving Invasive Mucinous Adenocarcinoma of the Lung. EMBO Mol. Med. 2017, 9, 462–481. [Google Scholar] [CrossRef]
- Vancheri, C.; Cottin, V.; Kreuter, M.; Hilberg, O. IPF, Comorbidities and Management Implications. Sarcoidosis Vasc. Diffuse Lung Dis. 2015, 32 (Suppl. S1), 17–23. [Google Scholar]
- Landi, C.; Carleo, A.; Vantaggiato, L.; Bergantini, L.; d’Alessandro, M.; Cameli, P.; Bargagli, E. Common Molecular Pathways Targeted by Nintedanib in Cancer and IPF: A Bioinformatic Study. Pulm. Pharmacol. Ther. 2020, 64, 101941. [Google Scholar] [CrossRef]
- Cameli, P.; Carleo, A.; Bergantini, L.; Landi, C.; Prasse, A.; Bargagli, E. Oxidant/Antioxidant Disequilibrium in Idiopathic Pulmonary Fibrosis Pathogenesis. Inflammation 2020, 43, 1–7. [Google Scholar] [CrossRef]
- Kinoshita, T.; Goto, T. Molecular Mechanisms of Pulmonary Fibrogenesis and Its Progression to Lung Cancer: A Review. Int. J. Mol. Sci. 2019, 20, E1461. [Google Scholar] [CrossRef]
- Karampitsakos, T.; Tzilas, V.; Tringidou, R.; Steiropoulos, P.; Aidinis, V.; Papiris, S.A.; Bouros, D.; Tzouvelekis, A. Lung Cancer in Patients with Idiopathic Pulmonary Fibrosis. Pulm. Pharmacol. Ther. 2017, 45, 1–10. [Google Scholar] [CrossRef]
- Landi, C.; Bargagli, E.; Carleo, A.; Refini, R.M.; Bennett, D.; Bianchi, L.; Cillis, G.; Prasse, A.; Bini, L.; Rottoli, P. Bronchoalveolar Lavage Proteomic Analysis in Pulmonary Fibrosis Associated with Systemic Sclerosis: S100A6 and 14-3-3ε as Potential Biomarkers. Rheumatology 2019, 58, 165–178. [Google Scholar] [CrossRef]
- Landi, C.; Bargagli, E.; Carleo, A.; Bianchi, L.; Gagliardi, A.; Cillis, G.; Perari, M.G.; Refini, R.M.; Prasse, A.; Bini, L.; et al. A Functional Proteomics Approach to the Comprehension of Sarcoidosis. J. Proteom. 2015, 128, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Landi, C.; Bargagli, E.; Magi, B.; Prasse, A.; Muller-Quernheim, J.; Bini, L.; Rottoli, P. Proteome Analysis of Bronchoalveolar Lavage in Pulmonary Langerhans Cell Histiocytosis. J. Clin. Bioinform. 2011, 1, 31. [Google Scholar] [CrossRef] [PubMed]
- Rottoli, P.; Bargagli, E.; Landi, C.; Magi, B. Proteomic Analysis in Interstitial Lung Diseases: A Review. Curr. Opin. Pulm. Med. 2009, 15, 470–478. [Google Scholar] [CrossRef]
- Carleo, A.; Bargagli, E.; Landi, C.; Bennett, D.; Bianchi, L.; Gagliardi, A.; Carnemolla, C.; Perari, M.G.; Cillis, G.; Armini, A.; et al. Comparative Proteomic Analysis of Bronchoalveolar Lavage of Familial and Sporadic Cases of Idiopathic Pulmonary Fibrosis. J. Breath Res. 2016, 10, 026007. [Google Scholar] [CrossRef]
- Carleo, A.; Landi, C.; Prasse, A.; Bergantini, L.; D’Alessandro, M.; Cameli, P.; Janciauskiene, S.; Rottoli, P.; Bini, L.; Bargagli, E. Proteomic Characterization of Idiopathic Pulmonary Fibrosis Patients: Stable versus Acute Exacerbation. Monaldi Arch. Chest Dis. 2020, 90, 2. [Google Scholar] [CrossRef]
