Single–Molecule Study of DNAzyme Reveals Its Intrinsic Conformational Dynamics
Abstract
1. Introduction
2. Results and Discussion
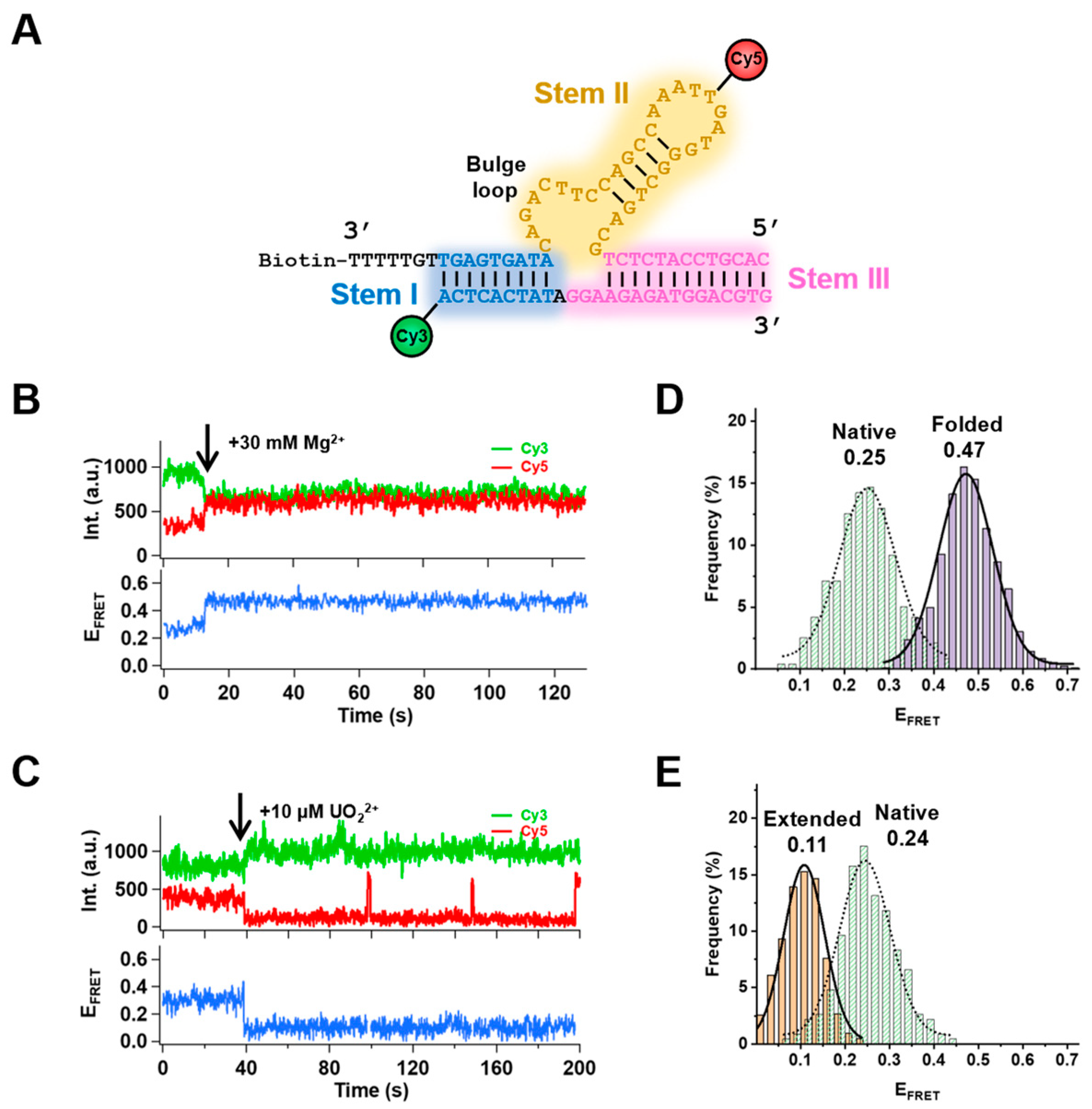
2.1. Metal–Ion–Dependent Folding Induced by Mg2+ and Extending by UO22+
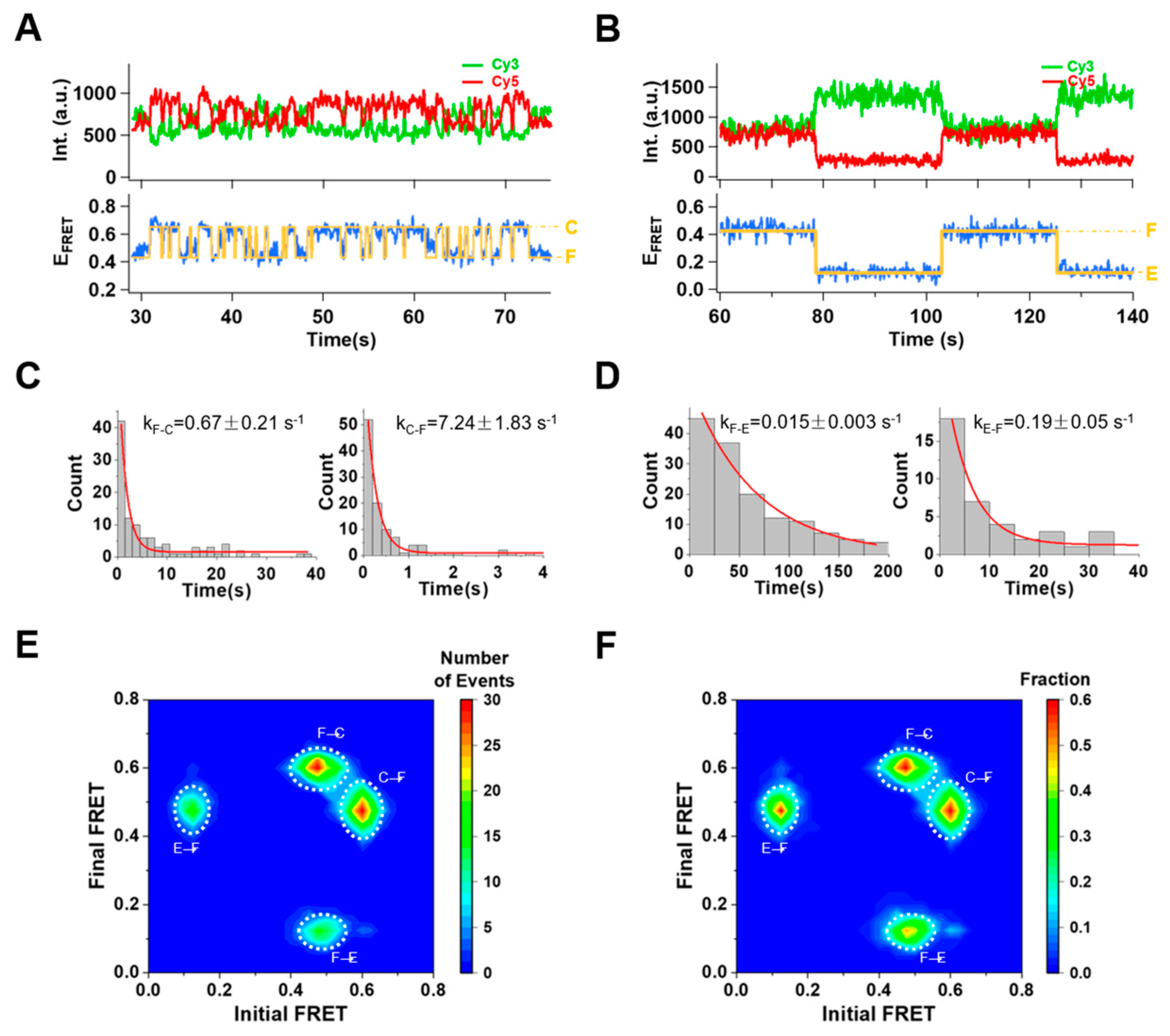
2.2. Conformational Switches with Mg2+ Presence
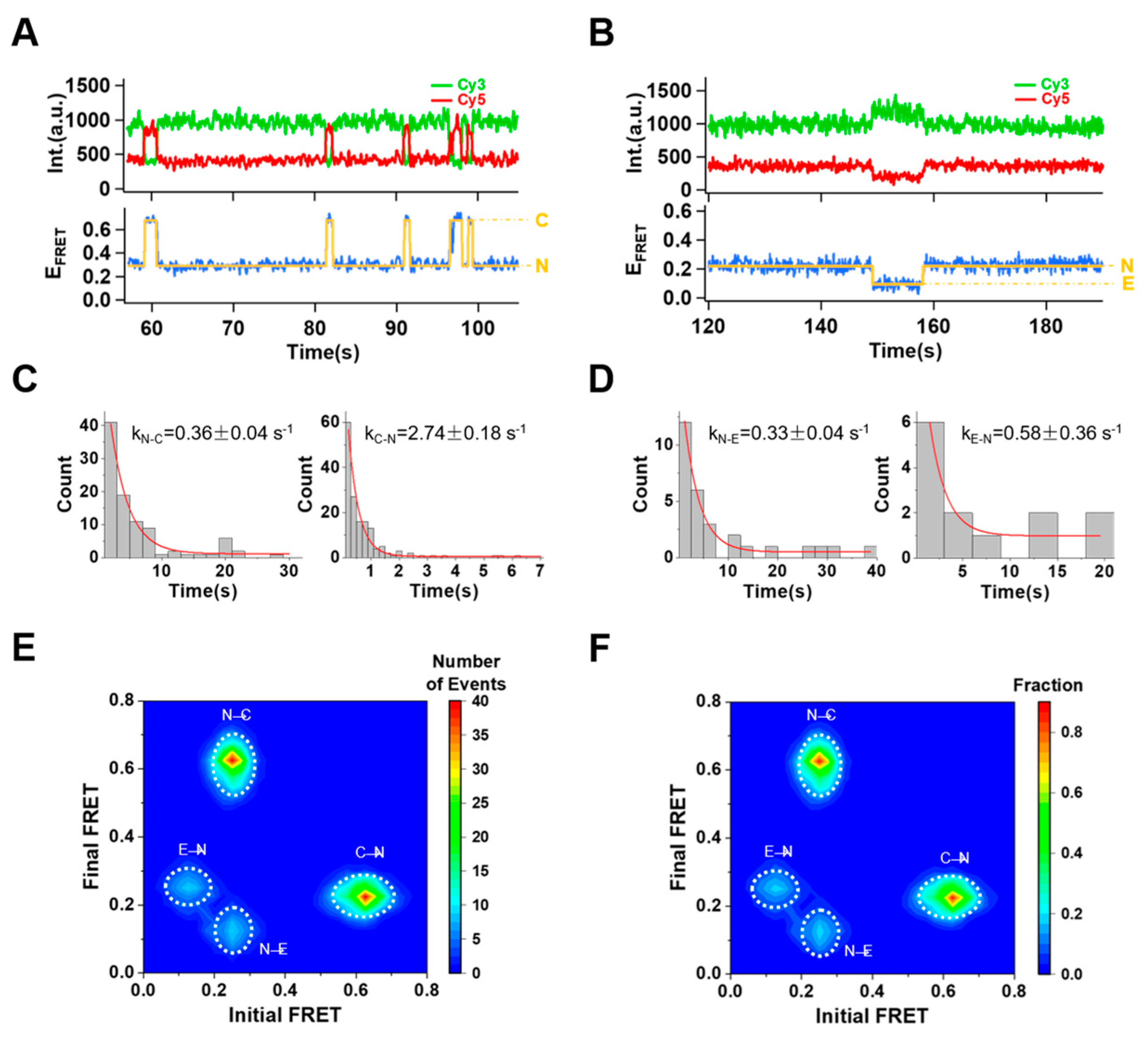
2.3. Conformational Changes without Divalent Metal Ions
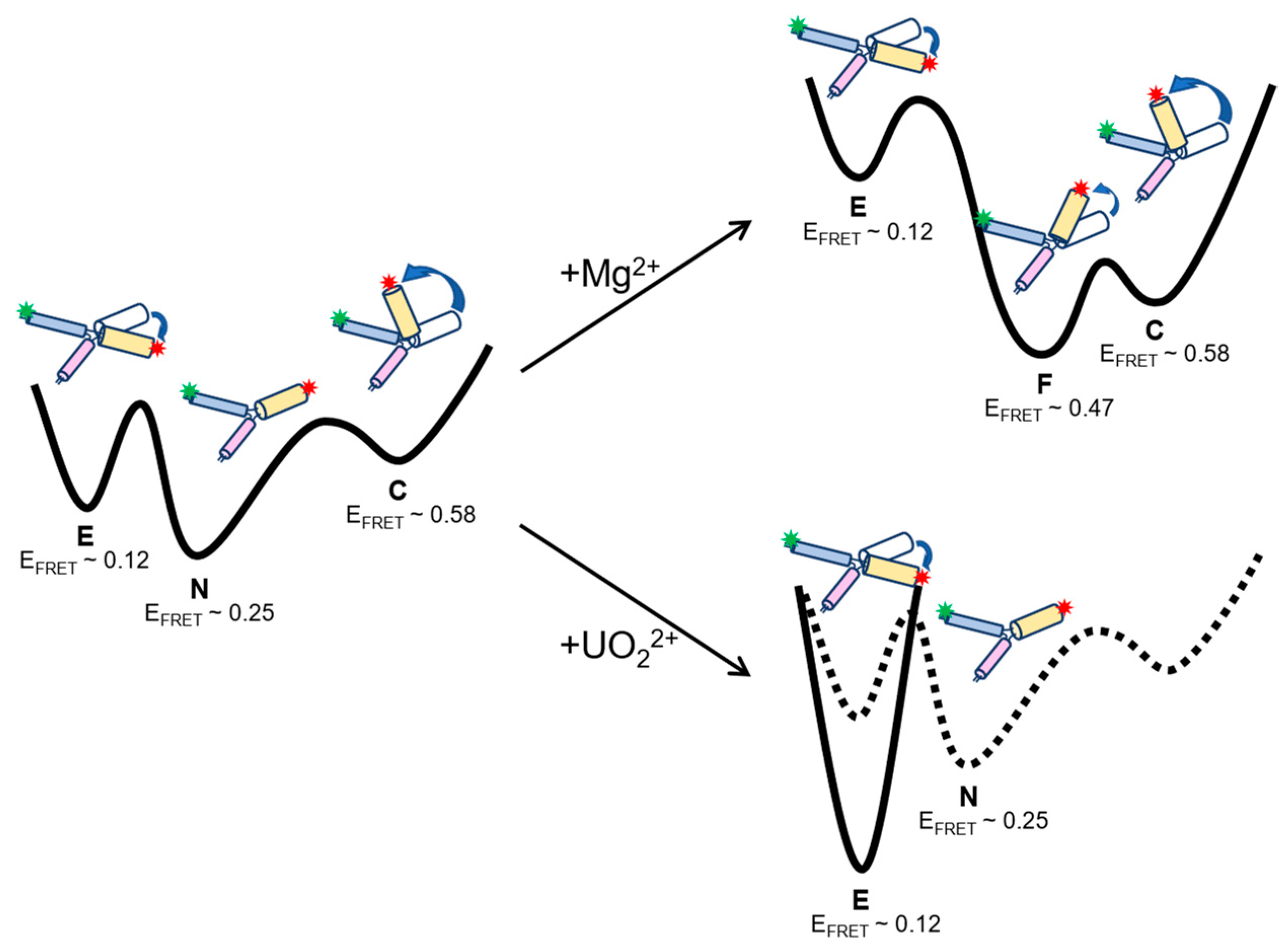
2.4. Free Energy Landscapes of 39E Conformation Dynamics with and without Divalent Metal Ions
3. Materials and Methods
3.1. Chemicals
3.2. smFRET Experiment
3.3. Free Energy Estimation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Breaker, R.R.; Joyce, G.F. A DNA enzyme that cleaves RNA. Chem. Biol. 1994, 1, 223–229. [Google Scholar] [CrossRef]
- Breaker, R.R. In Vitro Selection of Catalytic Polynucleotides. Chem. Rev. 1997, 97, 371–390. [Google Scholar] [CrossRef]
- Wilson, D.S.; Szostak, J.W. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999, 68, 611–647. [Google Scholar] [CrossRef]
- Joyce, G.F. Directed evolution of nucleic acid enzymes. Annu. Rev. Biochem. 2004, 73, 791–836. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y. New transition-metal-dependent DNAzymes as efficient endonucleases and as selective metal biosensors. Chemistry 2002, 8, 4588–4596. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cao, Z.; Lu, Y. Functional nucleic acid sensors. Chem. Rev. 2009, 109, 1948–1998. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Liu, J.; Li, J.; Bruesehoff, P.J.; Pavot, C.M.; Brown, A.K. New highly sensitive and selective catalytic DNA biosensors for metal ions. Biosens. Bioelectron. 2003, 18, 529–540. [Google Scholar] [CrossRef]
