The Temporal and Geographical Dynamics of Potato Virus Y Diversity in Russia
Abstract
1. Introduction
2. Results
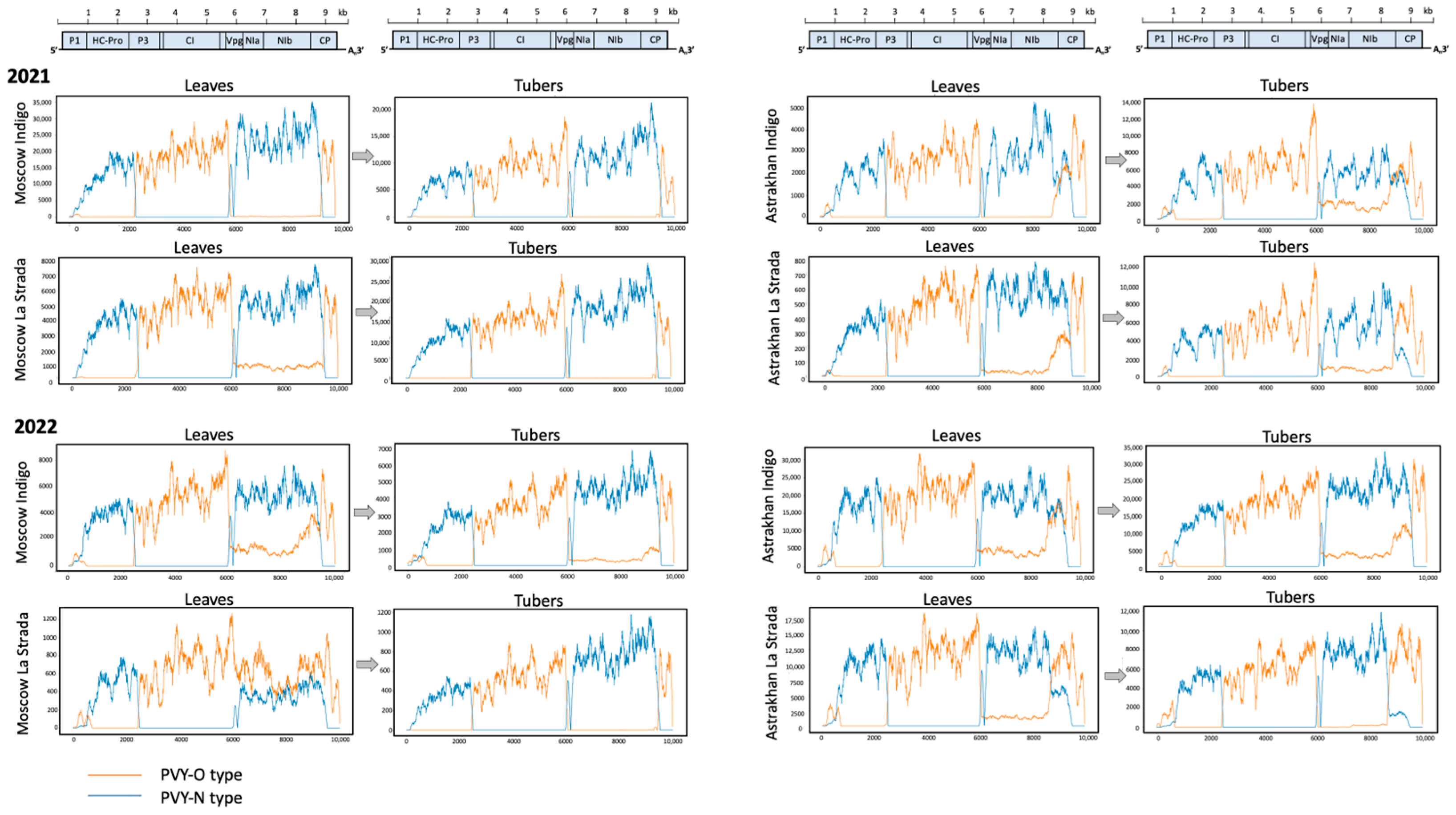
2.1. Field Spread and Tuber Transmission of PVY
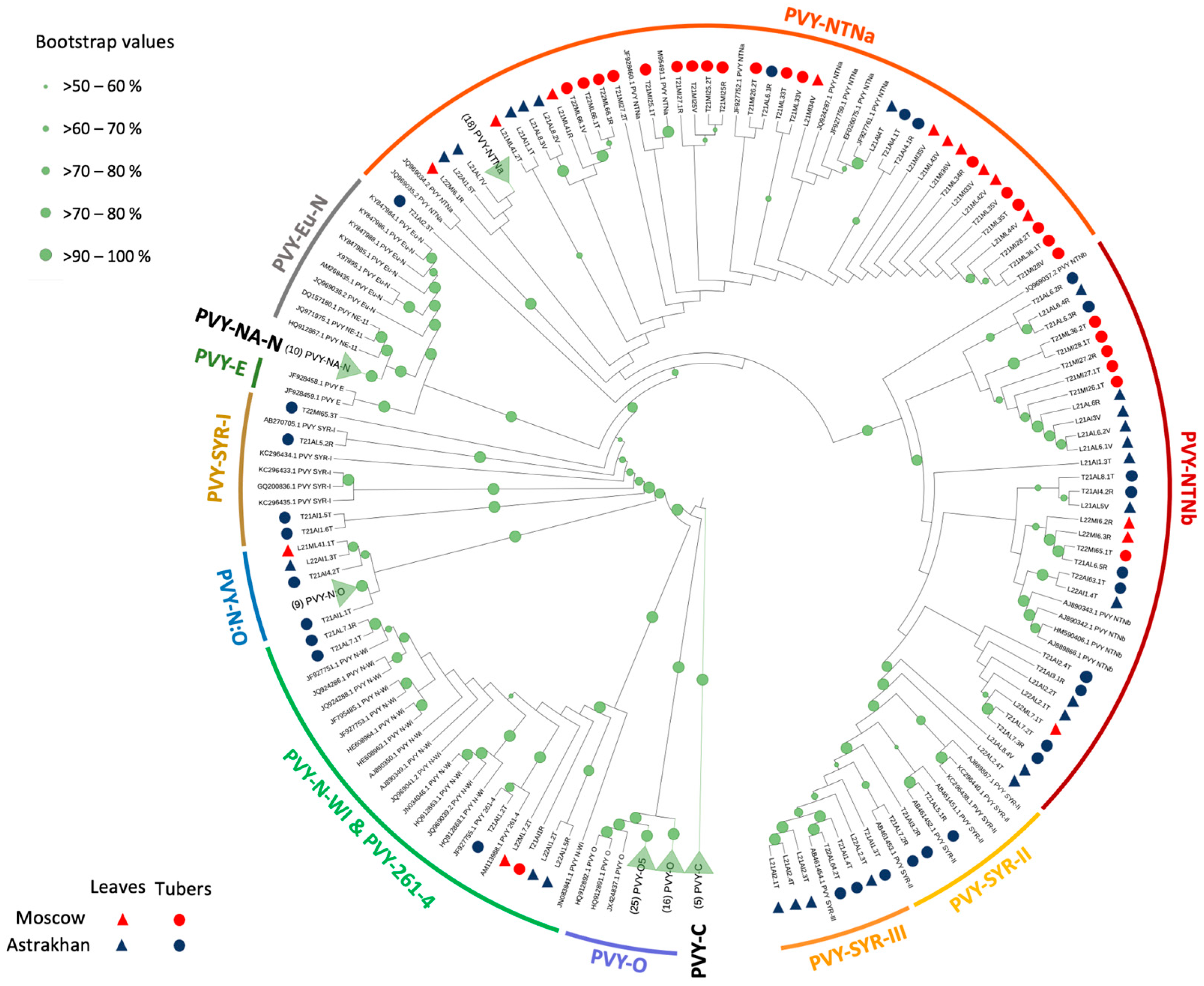
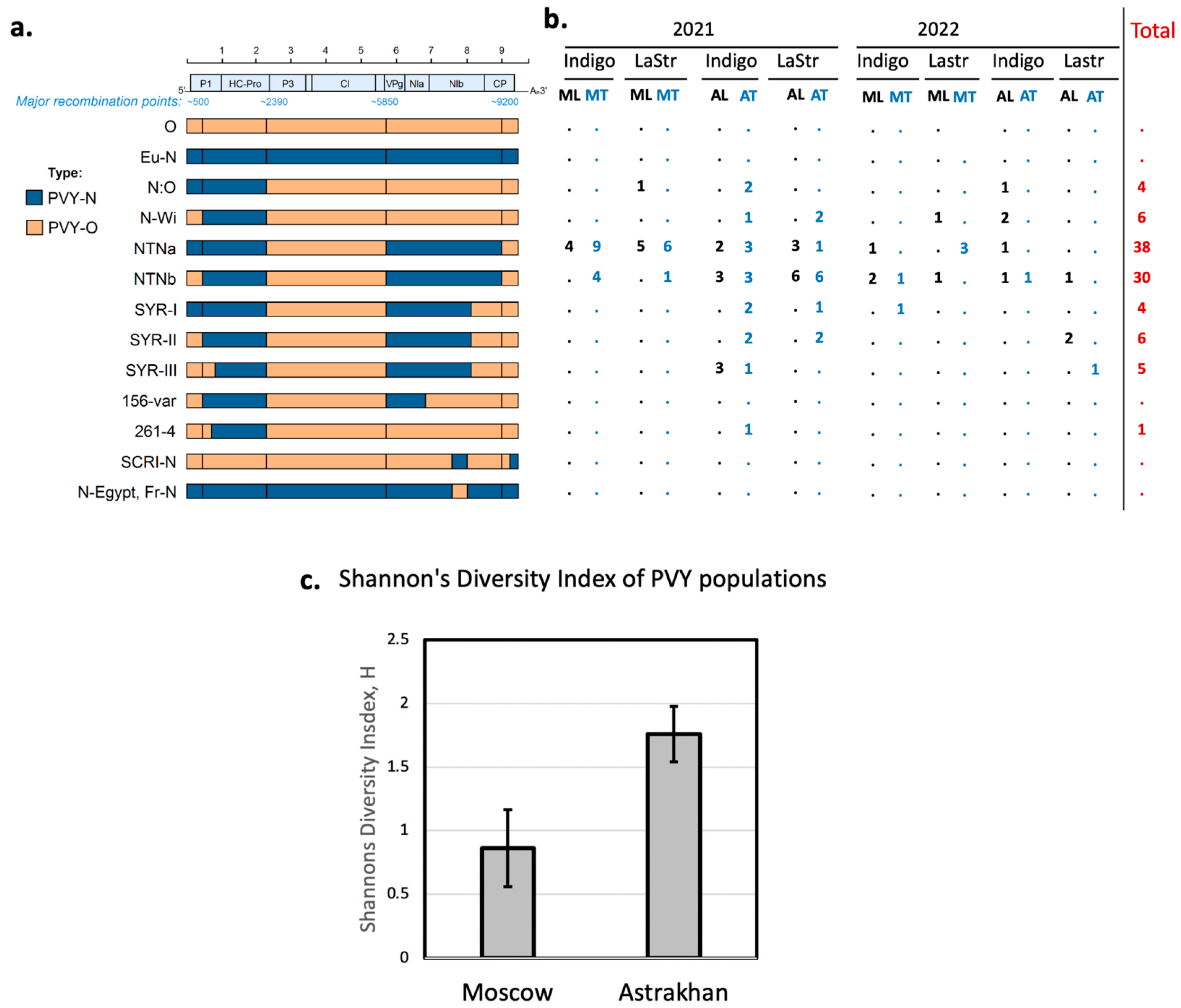
2.2. Phylogeny of De Novo-Assembled PVY Genomes
2.3. Factors Affecting the Diversity of PVY
3. Discussion
4. Materials and Methods
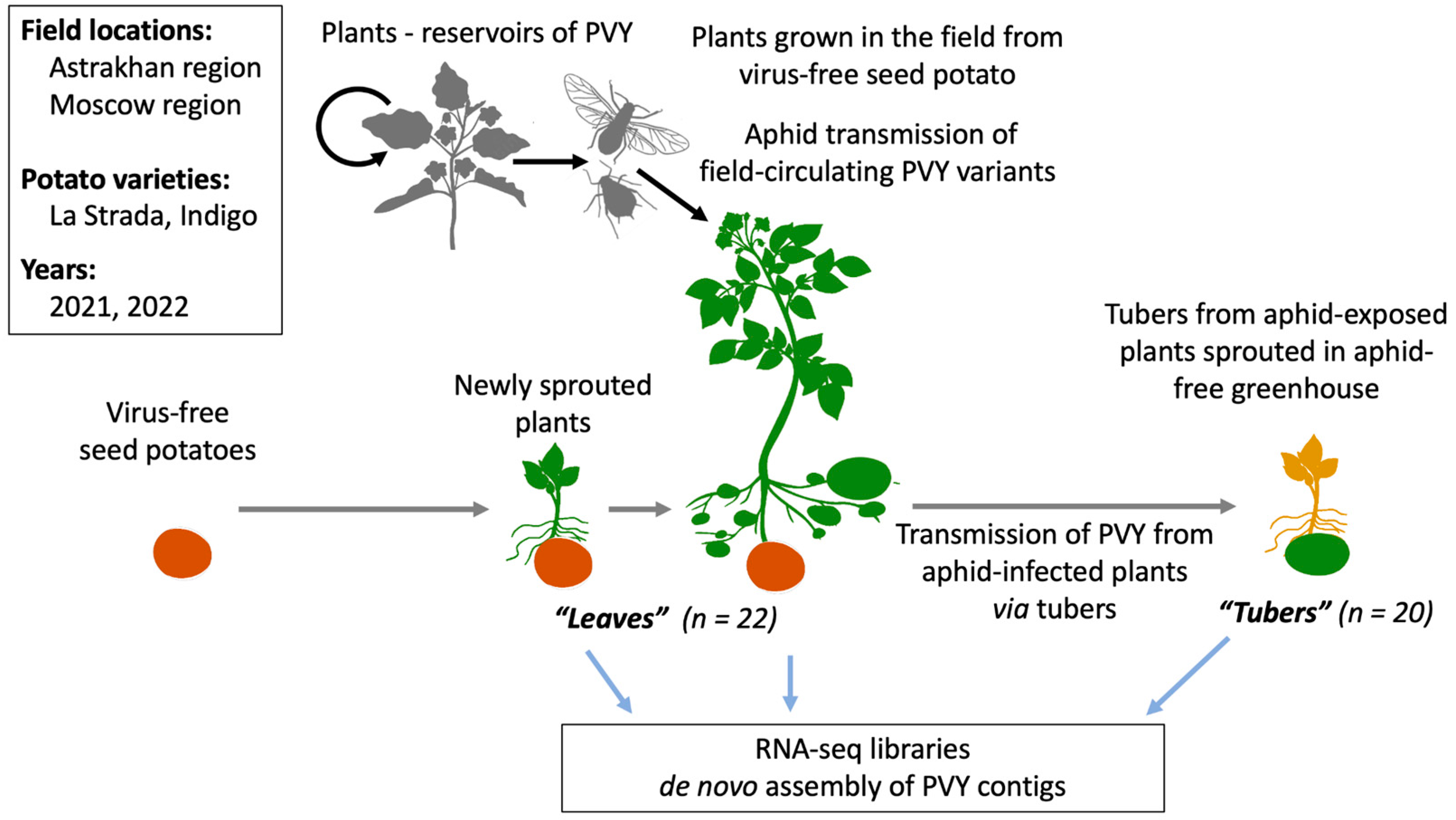
4.1. Plants and Experimental Design
4.2. RNA Extraction and High-Throughput Sequencing
4.3. De Novo Assembly and Sequence Sources
4.4. Phylogenetic and Comparative Analysis of PVY Sequences
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lacomme, C.; Jacquot, E. General Characteristics of Potato Virus Y (PVY) and Its Impact on Potato Production: An Overview. In Potato Virus Y: Biodiversity, Pathogenicity, Epidemiology and Management; Lacomme, C., Glais, L., Bellstedt, D.U., Dupuis, B., Karasev, A.V., Jacquot, E., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–19. ISBN 978-3-319-58860-5. [Google Scholar]
