Abstract
To redirect carbon flux from the γ-aminobutyric acid (GABA) shunt to the δ-aminolevulinic acid (ALA) biosynthetic pathway, we disrupted the GABA shunt route of the model cyanobacterium Synechocystis sp. PCC 6803 by inactivating Gdc, the gene-encoding glutamate decarboxylase. The generated ΔGdc strain exhibited lower intracellular GABA and higher ALA levels than the wild-type (WT) one. The ΔGdc strain’s ALA levels were ~2.8 times higher than those of the WT one when grown with levulinic acid (LA), a competitive inhibitor of porphobilinogen synthase. Abiotic stress conditions including salinity induced by 10 mM NaCl and cold at 4 °C increased the ALA levels in ΔGdc up to ~2.5 and 5 ng g−1 cell DW, respectively. The highest ALA production in the ΔGdc cyanobacteria grown in BG11 medium was triggered by glucose induction, followed by glutamate supplementation with 60 mM of LA, thereby resulting in ~360 ng g−1 cell DW of ALA, that is >300-fold higher ALA accumulation than that observed in ΔGdc cyanobacteria grown in normal medium. Increased levels of the gdhA (involved in the interconversion of α-ketoglutarate to glutamate) and the hemA (a major regulatory target of the ALA biosynthetic pathway) transcripts occurred in ΔGdc cyanobacteria grown under modified growth conditions. Our study provides critical insight into the facilitation of ALA production in cyanobacteria.
1. Introduction
Apart from proteinogenic amino acids that form the essential building blocks of proteins, many non-protein amino acids are also produced in animal, plant and microbial cells [1,2,3]. These metabolites take part in essential physiological and biochemical processes in biological systems. Amongst the numerous non-protein amino acids found in prokaryotic and eukaryotic living systems, δ-aminolevulinic acid (ALA) is considered particularly important due to its ubiquitous presence and multifunctional properties in the metabolism of bacteria, algae, plants and animals, as well as due to its use in cosmetic, agricultural and pharmaceutical industry products [4]. ALA is considered a significant intermediate in tetrapyrrole biosynthesis; it plays a critical role in respiration and photosynthesis by controlling chlorophyll metabolism in green organisms [5].
The biosynthesis of ALA inside the cells chiefly occurs via two distinct metabolic routes: the C4 pathway (Shemin pathway) and the C5 pathway (Beale pathway), as illustrated in Figure 1. The C4 pathway appears in mammalian, fungal, protozoan and bacterial cells, in which ALA forms from succinyl-CoA and glycine via a single-step catalytic action of ALA synthase (ALAS) [5]. In the C5 pathway (hereafter referred to as the ‘ALA pathway’), occurring in blue-green algae (cyanobacteria), plants and several bacterial species, ALA forms from glutamate in three irreversible steps and is catalysed consecutively by the action of glutamyl-tRNA synthetase (GluRS), glutamyl-tRNA reductase (GluTR) and glutamate-1-semialdehyde 2,1-aminomutase (GSA-AM) [6]. In both the C4 and the ALA pathways, ALA is further catabolised to porphobilinogen (PBG) via porphobilinogen synthase (Pbs) and is subsequently metabolised to chlorophyll, phycobilins or heme [7]. In photoautotrophic and non-nitrogen-fixing cyanobacterium Synechocystis sp. PCC 6803 (hereafter referred to as ‘Synechocystis’), the genes encoding the enzymes along the ALA pathway (glutamate → PBG) are gltX, hemA, hemL and hemB, with the respective gene IDs: sll0179, slr1808, sll0017 and sll1994 (http://genome.microbedb.jp/cyanobase/GCA_000009725.1 (accessed on 7 April 2022)). On the other hand, glutamate (a precursor metabolite in the ALA pathway) is also a chief substrate for an adjacent metabolic route known as the ‘GABA shunt pathway’ in Synechocystis [8]. Previously, ALA production has been achieved via pathway engineering or nutrients optimization in various microbial strains including Yarrowia lipolytica, E. coli and Corynebacterium glutamicum [9,10,11]. However, although Synechocystis has been widely used as a model cyanobacterium for research because of its ability to grow heterotrophically on glucose as well as its publicly available genome sequence [12,13], the study on ALA production in Synechocystis with reference to GABA shunt pathway is still lacking. The GABA shunt pathway is an alternative yet less energy-efficient metabolic route for completing the tricarboxylic acid (TCA) cycle in a few cyanobacterial species [14]. This metabolic route operates independently of the TCA cycle by converting α-ketoglutarate to glutamate, GABA and succinyl semialdehyde. Comprehensively, initial metabolic routes (from glycolysis to TCA cycle) are mutually shared by the GABA shunt and the ALA pathways in Synechocystis and any strategy aiming to inhibit the GABA shunt pathway might result in redirecting the carbon flux via glutamate towards other carbon reserve products, such as ALA. Hence, since both the GABA and the ALA pathways share a common precursor (glutamate), a Synechocystis mutant strain lacking a functional Gdc (gene ID: sll1641) to encode glutamate decarboxylase (GDC), named ‘ΔGdc’ that can accumulate higher intracellular glutamate levels, was employed to analyse the ALA synthesis associated with a disrupted GABA shunt route. This study also summarises the role of key influencing physicochemical factors in regulating ALA metabolism in terms of the relevant metabolites and gene expression profiles observed in the Synechocystis sp. PCC 6803 wild-type (WT) and ΔGdc strains. Furthermore, the effects of external nutrients and levulinic acid (LA; a competitive inhibitor of Pbs) supplementation on ALA synthesis in the ΔGdc cyanobacteria were analysed to assess potential ALA production in cyanobacteria, a feature that could be particularly useful for future applications.
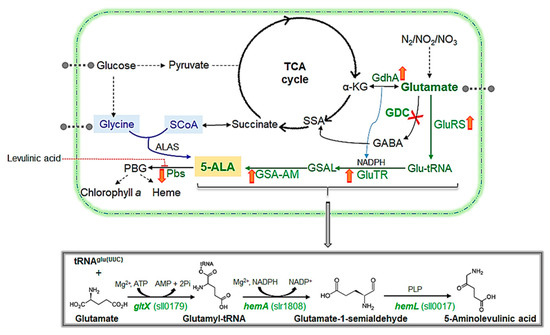
Figure 1.