- Bargagli, E.; Refini, R.M.; d’Alessandro, M.; Bergantini, L.; Cameli, P.; Vantaggiato, L.; Bini, L.; Landi, C. Metabolic Dysregulation in Idiopathic Pulmonary Fibrosis. Int. J. Mol. Sci. 2020, 21, E5663. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Gordon, E.M.; Figueroa, D.M.; Barochia, A.V.; Levine, S.J. Emerging Roles of Apolipoprotein E and Apolipoprotein A-I in the Pathogenesis and Treatment of Lung Disease. Am. J. Respir. Cell Mol. Biol. 2016, 55, 159–169. [Google Scholar] [CrossRef]
- Yoshino, J.; Mills, K.F.; Yoon, M.J.; Imai, S. Nicotinamide Mononucleotide, a Key NAD(+) Intermediate, Treats the Pathophysiology of Diet- and Age-Induced Diabetes in Mice. Cell Metab. 2011, 14, 528–536. [Google Scholar] [CrossRef]
- Cioffi, C.L.; Racz, B.; Varadi, A.; Freeman, E.E.; Conlon, M.P.; Chen, P.; Zhu, L.; Kitchen, D.B.; Barnes, K.D.; Martin, W.H.; et al. Design, Synthesis, and Preclinical Efficacy of Novel Nonretinoid Antagonists of Retinol-Binding Protein 4 in the Mouse Model of Hepatic Steatosis. J. Med. Chem. 2019, 62, 5470–5500. [Google Scholar] [CrossRef]
- Kotnik, P.; Fischer-Posovszky, P.; Wabitsch, M. RBP4: A Controversial Adipokine. Eur. J. Endocrinol. 2011, 165, 703–711. [Google Scholar] [CrossRef]
- Sun, W.; Shi, Y.; Yang, J.; Song, X.; Zhang, Y.; Zhang, W.; Zhou, X. Transthyretin and Retinol-Binding Protein as Discriminators of Diabetic Retinopathy in Type 1 Diabetes Mellitus. Int. Ophthalmol. 2022, 42, 1041–1049. [Google Scholar] [CrossRef]
- Pandey, G.K.; Balasubramanyam, J.; Balakumar, M.; Deepa, M.; Anjana, R.M.; Abhijit, S.; Kaviya, A.; Velmurugan, K.; Miranda, P.; Balasubramanyam, M.; et al. Altered Circulating Levels of Retinol Binding Protein 4 and Transthyretin in Relation to Insulin Resistance, Obesity, and Glucose Intolerance in Asian Indians. Endocr. Pract. 2015, 21, 861–869. [Google Scholar] [CrossRef]
- Kwanbunjan, K.; Panprathip, P.; Phosat, C.; Chumpathat, N.; Wechjakwen, N.; Puduang, S.; Auyyuenyong, R.; Henkel, I.; Schweigert, F.J. Association of Retinol Binding Protein 4 and Transthyretin with Triglyceride Levels and Insulin Resistance in Rural Thais with High Type 2 Diabetes Risk. BMC Endocr. Disord. 2018, 18, 26. [Google Scholar] [CrossRef]
- Jiao, C.; Cui, L.; Ma, A.; Li, N.; Si, H. Elevated Serum Levels of Retinol-Binding Protein 4 Are Associated with Breast Cancer Risk: A Case-Control Study. PLoS ONE 2016, 11, e0167498. [Google Scholar] [CrossRef]
- Wang, D.-D.; Zhao, Y.-M.; Wang, L.; Ren, G.; Wang, F.; Xia, Z.-G.; Wang, X.-L.; Zhang, T.; Pan, Q.; Dai, Z.; et al. Preoperative Serum Retinol-Binding Protein 4 Is Associated with the Prognosis of Patients with Hepatocellular Carcinoma after Curative Resection. J. Cancer Res. Clin. Oncol. 2011, 137, 651–658. [Google Scholar] [CrossRef]
- Wlodarczyk, B.; Gasiorowska, A.; Borkowska, A.; Malecka-Panas, E. Evaluation of Insulin-like Growth Factor (IGF-1) and Retinol Binding Protein (RBP-4) Levels in Patients with Newly Diagnosed Pancreatic Adenocarcinoma (PDAC). Pancreatology 2017, 17, 623–628. [Google Scholar] [CrossRef]
- Cheng, Y.; Liu, C.; Zhang, N.; Wang, S.; Zhang, Z. Proteomics Analysis for Finding Serum Markers of Ovarian Cancer. BioMed Res. Int. 2014, 2014, e179040. [Google Scholar] [CrossRef]