- Liu, J.; Lu, Y. Rational design of “turn-on” allosteric DNAzyme catalytic beacons for aqueous mercury ions with ultrahigh sensitivity and selectivity. Angew. Chem. Int. Ed. Engl. 2007, 46, 7587–7590. [Google Scholar] [CrossRef]
- Shimron, S.; Elbaz, J.; Henning, A.; Willner, I. Ion-induced DNAzyme switches. Chem. Commun. 2010, 46, 3250–3252. [Google Scholar] [CrossRef]
- Endo, M.; Takeuchi, Y.; Suzuki, Y.; Emura, T.; Hidaka, K.; Wang, F.; Willner, I.; Sugiyama, H. Single-Molecule Visualization of the Activity of a Zn(2+)-Dependent DNAzyme. Angew. Chem. Int. Ed. Engl. 2015, 54, 10550–10554. [Google Scholar] [CrossRef]
- Xiang, Y.; Lu, Y. DNA as sensors and imaging agents for metal ions. Inorg. Chem. 2014, 53, 1925–1942. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Hwang, K.; Lan, T.; Lu, Y. A DNAzyme-gold nanoparticle probe for uranyl ion in living cells. J. Am. Chem. Soc. 2013, 135, 5254–5257. [Google Scholar] [CrossRef]
- Li, L.; Feng, J.; Fan, Y.; Tang, B. Simultaneous imaging of Zn(2+) and Cu(2+) in living cells based on DNAzyme modified gold nanoparticle. Anal. Chem. 2015, 87, 4829–4835. [Google Scholar] [CrossRef]