- Genus: Potyvirus|ICTV. Available online: https://ictv.global/report/chapter/potyviridae/potyviridae/potyvirus (accessed on 17 August 2023).
- Karasev, A.V.; Gray, S.M. Continuous and Emerging Challenges of Potato Virus Y in Potato. Annu. Rev. Phytopathol. 2013, 51, 571–586. [Google Scholar] [CrossRef] [PubMed]
- Chung, B.Y.-W.; Miller, W.A.; Atkins, J.F.; Firth, A.E. An Overlapping Essential Gene in the Potyviridae. Proc. Natl. Acad. Sci. USA 2008, 105, 5897–5902. [Google Scholar] [CrossRef] [PubMed]
- Torrance, L.; Talianksy, M.E. Potato Virus Y Emergence and Evolution from the Andes of South America to Become a Major Destructive Pathogen of Potato and Other Solanaceous Crops Worldwide. Viruses 2020, 12, 1430. [Google Scholar] [CrossRef] [PubMed]
- Cervantes, F.A.; Alvarez, J.M. Within Plant Distribution of Potato Virus Y in Hairy Nightshade (Solanum Sarrachoides): An Inoculum Source Affecting PVY Aphid Transmission. Virus Res. 2011, 159, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, A.J.; Ohshima, K.; Yasaka, R.; Mohammadi, M.; Gibbs, M.J.; Jones, R.A.C. The Phylogenetics of the Global Population of Potato Virus Y and Its Necrogenic Recombinants. Virus Evol. 2017, 3, vex002. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, S.; Jones, R.A.C.; Matsuoka, H.; Ohshima, K.; Kreuze, J.; Gibbs, A.J. Potato Virus Y; the Andean Connection. Virus Evol. 2019, 5, vez037. [Google Scholar] [CrossRef] [PubMed]
- Karasev, A.V.; Hu, X.; Brown, C.J.; Kerlan, C.; Nikolaeva, O.V.; Crosslin, J.M.; Gray, S.M. Genetic Diversity of the Ordinary Strain of Potato Virus Y (PVY) and Origin of Recombinant PVY Strains. Phytopathology 2011, 101, 778–785. [Google Scholar] [CrossRef]
- Funke, C.N.; Nikolaeva, O.V.; Green, K.J.; Tran, L.T.; Chikh-Ali, M.; Quintero-Ferrer, A.; Cating, R.A.; Frost, K.E.; Hamm, P.B.; Olsen, N.; et al. Strain-Specific Resistance to Potato Virus Y (PVY) in Potato and Its Effect on the Relative Abundance of PVY Strains in Commercial Potato Fields. Plant Dis. 2017, 101, 20–28. [Google Scholar] [CrossRef]
- Green, K.J.; Brown, C.J.; Gray, S.M.; Karasev, A.V. Phylogenetic Study of Recombinant Strains of Potato Virus Y. Virology 2017, 507, 40–52. [Google Scholar] [CrossRef]