- Prapunpoj, P.; Leelawatwatana, L.; Schreiber, G.; Richardson, S.J. Change in Structure of the N-Terminal Region of Transthyretin Produces Change in Affinity of Transthyretin to T4 and T3. FEBS J. 2006, 273, 4013–4023. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Dhankhar, P.; Dalal, V. Structure-Based Mimicking of Hydroxylated Biphenyl Congeners (OHPCBs) for Human Transthyretin, an Important Enzyme of Thyroid Hormone System. J. Mol. Graph Model 2021, 105, 107870. [Google Scholar] [CrossRef] [PubMed]
- Nygård, M.; Wahlström, G.M.; Gustafsson, M.V.; Tokumoto, Y.M.; Bondesson, M. Hormone-Dependent Repression of the E2F-1 Gene by Thyroid Hormone Receptors. Mol. Endocrinol. 2003, 17, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Hughes, J.M.B.; Pride, N.B. Examination of the Carbon Monoxide Diffusing Capacity (DL(CO)) in Relation to Its KCO and VA Components. Am. J. Respir. Crit. Care Med. 2012, 186, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Liu, Y.; Wan, C.; Lu, C.; Cai, J.; He, S.; Ni, T.; Zhu, J.; Wei, L.; Zhang, Y.; et al. Decreased Expression of SERPINB1 Correlates with Tumor Invasion and Poor Prognosis in Hepatocellular Carcinoma. J. Mol. Hist. 2014, 45, 59–68. [Google Scholar] [CrossRef]
- Davalieva, K.; Kostovska, I.M.; Kiprijanovska, S.; Markoska, K.; Kubelka-Sabit, K.; Filipovski, V.; Stavridis, S.; Stankov, O.; Komina, S.; Petrusevska, G.; et al. Proteomics Analysis of Malignant and Benign Prostate Tissue by 2D DIGE/MS Reveals New Insights into Proteins Involved in Prostate Cancer. Prostate 2015, 75, 1586–1600. [Google Scholar] [CrossRef] [PubMed]
- Chou, R.-H.; Wen, H.-C.; Liang, W.-G.; Lin, S.-C.; Yuan, H.-W.; Wu, C.-W.; Chang, W.-S.W. Suppression of the Invasion and Migration of Cancer Cells by SERPINB Family Genes and Their Derived Peptides. Oncol. Rep. 2012, 27, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Hebert, J.D.; Myers, S.A.; Naba, A.; Abbruzzese, G.; Lamar, J.M.; Carr, S.A.; Hynes, R.O. Proteomic Profiling of the ECM of Xenograft Breast Cancer Metastases in Different Organs Reveals Distinct Metastatic Niches. Cancer Res. 2020, 80, 1475–1485. [Google Scholar] [CrossRef]
- Lerman, I.; Ma, X.; Seger, C.; Maolake, A.; Garcia-Hernandez, M. de la L.; Rangel-Moreno, J.; Ackerman, J.; Nastiuk, K.L.; Susiarjo, M.; Hammes, S.R. Epigenetic Suppression of SERPINB1 Promotes Inflammation-Mediated Prostate Cancer Progression. Mol. Cancer Res. 2019, 17, 845–859. [Google Scholar] [CrossRef]
- Goel, A.; Chhabra, R.; Ahmad, S.; Prasad, A.K.; Parmar, V.S.; Ghosh, B.; Saini, N. DAMTC Regulates Cytoskeletal Reorganization and Cell Motility in Human Lung Adenocarcinoma Cell Line: An Integrated Proteomics and Transcriptomics Approach. Cell Death Dis. 2012, 3, e402. [Google Scholar] [CrossRef]
- Meex, R.C.R.; Watt, M.J. Hepatokines: Linking Nonalcoholic Fatty Liver Disease and Insulin Resistance. Nat. Rev. Endocrinol. 2017, 13, 509–520. [Google Scholar] [CrossRef]
- Ding, Z.; Bae, Y.H.; Roy, P. Molecular Insights on Context-Specific Role of Profilin-1 in Cell Migration. Cell Adh. Migr. 2012, 6, 442–449. [Google Scholar] [CrossRef]