- Hwang, K.; Wu, P.; Kim, T.; Lei, L.; Tian, S.; Wang, Y.; Lu, Y. Photocaged DNAzymes as a general method for sensing metal ions in living cells. Angew. Chem. Int. Ed. Engl. 2014, 53, 13798–13802. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Zhang, J.; Yang, Z.; Mou, Q.; Ma, Y.; Xiong, Y.; Lu, Y. Functional DNA Regulated CRISPR-Cas12a Sensors for Point-of-Care Diagnostics of Non-Nucleic-Acid Targets. J. Am. Chem. Soc. 2020, 142, 207–213. [Google Scholar] [CrossRef]
- Gao, H.; Feng, M.; Li, F.; Zhang, K.; Zhang, T.; Zhang, Z.; Yang, C.; Deng, R.; Zhang, J.; Jiang, P. G-Quadruplex DNAzyme-Substrated CRISPR/Cas12 Assay for Label-Free Detection of Single-Celled Parasitic Infection. ACS Sens. 2022, 7, 2968–2977. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.K.; Li, J.; Pavot, C.M.; Lu, Y. A lead-dependent DNAzyme with a two-step mechanism. Biochemistry 2003, 42, 7152–7161. [Google Scholar] [CrossRef] [PubMed]
- Bonaccio, M.; Credali, A.; Peracchi, A. Kinetic and thermodynamic characterization of the RNA-cleaving 8-17 deoxyribozyme. Nucleic Acids Res. 2004, 32, 916–925. [Google Scholar] [CrossRef]
- Liu, J.; Lu, Y. FRET study of a trifluorophore-labeled DNAzyme. J. Am. Chem. Soc. 2002, 124, 15208–15216. [Google Scholar] [CrossRef]
- Kim, H.K.; Liu, J.; Li, J.; Nagraj, N.; Li, M.; Pavot, C.M.; Lu, Y. Metal-dependent global folding and activity of the 8-17 DNAzyme studied by fluorescence resonance energy transfer. J. Am. Chem. Soc. 2007, 129, 6896–6902. [Google Scholar] [CrossRef]