Schematic summary of the ALA biosynthetic pathways in prokaryotes. The C4 pathway (Shemin pathway) is marked in blue, while the C5 pathway (Beale pathway in Synechocystis) is marked in green. The native enzymes relevant to the ALA metabolism in Synechocystis are marked in green. The pathway engineered in this study is indicated with a red ‘X’ that symbolises the effect of the gene knockout. Cascade reactions are indicated with dashed arrows. The ALA pathway in Synechocystis comprises three enzymes: GluRS, GluTR and GSA-AM that are encoded by gltX, hemA and hemL, respectively. These enzymes facilitate the irreversible catalysis of glutamate to ALA, as shown in the overall reaction in the bottom box, along with the relevant genes and their IDs (in green). ALA is converted into PBG by the action of Pbs or of ALA dehydratase (hemB, sll1994), which is the main enzyme inhibited by levulinic acid (as indicated with a red dotted arrow). The red ‘up’ and ‘down’ arrows beside the gene indicate the up- or downregulation of the genes in the ΔGdc strain, resulting in ALA accumulation. Abbreviations used: 2Pi, diphosphate; α-KG, α-ketoglutarate; ALA, δ-aminolevulinic acid; ALAS, ALA synthase; ATP/AMP, adenosine tri(mono)phosphate, GDC, glutamate decarboxylase; GdhA, glutamate dehydrogenase; GluRS, glutamyl-tRNA synthetase; GluTR: glutamyl-tRNA reductase; Glut-tRNA, glutamyl-tRNA; GSA-AM, glutamate-1-semialdehyde 2,1-aminomutase; GSAL, glutamate-1-semialdehyde; NADPH, nicotinamide adenine dinucleotide phosphate; PBG, porphobilinogen, Pbs, porphobilinogen synthase, PLP, pyridoxal-5ʹ-phosphate; SCoA, succinyl coenzyme-A; SSA, succinic semialdehyde. Reproduced from [8,15,16].
2. Results and Discussion
2.1. Disruption of the GABA Shunt Pathway for the Production of ALA
In few cyanobacteria, including the model species Synechococcus PCC7002 and Synechocystis, the TCA cycle closes by an alternative bypass route via the GABA shunt. It is operated by a consecutive conversion of glutamate (chiefly deriving from the TCA cycle metabolite α-ketoglutarate) to GABA, subsequently closing the cycle through a succinyl semialdehyde formation (Figure 1). In Synechocystis, the GABA shunt was more active than the widely-distributed TCA cycle route and it was also studied for its possible role in overcoming physiological stresses [3]. The glutamate metabolism at this branch point forms a major connection between the carbon and nitrogen metabolism and provides the major metabolic precursors of the ALA pathway [15]. It has been shown previously that interruption in Gdc led to glutamate accumulation in plants and cyanobacteria [16,17]. However, redirecting carbon flux from the GABA shunt to the ALA biosynthetic pathway, in the form of glutamate utilisation, has not been previously studied in cyanobacteria. Hence, considering that glutamate availability to the ALA pathway might be enhanced in the absence of a functional GABA shunt in Synechocystis, we generated a mutant strain with a disrupted GABA shunt metabolic route through the inactivation of Gdc gene by using the strategy shown in Figure 2a and described in the experimental procedures (Section 3.2). The engineered strain, ΔGdc, was further analysed to determine its growth rate and chlorophyll a content. Firstly, comparing the growth patterns of the WT and the ΔGdc strains shows that GDC deactivation does not compromise organism growth, except for the period between day 2 to day 8 (until the establishment of the late-log phase), where the ΔGdc strain exhibits a slightly declined growth (Figure 3a). Later on, during the stationary phase, the growth of the ΔGdc strain was comparable to that of the WT one. Thus, in the absence of a functional GABA shunt, the organism could compete well for nutrient-depletion or pH change in the medium, two substantial factors influencing microbial cell growth during the stationary phase [18]. On the other hand, chlorophyll a content in the ΔGdc cyanobacteria, that was about 1.5, 2.5 and 2.4 µg mg−1 cells at mid-log, late-log and stationary phases of growth, respectively, was slightly lower than that observed in the WT ones (that was about 1.7, 2.6 and 3 µg mg−1 cells at mid-log, late-log and stationary phases of growth, respectively) (Figure 3b), thereby suggesting a clear impact of the disrupted GABA shunt on the C5 pathway of Synechocystis. As depicted in Figure 1, the ALA catabolism bifurcates into two metabolic routes, i.e., chlorophyll a or heme formation, so the decrease in chlorophyll a might be possibly due to increased flux to heme formation that is needed to investigate further as the ALA flux to chlorophyll a or heme synthesis is unknown in ΔGdc yet. These results indicate that despite the decreased photosynthetic pigment levels, the ΔGdc strain had efficient carbon assimilation to maintain its growth under normal photoautotrophic conditions.
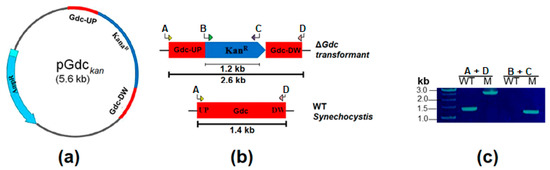
Figure 2.
Inactivation of Gdc in Synechocystis. (a) Design and plasmid construction for Gdc interrupted by a kanamycin antibiotic-resistant cassette and cloning into a pGEM-T easy vector. AmpR stands for the ampicillin-resistant cassette in pGEM-T easy vector backbone. (b) Simplified scheme of the wild-type (WT) Synechocystis and of the Synechocystis mutant (M) strain with the inactivated Gdc. Primers used for the performance of PCR are indicated (arrows labelled ‘A’–‘D’), with the expected PCR product sizes shown as bars beneath both maps. (c) Confirmation of the complete segregation of the mutant strain through PCR amplification. Lane WT stands for the wild-type, while lane M stands for the ΔGdc strain, with primer positions as indicated in (b).
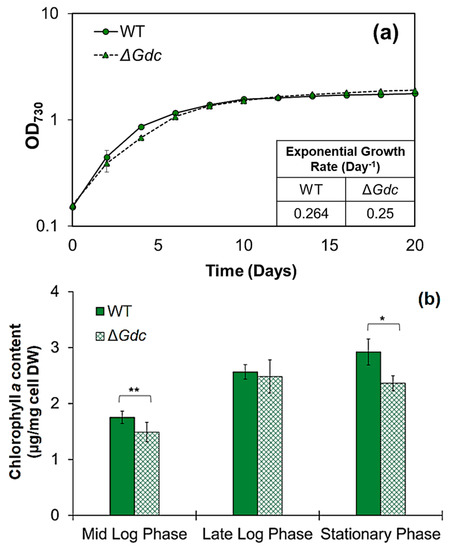
Figure 3.
Growth and pigment analysis. (a) Comparison of growth by measuring the optical density of cultures at 730 nm. (b) Chlorophyll a content at various growth phases of the Synechocystis wild-type (WT) and mutant (M) strain, grown in BG11 medium and in BG11 medium supplemented with kanamycin (25 µg mL−1), respectively. Results represent the mean ± standard deviation (SD) of experiments conducted in triplicate (n = 3). The error bars denoting the SD values in (a) cannot be seen since the SD is smaller than the size of the symbol. In (b), the differences between means of the individual groups were analysed using one-way analysis of variance (ANOVA) via GraphPad Prism 9.3.1 software (San Diego, CA, USA). * p < 0.05, ** p < 0.01, versus WT cells.