- Ali, R.A.; Gandhi, A.A.; Meng, H.; Yalavarthi, S.; Vreede, A.P.; Estes, S.K.; Palmer, O.R.; Bockenstedt, P.L.; Pinsky, D.J.; Greve, J.M.; et al. Adenosine Receptor Agonism Protects against NETosis and Thrombosis in Antiphospholipid Syndrome. Nat. Commun. 2019, 10, 1916. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Wan, R.; Hu, C.; Lu, Y. Profilin 1 Protein and Its Implications for Cancers. Oncology 2021, 35, 402–409. [Google Scholar] [CrossRef] [PubMed]
- Almatroodi, S.A.; McDonald, C.F.; Collins, A.L.; Darby, I.A.; Pouniotis, D.S. Quantitative Proteomics of Bronchoalveolar Lavage Fluid in Lung Adenocarcinoma. Cancer Genom. Proteom. 2015, 12, 39–48. [Google Scholar]
- Allen, A.; Gau, D.; Francoeur, P.; Sturm, J.; Wang, Y.; Martin, R.; Maranchie, J.; Duensing, A.; Kaczorowski, A.; Duensing, S.; et al. Actin-Binding Protein Profilin1 Promotes Aggressiveness of Clear-Cell Renal Cell Carcinoma Cells. J. Biol. Chem. 2020, 295, 15636–15649. [Google Scholar] [CrossRef]
- Shaba, E.; Landi, C.; Carleo, A.; Vantaggiato, L.; Paccagnini, E.; Gentile, M.; Bianchi, L.; Lupetti, P.; Bargagli, E.; Prasse, A.; et al. Proteome Characterization of BALF Extracellular Vesicles in Idiopathic Pulmonary Fibrosis: Unveiling Undercover Molecular Pathways. Int. J. Mol. Sci. 2021, 22, 5696. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, H.; Li, J.; Liu, Y.; Ding, Y. Overexpression of Rho GDP-Dissociation Inhibitor Alpha Is Associated with Tumor Progression and Poor Prognosis of Colorectal Cancer. J. Proteom. Res. 2008, 7, 3994–4003. [Google Scholar] [CrossRef] [PubMed]
- Rong, F.; Li, W.; Chen, K.; Li, D.M.; Duan, W.M.; Feng, Y.Z.; Li, F.; Zhou, X.W.; Fan, S.J.; Liu, Y.; et al. Knockdown of RhoGDIα Induces Apoptosis and Increases Lung Cancer Cell Chemosensitivity to Paclitaxel. Neoplasma 2012, 59, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.J.; Kim, J.-T.; Baek, K.E.; Kim, B.-Y.; Lee, H.G. Regulation of Rho GTPases by RhoGDIs in Human Cancers. Cells 2019, 8, 1037. [Google Scholar] [CrossRef]
- Bargagli, E.; Olivieri, C.; Prasse, A.; Bianchi, N.; Magi, B.; Cianti, R.; Bini, L.; Rottoli, P. Calgranulin B (S100A9) Levels in Bronchoalveolar Lavage Fluid of Patients with Interstitial Lung Diseases. Inflammation 2008, 31, 351–354. [Google Scholar] [CrossRef]
- Rottoli, P.; Magi, B.; Perari, M.G.; Liberatori, S.; Nikiforakis, N.; Bargagli, E.; Cianti, R.; Bini, L.; Pallini, V. Cytokine Profile and Proteome Analysis in Bronchoalveolar Lavage of Patients with Sarcoidosis, Pulmonary Fibrosis Associated with Systemic Sclerosis and Idiopathic Pulmonary Fibrosis. Proteomics 2005, 5, 1423–1430. [Google Scholar] [CrossRef]
- Grebhardt, S.; Veltkamp, C.; Ströbel, P.; Mayer, D. Hypoxia and HIF-1 Increase S100A8 and S100A9 Expression in Prostate Cancer. Int. J. Cancer 2012, 131, 2785–2794. [Google Scholar] [CrossRef]