- Lee, N.K.; Koh, H.R.; Han, K.Y.; Kim, S.K. Folding of 8-17 deoxyribozyme studied by three-color alternating-laser excitation of single molecules. J. Am. Chem. Soc. 2007, 129, 15526–15534. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Rasnik, I.; Liu, J.; Ha, T.; Lu, Y. Dissecting metal ion-dependent folding and catalysis of a single DNAzyme. Nat. Chem. Biol. 2007, 3, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Saran, R.; Yao, L.; Hoang, P.; Liu, J. Folding of the silver aptamer in a DNAzyme probed by 2-aminopurine fluorescence. Biochimie 2018, 145, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Kiy, M.M.; Jacobi, Z.E.; Liu, J. Metal-induced specific and nonspecific oligonucleotide folding studied by FRET and related biophysical and bioanalytical implications. Chemistry 2012, 18, 1202–1208. [Google Scholar] [CrossRef]
- Liu, J.; Brown, A.K.; Meng, X.; Cropek, D.M.; Istok, J.D.; Watson, D.B.; Lu, Y. A catalytic beacon sensor for uranium with parts-per-trillion sensitivity and millionfold selectivity. Proc. Natl. Acad. Sci. USA 2007, 104, 2056–2061. [Google Scholar] [CrossRef]
- Brown, A.K.; Liu, J.; He, Y.; Lu, Y. Biochemical characterization of a uranyl ion-specific DNAzyme. Chembiochem 2009, 10, 486–492. [Google Scholar] [CrossRef]
- He, Y.; Lu, Y. Metal-ion-dependent folding of a uranyl-specific DNAzyme: Insight into function from fluorescence resonance energy transfer studies. Chemistry 2011, 17, 13732–13742. [Google Scholar] [CrossRef]
- Liu, H.; Yu, X.; Chen, Y.; Zhang, J.; Wu, B.; Zheng, L.; Haruehanroengra, P.; Wang, R.; Li, S.; Lin, J.; et al. Crystal structure of an RNA-cleaving DNAzyme. Nat. Commun. 2017, 8, 2006. [Google Scholar] [CrossRef]