Under normal growth conditions (untreated), the maximal ALA levels in the Synechocystis mutant strain were about 1 ng g−1 DW of cells, that is approximately 7 times higher than those observed in the WT one at the late-log growth phase (after 6 days of culturing; Figure 4a). ALA accumulation was enhanced in both the WT and the ΔGdc strains in response to modified growth conditions. In an attempt to impose the smallest effect on the growth of cyanobacteria, a two-stage culture approach was established. The cells were first pre-grown under normal photoautotrophic conditions until their growth rate was comparable (at the late-log phase). They were subsequently subjected to the modified physicochemical conditions to generate high ALA levels. Numerous attempts have been made to enhance the feasibility and efficiency of ALA production in bacteria and fungi by modifying the nutrient supply or the growth conditions [19,20]. Among the nutrient factors, the presence of carbon or inorganic nitrogen sources, mineral salts, amino acids and ions, as well as the concentrations of precursors or inhibitors, were found to affect ALA production. There are several reports regarding the stress-relieving roles of ALA in plants in response to certain environmental factors (including abiotic signals such as salinity and drought) [21,22]. As a result, we examined whether various physicochemical factors could lead to an elevated ALA accumulation in the ΔGdc mutant. We observed that the cellular ALA contents were higher in the cold-treated WT strain and increased significantly in the ΔGdc strain compared to those in untreated cells. Moreover, salinity induced osmotic stress led to a 2-fold increase in the ALA levels in the examined Synechocystis mutant. As already mentioned, the exogenous ALA application can confer stress tolerance in higher plants; however, limited information is available regarding any such role of ALA in cyanobacteria or in the regulation of ALA metabolism and the in vivo synthesis of ALA, except for some research work that have been done decades ago by Beale [23], Troxler and Brown [24], Kipe-Nolt et al. [25] and by Rieble and Beale who have conducted studies on the characterisation of the GluTR in Synechocystis [26]. The effects of osmotic stress on the upregulation of the GDC pathway at the enzymatic activity and the transcription level in Synechocystis have been previously reported [27]. In this study, the increased ALA levels might be a response to the metabolic perturbations occurring in the absence of a GDC activity and the stress caused by the applied cold and salinity conditions. Transcriptional analyses were undertaken to obtain more information regarding the possible effect of those abiotic factors on gene regulation in the ALA pathway.
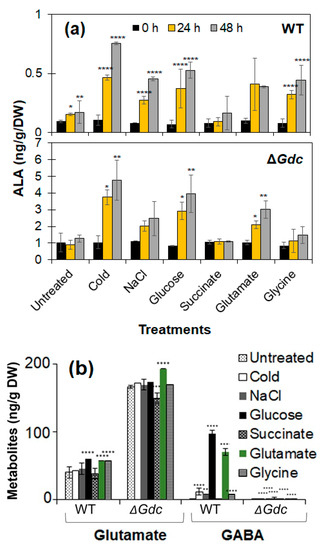
Figure 4.
Analysis of the metabolite alteration in Synechocystis strains under various treatments. (a) δ-Aminolevulinic acid (ALA) and (b) relevant metabolite (glutamate and GABA) levels in wild-type (WT) and ΔGdc strains of Synechocystis, after using the BG11 medium as a control (untreated). Results represent the mean ± standard deviation (SD) of experiments conducted in triplicate (n = 3). The differences between means of the individual groups were analysed using one-way analysis of variance (ANOVA) via GraphPad Prism 9.3.1 software (San Diego, CA, USA). In (a), * p < 0.05, ** p < 0.01, **** p < 0.001 versus 0 h, and in (b), *** p < 0.005, **** p < 0.001 versus untreated group.
We showed that external nutrients, such as glucose and glutamate, can increase the ALA levels in the ΔGdc strain up to ~3 and 2.1 times, respectively, in 24 h, compared to the cells grown in a normal medium. The increased duration of the exposure to the aforementioned nutrients (for up to 48 h) led to a further increase in the observed ALA accumulation. Synechocystis cells can uptake exogenous glucose as an efficient carbon source [28] and a major proportion of the assimilated carbon is known to be consumed by the TCA cycle metabolism (that is also a source of precursors for the GABA shunt and the ALA pathway) [29]. Glucose is the best carbon source for enhancing the growth and GABA levels of Synechocystis cells by upregulating the GDC transcript levels and catalytic activity [27]. On the other hand, inexpensive glucose has also been used as an efficient sole carbon source of the C5 pathway of bacteria for ALA production [20]. There are also reports of ALA overproduction via C5 pathway in various genetically engineered microorganisms grown under glutamate supplementation and achievement of higher ALA titers as a result of flux redistribution of the TCA cycle toward glutamate [30,31]. Further, we have observed that a Synechocystis mutant strain over expressing hemA requires glucose or glutamate supplementation essentially for its growth (data not published), hence complementing the results in present study that both nutrients serve as precursor substrates for ALA biosynthesis and the increase in ALA titers observed in the ΔGdc cyanobacteria were likely due to the availability of more substrate, owing to a lack of glucose and glutamate utilisation by the disrupted GABA shunt pathway. Succinate (a precursor for ALA synthesis in bacteria) did not seem to affect ALA production in Synechocystis. In contrast, there was a minor increase in ALA levels in glycine-treated Synechocystis cells compared to untreated cells. Glycine serves as the chief substrate for ALA synthesis via the ALAS catalysis in the C4 pathway (as described in introduction section and shown in Figure 1) and is largely restricted to a few bacteria, fungi and mammals and has not been demonstrated in plants and cyanobacteria. Synechocystis possesses the glycine uptake and degradation system [32]. Yet, there is no evidence of an ALA biosynthesis from glycine in cyanobacterial cells and there has been no direct synthesis link established between glycine and the ALA precursor glutamate. Hence, we assumed that the altered ALA levels observed in this scenario might result from the regulatory mechanisms of the C5 pathway aroused in response to the toxic cellular effects of glycine [32,33].
We subsequently analysed the effect of GDC inactivation on cellular metabolites that have a direct metabolic link to both the GABA shunt and the ALA pathway. We observed a significant increase and decrease in the cellular glutamate and GABA contents, respectively, in the ΔGdc strain (as compared to those of the WT one) (Figure 4b). Previous studies showed that a certain ATP-binding cassette-type transporter facilitates the glutamate uptake of Synechocystis cells. This glutamate is further utilised in various cellular metabolic processes, including the glutamine synthetase and glutamate synthase (GS/GO-GAT) cycle and the GABA shunt pathway, which are relevant to the interchangeable α-ketoglutarate and glutamate contents, respectively [34,35]. Fresh water cyanobacteria grow ideally under slightly alkaline pH conditions and tend to maintain their intracellular pH within the range of approximately 6.9 to 7.8 [36,37]. In this scenario GABA shunt pathway is one among other pathways that plays important role in buffering the cytosolic pH of cyanobacterial cells by maintaining the levels of acidic compound such as glutamate that is proved to be toxic to cells at higher concentration (>30 mM) and impedes cyanobacterial growth [8,38]. Our results indicate that an external glutamate supply had helped increase intracellular glutamate levels in the ΔGdc cyanobacteria. This glutamate is also further converted to α-ketoglutarate by glutamate dehydrogenase (GdhA) to regenerate the nicotinamide adenine dinucleotide phosphate (NADPH) required for ALA synthesis (Figure 1). These results were further supported by transcriptional analysis of relevant genes in these pathways and are discussed in the next section.