- Balamurugan, K. HIF-1 at the Crossroads of Hypoxia, Inflammation, and Cancer. Int. J. Cancer 2016, 138, 1058–1066. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Potdar, A.A.; Gong, Y.; Eswarappa, S.M.; Donnola, S.; Lathia, J.D.; Hambardzumyan, D.; Rich, J.N.; Fox, P.L. Profilin-1 Phosphorylation Directs Angiocrine Expression and Glioblastoma Progression through HIF-1α Accumulation. Nat. Cell Biol. 2014, 16, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Bargagli, E.; Olivieri, C.; Cintorino, M.; Refini, R.M.; Bianchi, N.; Prasse, A.; Rottoli, P. Calgranulin B (S100A9/MRP14): A Key Molecule in Idiopathic Pulmonary Fibrosis? Inflammation 2011, 34, 85–91. [Google Scholar] [CrossRef]
- Bennett, D.; Salvini, M.; Fui, A.; Cillis, G.; Cameli, P.; Mazzei, M.A.; Fossi, A.; Refini, R.M.; Rottoli, P. Calgranulin B and KL-6 in Bronchoalveolar Lavage of Patients with IPF and NSIP. Inflammation 2019, 42, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Xu, F.; Yu, J.; Yue, D.; Ren, X.; Wang, C. Up-regulation of the expression of S100A8 and S100A9 in lung adenocarcinoma and its correlation with inflammation and other clinical features. Chin. Med. J. 2010, 123, 2215–2220. [Google Scholar] [CrossRef]
- Bagheri, V.; Apostolopoulos, V. Pro-Inflammatory S100A9 Protein: A Double-Edged Sword in Cancer? Inflammation 2019, 42, 1137–1138. [Google Scholar] [CrossRef]
- Nacken, W.; Roth, J.; Sorg, C.; Kerkhoff, C. S100A9/S100A8: Myeloid Representatives of the S100 Protein Family as Prominent Players in Innate Immunity. Microsc. Res. Tech. 2003, 60, 569–580. [Google Scholar] [CrossRef]
- Hiratsuka, S.; Watanabe, A.; Aburatani, H.; Maru, Y. Tumour-Mediated Upregulation of Chemoattractants and Recruitment of Myeloid Cells Predetermines Lung Metastasis. Nat. Cell. Biol. 2006, 8, 1369–1375. [Google Scholar] [CrossRef]
- Luo, G.; Tang, M.; Zhao, Q.; Lu, L.; Xie, Y.; Li, Y.; Liu, C.; Tian, L.; Chen, X.; Yu, X. Bone Marrow Adipocytes Enhance Osteolytic Bone Destruction by Activating 1q21.3(S100A7/8/9-IL6R)-TLR4 Pathway in Lung Cancer. J. Cancer Res. Clin. Oncol. 2020, 146, 2241–2253. [Google Scholar] [CrossRef]
- He, X.; Wang, H.; Jin, T.; Xu, Y.; Mei, L.; Yang, J. TLR4 Activation Promotes Bone Marrow MSC Proliferation and Osteogenic Differentiation via Wnt3a and Wnt5a Signaling. PLoS ONE 2016, 11, e0149876. [Google Scholar] [CrossRef]
- Fang, W.-Y.; Chen, Y.-W.; Hsiao, J.-R.; Liu, C.-S.; Kuo, Y.-Z.; Wang, Y.-C.; Chang, K.-C.; Tsai, S.-T.; Chang, M.-Z.; Lin, S.-H.; et al. Elevated S100A9 Expression in Tumor Stroma Functions as an Early Recurrence Marker for Early-Stage Oral Cancer Patients through Increased Tumor Cell Invasion, Angiogenesis, Macrophage Recruitment and Interleukin-6 Production. Oncotarget 2015, 6, 28401–28424. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Li, D.; Wang, X.; Fang, E.; Yang, F.; Hu, A.; Wang, J.; Guo, Y.; Liu, Y.; Li, H.; et al. HNF4A-AS1/HnRNPU/CTCF Axis as a Therapeutic Target for Aerobic Glycolysis and Neuroblastoma Progression. J. Hematol. Oncol. 2020, 13, 24. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhang, K.; Yao, Y.; Liu, Y.; Ni, Y.; Liao, C.; Tu, Z.; Qiu, Y.; Wang, D.; Chen, D.; et al. Dual Nicotinamide Phosphoribosyltransferase and Epidermal Growth Factor Receptor Inhibitors for the Treatment of Cancer. Eur. J. Med. Chem. 2021, 211, 113022. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.; Gravel, M.; Bramoullé, A.; Bridon, G.; Avizonis, D.; Shore, G.C.; Roulston, A. Synergy between the NAMPT Inhibitor GMX1777(8) and Pemetrexed in Non-Small Cell Lung Cancer Cells Is Mediated by PARP Activation and Enhanced NAD Consumption. Cancer Res. 2014, 74, 5948–5954. [Google Scholar] [CrossRef]