- Gray, S.; De Boer, S.; Lorenzen, J.; Karasev, A.; Whitworth, J.; Nolte, P.; Singh, R.; Boucher, A.; Xu, H. Potato Virus Y: An Evolving Concern for Potato Crops in the United States and Canada. Plant Dis. 2010, 94, 1384–1397. [Google Scholar] [CrossRef]
- Green, K.J.; Brown, C.J.; Karasev, A.V. Genetic Diversity of Potato Virus Y (PVY): Sequence Analyses Reveal Ten Novel PVY Recombinant Structures. Arch. Virol. 2018, 163, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Baebler, Š.; Coll, A.; Gruden, K. Plant Molecular Responses to Potato Virus Y: A Continuum of Outcomes from Sensitivity and Tolerance to Resistance. Viruses 2020, 12, 217. [Google Scholar] [CrossRef] [PubMed]
- Solomon-Blackburn, R.M.; Bradshaw, J.E. Resistance to Potato Virus Y in a Multitrait Potato Breeding Scheme without Direct Selection in Each Generation. Potato Res. 2007, 50, 87–95. [Google Scholar] [CrossRef]
- Zhu, Y.; Qian, W.; Hua, J. Temperature Modulates Plant Defense Responses through NB-LRR Proteins. PLoS Pathog. 2010, 6, e1000844. [Google Scholar] [CrossRef]
- Szittya, G.; Silhavy, D.; Molnár, A.; Havelda, Z.; Lovas, Á.; Lakatos, L.; Bánfalvi, Z.; Burgyán, J. Low Temperature Inhibits RNA Silencing-Mediated Defence by the Control of SiRNA Generation. EMBO J. 2003, 22, 633–640. [Google Scholar] [CrossRef]