2.2. Transcriptional Regulation of the ALA Biosynthesis Pathway in the ΔGdc Mutant Strain
In an attempt to gain a better understanding of the underlying biology behind the effect of the GDC inactivation on the ALA biosynthesis pathway, we quantified the mRNA expression of gltX, hemA, hemL, hemB, Gdc and GdhA in the WT and the ΔGdc strains, under normal and modified growth conditions. The results demonstrated that the transcript levels of genes of the ALA pathway were upregulated in the ΔGdc strain compared to the WT one. In contrast, certain modified growth factors had substantially induced the expression of each gene of the ALA biosynthesis pathway in Synechocystis (Figure 5a). While comparing the effects of two different abiotic stress conditions on the mRNA levels of these genes, we found that the cold condition had increased the expression of gltX, hemA, hemL and hemB in the WT strain compared to the osmotic stress condition. The expression of those genes was further enhanced in the ΔGdc cyanobacteria, consistent with the observed increased ALA production (Figure 4a). We observed that both glucose and glutamate increased the expression of gltX, hemA, hemL and hemB several times in the ΔGdc strain compared to the control condition. According to previous studies, overall, the ALA metabolic pathway is regulated at various steps in response to changes in gene expression and intermediate products of the downstream metabolic routes [7]. Based on the transcriptional and enzymatic analysis performed in plants and bacteria, hemA and hemB (encoding GluTR and Pbs, respectively) are the major regulatory targets of the ALA biosynthesis pathway [7,39]. Since ALA is not the end-product in this metabolic route, GluTR (that catalyses the rate-limiting step in ALA synthesis) is feedback-inhibited by the end-product (heme). On the other hand, Pbs (that catalyses the major ALA catabolic reaction) is also feedback-inhibited by the downstream intermediate protoporphyrinogen IX in the heme biosynthesis pathway, while the overexpression of hemB has adversely affected ALA accumulation. LA, a structural analogue of ALA, can act as a competitive inhibitor of Pbs and has been used in many biotechnological studies to trigger ALA accumulation in microbes [23,40]. Hence, both hemA and hemB act as key regulatory nodes of the ALA metabolism. Furthermore, we observed that the glycine supplementation had intriguingly upregulated the transcript amounts of all four genes involved in the ALA metabolic pathway in both the WT and the mutant strains, in contrast to those of the control growth condition. As in higher plants, the transcriptional and post-transcriptional regulatory mechanisms as well as posttranslational enzymatic regulations of important metabolic processes in Synechocystis have been indicated in several studies [41,42]. Although ALA accumulation did not increase significantly after a 24 h glycine treatment (Figure 4a) in this study, the obvious transcriptional alterations of the pathway genes indicate a post-transcriptional regulation or feedback-inhibition of the ALA pathway by the downstream intermediate products that will be worth investigating in future.
Interestingly, the ΔGdc cells contained higher transcript levels of gdhA (the gene that encodes GdhA) in response to cold, glucose, glutamate and glycine supplementation when compared to those of the WT ones. In cyanobacteria, the carbon/nitrogen homeostasis is attained mostly by regulating the metabolite levels of α-ketoglutarate through various enzymatic pathways, including the GdhA (which catalyses the reversible conversion of α-ketoglutarate and glutamate) [43]. Moreover, the GdhA catalysed reaction acts as a connecting link between the TCA cycle and the ALA pathway. The upregulation of the gdhA transcription in the ΔGdc cells grown under modified nutrients and abiotic stress is, hence, another finding that acts as evidence of the glutamate flow to neighbouring pathways when the GABA shunt pathway is not functioning in cyanobacteria. The inactivation of Gdc had no apparent effect on the pathway genes in the Synechocystis that was exposed to succinate supplementation.
The above analysis suggested that the upregulation of hemB might have increased Pbs activity, consequently limiting ALA accumulation. We subsequently investigated the effect of various LA concentrations on the expression levels of pathway genes in Synechocystis WT and ΔGdc strains. As expected, a significant increase in the transcription of ALA biosynthesis genes, especially of hemA and hemL, was observed in Synechocystis, with the highest mRNA levels observed in ΔGdc cells in the presence of 60 mM LA (as shown in Figure 5b). Moreover, the transcript levels of gdhA were also markedly increased in Synechocystis in the presence of 60 mM LA, whereas the Gdc transcripts were not considerably upregulated in WT cells exposed to 60 mM LA, thereby suggesting that the LA inhibition of Pbs results in an upregulation of the GdhA and the ALA synthesis route to benefit the ALA accumulation. However, the mRNA levels of hemB (whose product is inhibited by LA) were increased in LA-treated cells, indicating the inadequate feedback-inhibition occurring from the downstream metabolic products. This inadequacy is due to an interruption in the ALA catabolism that results in an upregulation of hemB, but not of the activity of Pbs, as LA blocks the latter. This biochemical change might also be responsible for the increased hemA and hemL transcription, whose products are directly involved in the catalysis of the ALA biosynthesis. The effect of LA on the cellular ALA levels of Synechocystis was assessed to validate these results further.
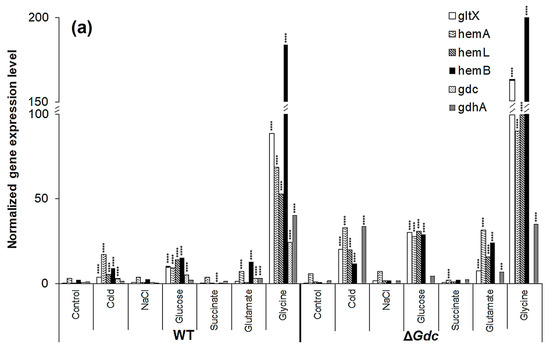
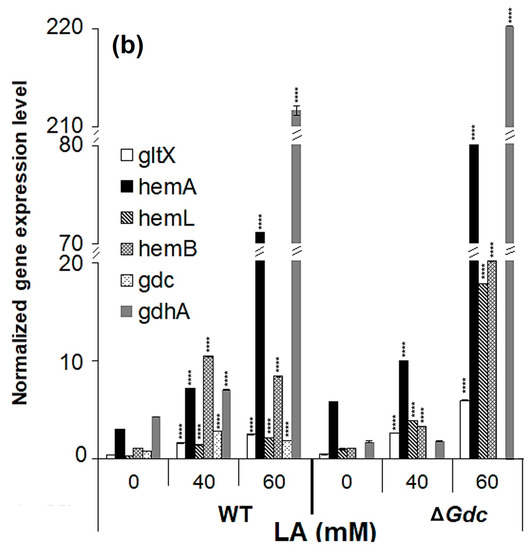
Figure 5.
Transcriptional regulation of pathway genes in the Synechocystis wild-type (WT) and the Gdc knockout mutant (ΔGdc) strains. Analysis of the gene expression profile of various genes related to δ-aminolevulinic acid (ALA) and glutamate metabolism in Synechocystis sp. PCC6803 (WT) and the ΔGdc strain grown under (a) various treatments and (b) different concentrations of levulinic acid (LA). Results were normalised to 16S rRNA levels and bars represent the mean ± standard errors of three independent real-time RT-PCR reactions. If the error bar cannot be seen, the deviation is very small. *** p < 0.01, **** p < 0.001 versus untreated group. The respective gene IDs and the descriptions of the targeted genes are provided in Table 1 and Figure 1.