- Li, W.; Liu, Y.; Li, Z.J.; Shi, Y.; Deng, J.; Bai, J.; Ma, L.; Zeng, X.X.; Feng, S.S.; Ren, J.L.; et al. Unravelling the Role of LncRNA WT1-AS/MiR-206/NAMPT Axis as Prognostic Biomarkers in Lung Adenocarcinoma. Biomolecules 2021, 11, 203. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE Database Resources in 2022: A Hub for Mass Spectrometry-Based Proteomics Evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef]
- Landi, C.; Cameli, P.; Vantaggiato, L.; Bergantini, L.; d’Alessandro, M.; Perruzza, M.; Carleo, A.; Shaba, E.; Di Giuseppe, F.; Angelucci, S.; et al. Ceruloplasmin and Oxidative Stress in Severe Eosinophilic Asthma Patients Treated with Mepolizumab and Benralizumab. Biochim. Biophys. Acta Proteins Proteom. 2021, 1869, 140563. [Google Scholar] [CrossRef]
- Towbin, H.; Staehelin, T.; Gordon, J. Electrophoretic Transfer of Proteins from Polyacrylamide Gels to Nitrocellulose Sheets: Procedure and Some Applications. Proc. Natl. Acad. Sci. USA 1979, 76, 4350–4354. [Google Scholar] [CrossRef]
| Parameters | IPF | LC-IPF | Healthy Controls | p-Value |
|---|---|---|---|---|
| N° | 23 | 7 | 15 | |
| Age (yrs) | 75 ± 6.1 | 63 ± 9 | 54.3 ± 12.9 | 0.0010 |
| Male gender (%) | 17 | 6 | 5 | 0.177 |
| Smoking status | ||||
| - Current | 0 | 0 | 1 | 0.854 |
| - Former | 12 | 3 | 4 | |
| - Never | 11 | 4 | 10 | |
| HRCT pattern | ||||
| - UIP | 20 | 6 | n.a | 0.954 |
| - Probable UIP | 3 | 1 | n.a | |
| PFTs | ||||
| - FVC (mL) | 2482 ± 804.2 | 2505 ± 688 | n.a | 0.8611 |
| - FVC (% p.v.) | 75.6 ± 18.6 | 63.5 ± 15.8 | n.a | 0.1994 |
| - FEV1 (mL) | 1952 ± 712.1 | 1920 ± 420.9 | n.a | 0.8013 |
| - FEV1 (% p.v.) | 78 ± 21.4 | 64.3 ± 17.4 | n.a | 0.179 |
| - DLCO (% p.v.) | 49.1 ± 21.4 | 36.3 ± 13.5 | n.a | 0.2772 |
| - KCO (% p.v.) | 89.4 ± 24.8 | 68.8 ± 23.5 | n.a | 0.272 |
| BAL cellular analysis | ||||
| - % Macrophage | 74.5 ± 21.1 | 65 ± 21.9 | 82 ± 15.3 | 0.0257 |
| - % Lymphocytes | 12.2 ± 9.5 | 12 ± 15.5 | 15.6 ± 12.1 | 0.2598 |
| - % Neutrophils | 11.2 ± 12.6 | 18 ± 17.1 | 5.4 ± 6.3 | 0.0140 |
| - % Eosinophils | 2.7 ± 4.1 | 5.6 ± 6.2 | 2.2 ± 1.6 | 0.1568 |
| Mascot Search Results | ||||||||
|---|---|---|---|---|---|---|---|---|
| Spot n° | Protein Name | Entry Name | Accession Number | MetaCore Name | pI MW | Score | Matched Peptides | Coverage (%) |
| 3 | Immunoglobulin heavy constant alpha 1 Fragment N | IGHA1_HUMAN | P01876 | IGHA1, IgA, IgA1 | 6.08–38,486 | 184 | 14/18 | 28 |
| 5 | Ferritin light chain | FRIL_HUMAN | P02792 | FTL | 5.51–20,064 | 187 | 18/47 | 68 |
| 7 | Hemoglobin subunit alpha | HBA_HUMAN | P69905 | Alpha1-globin | 8.72–15,305 | 181 | 10/12 | 69 |