- Feng, M.; Gu, C.; Sun, Y.; Zhang, S.; Tong, A.; Xiang, Y. Enhancing Catalytic Activity of Uranyl-Dependent DNAzyme by Flexible Linker Insertion for More Sensitive Detection of Uranyl Ion. Anal. Chem. 2019, 91, 6608–6615. [Google Scholar] [CrossRef]
- Parra-Meneses, V.; Rojas-Hernandez, F.; Cepeda-Plaza, M. The role of Na(+) in catalysis by the 8-17 DNAzyme. Org. Biomol. Chem. 2022, 20, 6356–6362. [Google Scholar] [CrossRef]
- Cortes-Guajardo, C.; Rojas-Hernandez, F.; Paillao-Bustos, R.; Cepeda-Plaza, M. Hydrated metal ion as a general acid in the catalytic mechanism of the 8-17 DNAzyme. Org. Biomol. Chem. 2021, 19, 5395–5402. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Hong, H.; Yuan, G.; Qian, H.; Li, B.; Cao, Y.; Wang, W.; Wu, C.X.; Chen, H. Hidden Intermediate State and Second Pathway Determining Folding and Unfolding Dynamics of GB1 Protein at Low Forces. Phys. Rev. Lett. 2020, 125, 198101. [Google Scholar] [CrossRef] [PubMed]
- Bustamante, C.; Alexander, L.; Maciuba, K.; Kaiser, C.M. Single-Molecule Studies of Protein Folding with Optical Tweezers. Annu. Rev. Biochem. 2020, 89, 443–470. [Google Scholar] [CrossRef]
- Zhang, J.L.H.; Qin, M.; Wang, W.; Cao, Y. Quantifying cation-π interactions in marine adhesive proteins using single-molecule force spectroscopy. Supramol. Mater. 2022, 1, 100005. [Google Scholar] [CrossRef]