Table 1.
Oligonucleotides used in this study and their relative IDs as registered in the Cyanobase (http://genome.microbedb.jp/cyanobase/GCA_000009725.1, accessed on 7 April 2022); restriction sites are underlined.
2.3. Cyanobacterial Growth and ALA Production in the Presence of LA
We investigated the effect of LA on the ALA synthesis and the growth of cyanobacteria cultured under normal photoautotrophic conditions. From the results shown in Figure 6a,b, it is obvious that LA at higher concentrations (40–60 mM) had significantly impeded the cell growth (biomass levels) of both the WT and the ΔGdc mutant strains, as there was a continuous decline in biomass levels within 48 h of the LA supplementation. The maximum amount of ALA was produced in the presence of 60 mM of LA in the Synechocystis cultures. In cultures supplemented with a lower LA concentration (20 mM), the growth of the ΔGdc cells was affected more than that of the WT ones, yet no increase in the ALA content was observed. Our results are supported by previous studies reporting the growth inhibitory effect of LA supplementation on microalgae, which is a possible consequence of the inhibition of chlorophyll synthesis [23,25]. Previously, it was noticed that cell growth of cyanobacterial cultures was completely inhibited in response to LA concentrations above saturation point and also that just 10 mM LA caused nearly 50% inhibition in chlorophyll synthesis of an algal culture [23,25]. In this regard, further studies are needed to be undertaken in future to observe the saturation-type kinetics of ALA production at different LA concentrations in Synechocystis. In order to further validate the notion that ALA accumulation in the glucose- or the glutamate-supplemented ΔGdc cells may be limited owing to a higher Pbs activity that further breakdowns ALA into downstream metabolites, the levels of ALA were quantified in LA-treated ΔGdc cyanobacteria grown in a medium that was supplemented with 10 mM of glucose or glutamate. The LA treatment benefited the ALA accumulation in ΔGdc cells, thereby resulting in a nearly 6- and 7-fold increased ALA content in the presence of glutamate and glucose (that were correspondingly ~290 and 360 ng g−1 cell DW), respectively, as compared to the ALA content in ΔGdc cells treated with LA alone (Figure 6c). Bioproduction of ALA has been achieved previously from harmless and non-pathogenic microorganisms including Yarrowia lipolytica, E. coli and Corynebacterium glutamicum, with the maximum ALA yield reaching 2.217, 11.5 and 1.79 g L−1, respectively [9,10,11]. Although the ALA production in cyanobacterial cells from current study is far less than the other studies reported for photosynthetic microorganisms (Table 2), our results suggest that optimising ALA production in Synechocystis could be achieved by balancing the objective metabolic routes by targeting the metabolic engineering and precursors/inhibitors.
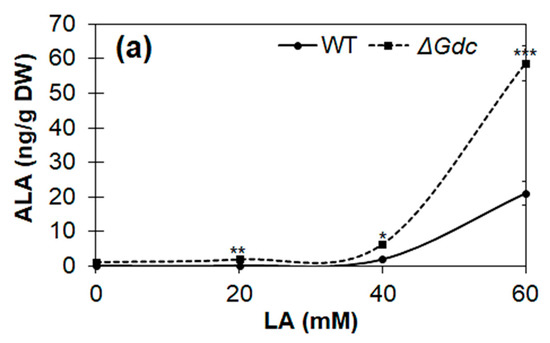
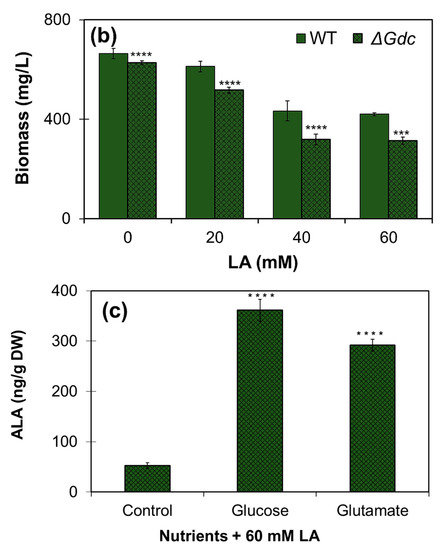
Figure 6.
Quantification of the δ-aminolevulinic acid (ALA) production under a levulinic acid (LA) supply in the wild-type (WT) and the mutant ΔGdc strains of Synechocystis. Effects of LA on (a) ALA production and (b) on the growth of cultures (biomass levels) in WT and ΔGdc Synechocystis strains. Strains were grown until they reached the late-log phase, followed by a 48 h LA supplementation at the indicated concentrations. Results represent the mean ± standard deviation (SD) of experiments conducted in triplicate (n = 3). The differences between means of the individual groups were analysed using one-way analysis of variance (ANOVA) via GraphPad Prism 9.3.1 software (San Diego, CA, USA). * p < 0.05, ** p < 0.01 *** p < 0.005, **** p < 0.001 versus WT group. (c) Effect of the supplementation of various nutrients (10 mM of glucose and glutamate) in addition to 60 mM LA on ALA production in ΔGdc strain cultures. The mutant strain was grown until the late-log phase and was subsequently supplied with LA + glucose or glutamate for 48 h. A culture growing without the supplemented nutrients but with the addition of LA, which was used as a control. Results represent the mean ± standard deviation (SD) of experiments conducted in triplicate (n = 3). The differences between means of the individual groups were analysed using one-way analysis of variance (ANOVA) via GraphPad Prism 9.3.1 software (San Diego, CA, USA). **** p < 0.001 versus control group.

Table 2.
Production of ALA by photosynthetic microbes.
We also considered that the ALA biosynthesis in Synechocystis is tightly regulated at both the transcriptional and the post-transcriptional levels in response to the fluctuations in neighbouring/competing pathways, external growth factors and the downstream intermediates’ flux. By looking into the important regulatory role of hemB in ALA accumulation in Synechocystis, a detailed characterisation of its product would be desirable, along with further research on the inhibition kinetics caused by downstream products, LA and other protein inhibitors; such an insight might prove beneficial for the utilisation of cyanobacteria as cell factories for ALA biosynthesis, particularly from the perspective of their utility in future applications.
3. Materials and Methods
3.1. Strains and Culture Conditions
The WT strain of Synechocystis sp. PCC 6803 was obtained from the Pasteur culture collection (Paris, France) and the ΔGdc mutant strain was grown in BG11 medium supplemented with Na2CO3 (BDH, Poole, Dorset England) as a carbon source and 20 mM HEPES-NaOH (pH 7.5) (Sigma-Aldrich, Saint Louis, USA) [47] in 500 mL cotton-plugged Erlenmeyer flasks with a working volume of 200 mL [17,27]. Kanamycin (25 μg mL−1) was added into the BG11 medium for the growth of the ΔGdc strain. Cells were cultured at 28 °C ± 2 °C under continuous illumination at 50 μmol photons m−2 s−1. Illumination was provided by means of cool white LED lamps (LUMAX Ecosaveplus LED, Shanghai, China) from two sides and the growth rate was determined by measuring the optical density (OD) at 730 nm (OD730). Cultures were maintained on solid agar plates containing BG11 media that were supplemented with 10 mM TES (pH 7.5) (Sigma-Aldrich, Saint Louis, USA), 3 g L−1 Na-thiosulfate (Ajax Finechem, Sydney, Australia) and 1.5% agar (Difco, Sparks, USA), with or without antibiotic for the growth of the mutant or the WT strain, respectively. The E. coli strain DH5α (stored in the lab) was a host for constructing and storing the recombinant plasmids used in this study. Table 3 summarises all constructs and strains used in this study.