| 10 | Retinol-binding protein 4 | RET4_HUMAN | P02753 | RBP4 | 5.76–23,337 | 136 | 8/14 | 52 |
| 13 | Immunoglobulin heavy constant alpha 1 Fragment N | IGHA1_HUMAN | P01876 | IGHA1, IgA, IgA1 | 6.08–38,486 | 163 | 11/13 | 25 |
| 14 | Haptoglobin | HPT_HUMAN | P00738 | HP, HP-alpha, HP-beta | 6.13–45,861 | 123 | 9/17 | 20 |
| 15 | Hemoglobin subunit beta | HBB_HUMAN | P68871 | VV-Hemorphin-7 | 6.75–16,102 | 138 | 8/10 | 61 |
| 16 | Hemoglobin subunit alpha | HBA_HUMAN | P69905 | Alpha1-globin | 8.72–15,305 | 135 | 7/7 | 61 |
| 17 | Beta-2-microglobulin | B2MG_HUMAN | P61769 | Beta-2-microglobulin | 6.06–13,820 | 250 | 11/11 | 87 |
| 19 | Fatty acid-binding protein, adipocyte | FABP4_HUMAN | P15090 | Fatty acid-binding protein | 6.59–14,824 | 207 | 13/18 | 68 |
| 22 | Beta-2-microglobulin | B2MG_HUMAN | P61769 | Beta-2-microglobulin | 6.06–13,820 | 84 | 4/5 | 44 |
| 25 | Annexin A3 | ANXA3_HUMAN | P12429 | Annexin III | 5.63–36,524 | 86 | 6/6 | 17 |
| 28 | Hemoglobin subunit alpha Fragment N | HBA_HUMAN | P69905 | Alpha1-globin | 8.72–15,305 | 81 | 5/6 | 34 |
| 29 | Protein S100-A9 | S10A9_HUMAN | P06702 | Calgranulin B | 5.71–13,291 | 151 | 7/9 | 84 |
| 30 | Protein S100-A6 | S10A6_HUMAN | P06703 | Calcyclin | 5.33–10,230 | 111 | 10/13 | 40 |
| 31 | Albumin fragment C | ALBU_HUMAN | P02768 | Albumin | 5.92–71,317 | 135 | 11/18 | 18 |
| 32 | Immunoglobulin kappa constant | IGKC_HUMAN | P01834 | IGKC | 6.11–11,929 | 132 | 6/7 | 59 |
| 33 | Rho GDP-dissociation inhibitor 1 | GDIR1_HUMAN | P52565 | RhoGDI alpha | 5.01–23,207 | 109 | 7/12 | 31 |
| 34 | Apolipoprotein A-I | APOA1_HUMAN | P02647 | HDL, Apoa1 | 5.56–30,759 | 268 | 20/28 | 53 |
| 35 | Immunoglobulin kappa constant | IGKC_HUMAN | P01834 | IGKC | 6.11–11,929 | 101 | 5/8 | 53 |
| 36 | Alpha-2-HS-glycoprotein | FETUA_HUMAN | P02765 | Fetuin-A, AHSG-A, AHSG-B | 5.43–40,114 | 168 | 16/32 | 40 |
| 37 | Transthyretin | TTHY_HUMAN | P02766 | Transthyretin | 5.52–15,991 | 130 | 7/13 | 72 |
| 38 | Transthyretin | TTHY_HUMAN | P02766 | Transthyretin | 5.52–15,991 | 131 | 7/13 | 57 |
| 40 | Transthyretin | TTHY_HUMAN | P02766 | Transthyretin | 5.52–15,991 | 147 | 7/9 | 64 |
| 42 | Apolipoprotein A-I | APOA1_HUMAN | P02647 | HDL, Apoa1 | 5.56–30,759 | 174 | 13/21 | 37 |
| 43 | Protein S100-A11 | S10AB_HUMAN | P31949 | S100 | 6.56–11,847 | 138 | 8/9 | 58 |
| 46 | Haptoglobin | HPT_HUMAN | P00738 | HP, HP-alpha, HP-beta | 6.13–45,861 | 149 | 9/11 | 24 |
| 53 | Peroxiredoxin-1 | PRDX1_HUMAN | Q06830 | Peroxiredoxin | 8.25–223,225 | 286 | 16/18 | 62 |
| 55 | Immunoglobulin heavy constant gamma | IGHG1_HUMAN | P01857 | IGHG1, IgG, IgG1 | 8.46–36,596 | 130 | 8/11 | 37 |
| 57 | Albumin Fragment C | ALBU_HUMAN | P02768 | Albumin | 5.92–71,317 | 129 | 11/13 | 19 |
| 60 | Immunoglobulin kappa constant | IGKC_HUMAN | P01834 | IGKC | 6.11–11,929 | 76 | 4/9 | 48 |
| 61 | Immunoglobulin kappa constant | IGKC_HUMAN | P01835 | IGKC | 6.11–11,929 | 137 | 7/12 | 78 |
| 63 | Profilin-1 | PROF1_HUMAN | P07737 | Profilin | 8.44–15,216 | 261 | 17/19 | 75 |
| 65 | Cystatin | CYTB_HUMAN | P04080 | Cystatin B | 6.96–11,190 | 93 | 5/10 | 58 |