- Ha, T. Single-molecule fluorescence resonance energy transfer. Methods 2001, 25, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Lerner, E.; Cordes, T.; Ingargiola, A.; Alhadid, Y.; Chung, S.; Michalet, X.; Weiss, S. Toward dynamic structural biology: Two decades of single-molecule Forster resonance energy transfer. Science 2018, 359, eaan1133. [Google Scholar] [CrossRef]
- Lehn, J. Supramolecular materials: Dynamic, responsive, adaptive. Supramol. Mater. 2022, 1, 100007. [Google Scholar] [CrossRef]
- Ha, T.; Enderle, T.; Ogletree, D.F.; Chemla, D.S.; Selvin, P.R.; Weiss, S. Probing the interaction between two single molecules: Fluorescence resonance energy transfer between a single donor and a single acceptor. Proc. Natl. Acad. Sci. USA 1996, 93, 6264–6268. [Google Scholar] [CrossRef]
- Leveille, M.P.; Tran, T.; Dingillo, G.; Cannon, B. Detection of Mg(2+)-dependent, coaxial stacking rearrangements in a bulged three-way DNA junction by single-molecule FRET. Biophys. Chem. 2019, 245, 25–33. [Google Scholar] [CrossRef]
- McKinney, S.A.; Declais, A.C.; Lilley, D.M.; Ha, T. Structural dynamics of individual Holliday junctions. Nat. Struct. Biol. 2003, 10, 93–97. [Google Scholar] [CrossRef]
- Steiner, M.; Karunatilaka, K.S.; Sigel, R.K.; Rueda, D. Single-molecule studies of group II intron ribozymes. Proc. Natl. Acad. Sci. USA 2008, 105, 13853–13858. [Google Scholar] [CrossRef] [PubMed]
- Suddala, K.C.; Wang, J.; Hou, Q.; Walter, N.G. Mg(2+) shifts ligand-mediated folding of a riboswitch from induced-fit to conformational selection. J. Am. Chem. Soc. 2015, 137, 14075–14083. [Google Scholar] [CrossRef] [PubMed]