Table 3.
Strains and plasmids used in this study.
3.2. Construction of the ΔGdc Strain
In order to construct the plasmid to inactivate the Gdc in the Synechocystis genome, the 1.4-kb gene of the Synechocystis Gdc was amplified by polymerase chain reaction (PCR) with gene-specific primers (Macrogen, Seoul, Korea), as shown in Table 1. The PCR product was ligated into a pGEM®-T Easy Vector (Promega), creating a pGdc plasmid containing the Gdc gene. In order to knockout Gdc, a 1.2-kb kanamycin-resistant gene from the pUC4K vector was cloned into the KpnI site of the pGdc, producing a pGdckan plasmid. The design and the construction of the resulting plasmid pGdckan is shown in Figure 2a,b. The obtained plasmid was used to transform Synechocystis WT cells by natural transformation [48] and generate the mutant strain with the disrupted Gdc (ΔGdc). The transformant cells were selected on a BG11-containing plate supplemented with kanamycin at a concentration of 25 μg mL−1, within three weeks. To ensure the insertion and complete segregation of genes, transformants were subcultured for up to five generations. The complete segregation of the obtained Gdc knockout was analysed by a colony PCR (Figure 2c and Figure S1).
3.3. Experimental Treatments and Analysis of Metabolites
The chlorophyll a content in the Synechocystis WT and ΔGdc strains was determined by an N,N dimethylformamide (DMF) (EMSURE® ACS) extraction, according to a protocol adapted from Jantaro et al. [49]. Briefly, liquid cultures of the WT and ΔGdc strains were initiated at an OD730 of ~0.1 and were grown until the mid-log (OD730 of 0.3–0.4), late-log (OD730 of 0.8–1.0) or stationary (OD730 of 1.5–2.0) growth phases. Cell suspensions (corresponding to 10 mg of biomass) from various growth phases were centrifuged at 2500× g for 10 min and cell pellets were extracted by using DMF and by vigorous mixing at room temperature, followed by a measurement of their chlorophyll a content.
Cellular ALA, glutamate and GABA were extracted from the pelleted liquid cultures of the WT and the ΔGdc Synechocystis strains grown until the late-log phase in 200 mL of BG11 medium, followed by various physicochemical treatments for 24 and 48 h. The physicochemical treatments included supplementing the BG11 medium with 10 mM of NaCl, glutamate, glucose, glycine or succinate (Sigma-Aldrich, Saint Louis, MO, USA) or exposing the cultures to a cold temperature (4 °C). LA (Sigma, Aldrich) was prepared as a stock solution of 1 M and neutralized to pH 7.6 with NaOH (Ajax Finechem, Sydney, Australia). The filter sterilized (0.45-µm Millipore filter) LA solution was added at various concentrations (20, 40 and 60 mM), for 48 h, in late-log phase grown cultures. The extraction and quantification of cellular ALA, glutamate and GABA contents were performed as previously described [44]. In brief, ALA, glutamate and GABA were extracted from dry cells, derivatised with o-phthalaldehyde or 9-fluorenylmethyl chloroformate (Agilent, California, USA) and analysed by an HPLC system (Shimadzu Scientific In-struments Inc., Columbia, IN, USA) at 262 and 338 nm. The quantification of ALA and glutamate was done by comparing the peak areas with standards (purity > 99%; Sigma-Aldrich, Saint Louis, USA). All experiments were performed in triplicate.
3.4. Total RNA Extraction and Gene Expression Analysis
Synechocystis WT and ΔGdc strains were grown under the experimental conditions described earlier to undertake the ALA extraction step. Cells (at an OD730 of ~0.8–1.0) treated under various physicochemical conditions for 24 h were harvested by centrifugation at 2500× g, for 10 min, at 25 °C. The cell pellet was used for RNA extraction using GenUP™ Total RNA Mini Kit (biotechrabbit, Berlin, Germany) and reverse transcription-quantitative PCR (RT-qPCR) by using the methods described before [44]. The cDNAs of the WT and the ΔGdc strains were synthesized using RevertUP™ reverse Transcriptase (biotechrabbit), according to the manufacturer’s protocol and used to determine the transcript levels of the genes targeted by corresponding primers (Macrogen, Seoul, Korea); both the primer sequences and the target sequence IDs are shown in Table 1. Experiments were performed by using three independent RT-PCR/qPCR assays.
3.5. Statistics
Unless otherwise stated, the data presented represent independent experimental triplicates of the mean and are presented as mean ± standard deviation (SD). The differences between means of the individual groups were analyzed using one-way analysis of variance (ANOVA) via GraphPad Prism 9.3.1 software (San Diego, CA, USA). * p < 0.05, ** p < 0.01 *** p < 0.005, **** p < 0.001 versus control groups.
4. Conclusions
To the best of our knowledge, increased ALA production in cyanobacterium Synechocystis sp. PCC 6803 with disrupted GABA shunt route via Gdc inactivation (ΔGdc) has not been previously reported in cyanobacteria. The ΔGdc mutant had increased cellular levels of precursor metabolites for ALA biosynthesis which may then maximize the pathway flux towards ALA synthesis. The carbon flux redirected from GABA shunt pathway towards ALA biosynthesis was further improved under specific nutrients conditions in engineered strain. However, further research regarding engineering of the key regulatory metabolic steps, characterization of enzymes involved and analysis on extracellular ALA secretion shall provide insights into the detailed mechanism of ALA production in Synechocystis, that might prove beneficial in utilizing cyanobacteria as cell factories for ALA biosynthesis from the perspectives of potential applications in agriculture as an herbicide, insecticide and a growth-promoting factor for plants and medical field for cancer treatment and other clinical uses.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24021213/s1.