| 67 | Haptoglobin | HPT_HUMAN | P00738 | HP, HP-alpha, HP-beta | 6.13–45,861 | 232 | 18/23 | 34 |
| 68 | Apolipoprotein A-I | APOA1_HUMAN | P02647 | HDL, Apoa1 | 5.56–30,759 | 248 | 19/28 | 49 |
| 69 | MIX: Pulmonary surfactant-associated protein A2 | SFPA2_HUMAN | Q8IWL1 | SP-A2 | 5.07–26,622 | 146 | 10/22 | 45 |
| Pulmonary surfactant-associated protein A1 | SFTA1_HUMAN | Q8IWL2 | SP-A | 5.07–26,623 | 146 | 10/22 | 45 | |
| Transthyretin | TTHY_HUMAN | P02766 | Transthyretin | 5.52–15,991 | 90 | 6/22 | 71 | |
| 72 | Complement factor B | CFAB_HUMAN | P00751 | Factor B, Factor Ba, Factor Bb | 6.67–86,847 | 273 | 21/25 | 30 |
| 73 | Alpha-2-HS-glycoprotein | FETUA_HUMAN | P02765 | Fetuin-A, AHSG-A, AHSG-B | 5.43–40,114 | 94 | 7/13 | 19 |
| 74 | Calcyphosin | CAYP1_HUMAN | Q13938 | CAPS, Calcyphosin | 6.90–23,650 | 346 | 22/26 | 53 |
| 76 | Immunoglobulin kappa constant | IGKC_HUMAN | P01834 | 6.11–11,929 | 98 | 5/9 | 55 | |
| 77 | Leukocyte elastase inhibitor | ILEU_HUMAN | P30740 | SERPINB1 | 5.90–42,829 | 76 | 5/7 | 15 |
| 78 | Serotransferrin | TRFE_HUMAN | P02787 | Holotransferrin | 6.81–79,294 | 506 | 42/53 | 49 |
| 82 | Immunoglobulin kappa constant | IGKC_HUMAN | P01834 | IGKC | 6.11–11,929 | 118 | 6/10 | 59 |
| 86 | Apolipoprotein A-I | APOA1_HUMAN | P02647 | HDL, Apoa1 | 5.56–30,759 | 175 | 12/16 | 28 |
| 87 | Immunoglobulin heavy constant gamma | IGHG1_HUMAN | P01857 | IGHG1, IgG, IgG1 | 8.46–36,596 | 94 | 6/9 | 27 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vantaggiato, L.; Shaba, E.; Cameli, P.; Bergantini, L.; d’Alessandro, M.; Carleo, A.; Montuori, G.; Bini, L.; Bargagli, E.; Landi, C. BAL Proteomic Signature of Lung Adenocarcinoma in IPF Patients and Its Transposition in Serum Samples for Less Invasive Diagnostic Procedures. Int. J. Mol. Sci. 2023, 24, 925. https://doi.org/10.3390/ijms24020925
Vantaggiato L, Shaba E, Cameli P, Bergantini L, d’Alessandro M, Carleo A, Montuori G, Bini L, Bargagli E, Landi C. BAL Proteomic Signature of Lung Adenocarcinoma in IPF Patients and Its Transposition in Serum Samples for Less Invasive Diagnostic Procedures. International Journal of Molecular Sciences. 2023; 24(2):925. https://doi.org/10.3390/ijms24020925
Chicago/Turabian StyleVantaggiato, Lorenza, Enxhi Shaba, Paolo Cameli, Laura Bergantini, Miriana d’Alessandro, Alfonso Carleo, Giusy Montuori, Luca Bini, Elena Bargagli, and Claudia Landi. 2023. "BAL Proteomic Signature of Lung Adenocarcinoma in IPF Patients and Its Transposition in Serum Samples for Less Invasive Diagnostic Procedures" International Journal of Molecular Sciences 24, no. 2: 925. https://doi.org/10.3390/ijms24020925
APA StyleVantaggiato, L., Shaba, E., Cameli, P., Bergantini, L., d’Alessandro, M., Carleo, A., Montuori, G., Bini, L., Bargagli, E., & Landi, C. (2023). BAL Proteomic Signature of Lung Adenocarcinoma in IPF Patients and Its Transposition in Serum Samples for Less Invasive Diagnostic Procedures. International Journal of Molecular Sciences, 24(2), 925. https://doi.org/10.3390/ijms24020925