- McKinney, S.A.; Joo, C.; Ha, T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys. J. 2006, 91, 1941–1951. [Google Scholar] [CrossRef]
- Blanco, M.; Walter, N.G. Analysis of complex single-molecule FRET time trajectories. Methods Enzymol. 2010, 472, 153–178. [Google Scholar] [CrossRef]
- Pan, H.; Xia, Y.; Qin, M.; Cao, Y.; Wang, W. A simple procedure to improve the surface passivation for single molecule fluorescence studies. Phys. Biol. 2015, 12, 045006. [Google Scholar] [CrossRef]
- Roy, R.; Hohng, S.; Ha, T. A practical guide to single-molecule FRET. Nat. Methods 2008, 5, 507–516. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Ji, Z.; Wang, X.; Cao, Y.; Pan, H. Single–Molecule Study of DNAzyme Reveals Its Intrinsic Conformational Dynamics. Int. J. Mol. Sci. 2023, 24, 1212. https://doi.org/10.3390/ijms24021212
Zhang Y, Ji Z, Wang X, Cao Y, Pan H. Single–Molecule Study of DNAzyme Reveals Its Intrinsic Conformational Dynamics. International Journal of Molecular Sciences. 2023; 24(2):1212. https://doi.org/10.3390/ijms24021212
Chicago/Turabian StyleZhang, Yiming, Zongzhou Ji, Xin Wang, Yi Cao, and Hai Pan. 2023. "Single–Molecule Study of DNAzyme Reveals Its Intrinsic Conformational Dynamics" International Journal of Molecular Sciences 24, no. 2: 1212. https://doi.org/10.3390/ijms24021212
APA StyleZhang, Y., Ji, Z., Wang, X., Cao, Y., & Pan, H. (2023). Single–Molecule Study of DNAzyme Reveals Its Intrinsic Conformational Dynamics. International Journal of Molecular Sciences, 24(2), 1212. https://doi.org/10.3390/ijms24021212