Author Contributions
Conceptualization, S.K. and W.D.-E.; methodology, S.K.; validation, S.K.; formal analysis, S.K.; investigation, S.K.; resources, S.K. and W.D.-E.; writing—original draft preparation, S.K.; writing—review and editing, W.D.-E.; visualization, S.K. and W.D.-E.; supervision, W.D.-E.; project administration, W.D.-E.; funding acquisition, W.D.-E. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Second Century Fund (C2F), Chulalongkorn University, Bangkok to S.K. Chulalongkorn University supported financially the Natural Product Biotechnology Research Unit GRU 6205633003-1 and the Competitive Track career path for W.D.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors would like to thank the Faculty of Pharmaceutical Sciences, Chulalongkorn University, Thailand, for providing the facilities and services enabling us to undertake this research work. Authors are grateful to Ponsawan Netcharoensirisuk for her help in conducting the statistical analysis.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Vranova, V.; Rejsek, K.; Skene, K.R.; Formanek, P. Non-protein amino acids: Plant, soil and ecosystem interactions. Plant Soil. 2011, 342, 31–48. [Google Scholar] [CrossRef]
- Kang, Z.; Zhang, J.; Zhou, J.; Qi, Q.; Du, G.; Chen, J. Recent advances in microbial production of δ-aminolevulinic acid and vitamin B12. Biotechnol. Adv. 2012, 30, 1533–1542. [Google Scholar] [CrossRef] [PubMed]
- Jantaro, S.; Kanwal, S. Low molecular weight nitrogenous compounds (GABA and polyamines) in blue green algae, In Algal green Chemistry: Recent Progress in Biotechnology; Rastogi, R.P., Madamwar, D., Pandey, A., Eds.; Elsevier B.V.: Amsterdam, The Netherlands, 2017; pp. 149–169. [Google Scholar]
- Su, A.; Yu, Q.; Luo, Y.; Yang, J.; Wang, E.; Yua, H. Metabolic engineering of microorganisms for the production of multifunctional non-protein amino acids: γ-aminobutyric acid and δ-aminolevulinic acid. Microb. Biotechnol. 2021, 14, 2279–2290. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Hong, K.; Mao, Y.; Ma, H.; Chen, T.; Wang, Z. Natural 5-aminolevulinic acid: Sources, biosynthesis, detection and applications. Front. Bioeng. Biotechnol. 2022, 10, 841443. [Google Scholar] [CrossRef]
- Rieble, S.; Beale, S.I. Purification of glutamyl-tRNA reductase from Synechocystis sp. PCC 6803. J. Biol. Chem. 1991, 266, 9740–9745. [Google Scholar] [CrossRef]
- Zhang, J.; Kang, Z.; Chen, J.; Du, G. Optimization of the heme biosynthesis pathway for the production of 5-aminolevulinic acid in Escherichia coli. Sci. Rep. 2015, 5, 8584. [Google Scholar] [CrossRef]
- Kanwal, S.; Incharoensakdi, A. The role of GAD pathway for regulation of GABA accumulation and C/N balance in Synechocystis sp. PCC6803. J. Appl. Phycol. 2019, 31, 3503–3514. [Google Scholar] [CrossRef]
- Cui, Z.; Zhu, Z.; Zhang, J.; Jiang, Z.; Liu, Y.; Wang, Q.; Hou, J.; Qi, Q. Efficient 5-aminolevulinic acid production through reconstructing the metabolic pathway in SDH-deficient Yarrowia lipolytica. Biochem. Eng. J. 2021, 174, 108125. [Google Scholar] [CrossRef]
- Zhu, C.; Chen, J.; Wang, Y.; Wang, L.; Guo, X.; Chen, N.; Zheng, P.; Sun, J.; Ma, Y. Enhancing 5-aminolevulinic acid tolerance and production by engineering the antioxidant defense system of Escherichia coli. Biotechnol. Bioeng. 2019, 116, 2018–2028. [Google Scholar] [CrossRef]
- Yu, X.; Jin, H.; Liu, W.; Wang, Q.; Qi, Q. Engineering Corynebacterium glutamicum to produce 5-aminolevulinic acid from glucose. Microb. Cell Fact. 2015, 14, 183. [Google Scholar] [CrossRef]
- Anderson, S.L.; McIntosh, L. Light-activated heterotrophic growth of the cyanobacterium Synechocystis sp. strain PCC 6803: A blue-light-requiring process. J. Bacteriol. 1991, 9, 2761–2767. [Google Scholar] [CrossRef]
- Morris, J.N.; Crawford, T.S.; Jeffs, A.; Stockwell, P.A.; Eaton-Rye, J.J.; Summerfield, T.C. Whole genome re-sequencing of two “wild-type” strains of the model cyanobacterium Synechocystis sp. PCC 6803. N. Z. J. Bot. 2014, 52, 36–47. [Google Scholar] [CrossRef]
- Zhang, S.; Bryant, D.A. The tricarboxylic acid cycle in cyanobacteria. Science 2012, 334, 1551–1553. [Google Scholar] [CrossRef]
- Kipe-Nolt, J.A.; Stevens, S.E. Biosynthesis of δ-aminolevulinic acid from glutamate in Agmenellum quadruplicatum. Plant Physiol. 1980, 65, 126–128. [Google Scholar] [CrossRef] [PubMed]
- Kisaka, H.; Kida, T.; Miwa, T. Antisense suppression of glutamate decarboxylase in tomato (Lycopersicon esculentum L.) results in accumulation of glutamate in transgenic tomato fruits. Plant Biotech. 2006, 23, 267–274. [Google Scholar] [CrossRef]
- Kanwal, S.; Khetkorn, W.; Incharoensakdi, A. GABA accumulation in response to different nitrogenous compounds in unicellular cyanobacterium Synechocystis sp. PCC 6803. Curr. Microbiol. 2015, 70, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Jaishankar, J.; Srivastava, P. Molecular Basis of Stationary Phase Survival and Applications. Front. Microbiol. 2017, 8, 2000. [Google Scholar] [CrossRef]
- Nishikawa, S.; Murooka, Y. 5-Aminolevulinic acid: Production by fermentation, and agricultural and biomedical applications. Biotechnol. Genet. Eng. Rev. 2001, 18, 149–170. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhang, G.; Li, X.; Zhang, J. Microbial production and applications of 5-aminolevulinic acid. Appl. Microbiol. Biotechnol. 2014, 98, 7349–7357. [Google Scholar] [CrossRef]
- Wu, Y.; Jin, X.; Liao, W.; Hu, L.; Dawuda, M.M.; Zhao, X.; Tang, Z.; Gong, T.; Yu, J. 5-Aminolevulinic acid (ALA) alleviated salinity stress in cucumber seedlings by enhancing chlorophyll synthesis pathway. Front. Plant Sci. 2018, 9, 635. [Google Scholar] [CrossRef]
- Liu, D.; Wu, L.; Naeem, M.S.; Liu, H.; Deng, X.; Xu, L.; Zhang, F. 5-Aminolevulinic acid enhances photosynthetic gas exchange, chlorophyll fluorescence and antioxidant system in oilseed rape under drought stress. Acta Physiol. Plant. 2013, 35, 2747–2759. [Google Scholar] [CrossRef]
- Beale, S.I. The biosynthesis of δ-aminolevulinic acid in Chlorella. Plant Physiol. 1970, 45, 504–506. [Google Scholar] [CrossRef]
- Troxler, R.F.; Brown, A.S. Metabolism of δ-aminolevulinic acid in red and blue-green algae. Plant Physiol. 1975, 55, 463–467. [Google Scholar] [CrossRef] [PubMed]
- Kipe-Nolt, J.A.; Stevens, S.J.; Stevens, C.L. Biosynthesis of δ-aminolevulinic acid by blue-green algae (cyanobacteria). J. Bacteriol. 1978, 135, 286–288. [Google Scholar] [CrossRef] [PubMed]
- Rieble, S.; Beale, S.I. Transformation of glutamate to δ-aminolevulinic acid by soluble extracts of Synechocystis sp. PCC 6803 and other oxygenic prokaryotes. J. Biol. Chem. 1988, 263, 8861–8871. [Google Scholar] [CrossRef]
- Kanwal, S.; Rastogi, R.P.; Incharoensakdi, A. Glutamate decarboxylase activity and gamma-aminobutyric acid content in Synechocystis sp. PCC 6803 under osmotic stress and different carbon sources. J. Appl. Phycol. 2014, 26, 2327–2333. [Google Scholar] [CrossRef]
- Mikkat, S.; Effmert, U.; Hagemann, M. Uptake and use of the osmoprotective compounds trehalose, glucosylglycerol and sucrose by the cyanobacterium Synechocystis sp. PCC6803. Arch. Microbiol. 1997, 167, 112–118. [Google Scholar] [CrossRef]
- Hendry, J.I.; Prasannan, C.; Ma, F.; Möllers, K.B.; Jaiswal, D.; Digmurti, M.; Allen, D.K.; Frigaard, N.-U.; Dasgupta, S.; Wangikar, P.P. Rerouting of carbon flux in a glycogen mutant of cyanobacteria assessed via isotopically non-stationary 13 C metabolic flux analysis. Biotechnol. Bioeng. 2017, 114, 2298–2308. [Google Scholar] [CrossRef]
- Yi, Y.C.; Shih, I.T.; Yu, T.H.; Lee, Y.J.; Ng, I.S. Challenges and opportunities of bioprocessing 5-aminolevulinic acid using genetic and metabolic engineering: A critical review. Bioresour. Bioprocess. 2021, 8, 100. [Google Scholar] [CrossRef]
- Zhang, B.; Ye, B.C. Pathway engineering in Corynebacterium glutamicum S9114 for 5-aminolevulinic acid production. 3 Biotech 2018, 8, 247. [Google Scholar] [CrossRef]
- Eisenhut, M.; Bauwe, H.; Hagemann, M. Glycine accumulation is toxic for the cyanobacterium Synechocystis sp. strain PCC 6803, but can be compensated by supplementation with magnesium ions. FEMS Microbiol. Lett. 2007, 277, 232–237. [Google Scholar] [CrossRef] [PubMed]
- Martínez, J.L.; Petranovic, D.; Nielsen, J. Heme metabolism in stress regulation and protein production: From Cinderella to a key player. Bioengineered 2016, 7, 112–115. [Google Scholar] [CrossRef]
- Steward, F.C.; Durzan, D.J. Metabolism of nitrogenous compounds. In Plant Physiology; Steward, F.C., Ed.; Academic Press: New York, NY, USA, 1965; pp. 379–686. [Google Scholar]
- Quintero, M.J.; Montesinos, M.L.; Herrero, A.; Flores, E. Identification of genes encoding amino acid permeases by inactivation of selected ORFs from the Synechocystis genomic sequence. Genome Res. 2001, 11, 2034–2040. [Google Scholar] [CrossRef] [PubMed]
- Knoop, H.; Gründel, M.; Zilliges, Y.; Lehmann, R.; Hoffmann, S.; Lockau, W.; Steuer, R. Flux balance analysis of cyanobacterial metabolism: The metabolic network of Synechocystis sp. PCC 6803. PLoS Comput. Biol. 2013, 9, e1003081. [Google Scholar] [CrossRef] [PubMed]
- Falkner, G.; Horner, F. pH Changes in the cytoplasm of the blue-green alga Anacystis nidulans caused by light-dependent proton flux into the thylakoid space. Plant Physiol. 1976, 58, 717–718. [Google Scholar] [CrossRef]
- Shelp, B.J.; Bown, A.W.; McLean, M.D. Metabolism and functions of gamma-aminobutyric acid. Trends Plant Sci. 1999, 4, 446–452. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, R.; Tanaka, A. Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 2007, 58, 321–346. [Google Scholar] [CrossRef]
- Saikeur, A.; Choorit, W.; Prasertsan, P.; Kantachote, D.; Sasaki, K. Influence of precursors and inhibitor on the production of extracellular 5-aminolevulinic acid and biomass by Rhodopseudomonas palustris KG31. Biosci. Biotechnol. Biochem. 2009, 73, 987–992. [Google Scholar] [CrossRef]
- Lee, S.; Ryu, J.; Kim, S.Y.; Jeon, J.; Song, J.Y.; Cho, H.-T.; Choi, S.-B.; Choi, D.; de Marsac, N.T.; Park, Y.-I. Transcriptional regulation of the respiratory genes in the cyanobacterium Synechocystis sp. PCC 6803 during the early response to glucose feeding. Plant Physiol. 2007, 145, 1018–1030. [Google Scholar] [CrossRef]
- Wilde, A.; Hihara, Y. Transcriptional and posttranscriptional regulation of cyanobacterial photosynthesis. Biochim. Biophys. Acta (BBA)-Bioenerg. 2016, 1857, 296–308. [Google Scholar] [CrossRef] [PubMed]
- Forchhammer, K.; Selim, K.A. Carbon/nitrogen homeostasis control in cyanobacteria. FEMS Microbiol. Rev. 2020, 44, 33–53. [Google Scholar] [CrossRef] [PubMed]
- Kanwal, S.; Incharoensakdi, A. Characterization of glutamate decarboxylase from Synechocystis sp. PCC6803 and its role in nitrogen metabolism. Plant Physiol. Biochem. 2016, 99, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Ano, A.; Funahashi, H.; Nakao, K.; Nishizawa, Y. Effects of Levulinic acid on 5-aminolevulinic acid production in heterotrophic cultures on Chlorella regularis YA-603. J. Biosci. Bioeng. 2000, 89, 176–180. [Google Scholar] [CrossRef] [PubMed]
- Anderson, T.; Briseid, T.; Nesbakken, T.; Ormerod, J.; Sirevaag, R.; Thorud, M. Mechanism of systhesis of 5-aminolevulinic acid in purple, green and blue-green bacteria. FEMS Microbiol. Lett. 1983, 19, 303–306. [Google Scholar] [CrossRef]
- Rippka, R.; Deruelles, J.; Waterbury, J.B.; Herdman, M.; Stanier, R.Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1979, 111, 1–61. [Google Scholar] [CrossRef]
- Kanwal, S.; Incharoensakdi, A. GABA synthesis mediated by γ-aminobutanal dehydrogenase in Synechocystis sp. PCC6803 with disrupted glutamate and α-ketoglutarate decarboxylase genes. Plant Sci. 2020, 290, 110287. [Google Scholar] [CrossRef]
- Jantaro, S.; Baebprasert, W.; Piyamawadee, C.; Sodsuay, O.; Incharoensakdi, A. Exogenous spermidine alleviates UV-induced growth inhibition of Synechocystis sp. PCC 6803 via reduction of hydrogen peroxide and malonaldehyde levels. Appl. Biochem. Biotechnol. 2014, 173, 1145–1156. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).