- Shams-Bakhsh, M.; Canto, T.; Palukaitis, P. Enhanced Resistance and Neutralization of Defense Responses by Suppressors of RNA Silencing. Virus Res. 2007, 130, 103–109. [Google Scholar] [CrossRef][Green Version]
- Fesenko, I.; Spechenkova, N.; Mamaeva, A.; Makhotenko, A.V.; Love, A.J.; Kalinina, N.O.; Taliansky, M. Role of the Methionine Cycle in the Temperature-Sensitive Responses of Potato Plants to Potato Virus Y. Mol. Plant Pathol. 2021, 22, 77–91. [Google Scholar] [CrossRef]
- Hofius, D.; Maier, A.T.; Dietrich, C.; Jungkunz, I.; Börnke, F.; Maiss, E.; Sonnewald, U. Capsid Protein-Mediated Recruitment of Host DnaJ-like Proteins Is Required for Potato Virus Y Infection in Tobacco Plants. J. Virol. 2007, 81, 11870–11880. [Google Scholar] [CrossRef]
- Mäkinen, K.; Hafren, A. Intracellular Coordination of Potyviral RNA Functions in Infection. Front. Plant Sci. 2014, 5, 110. [Google Scholar] [CrossRef]
- Makarova, S.; Makhotenko, A.; Spechenkova, N.; Love, A.J.; Kalinina, N.O.; Taliansky, M. Interactive Responses of Potato (Solanum Tuberosum L.) Plants to Heat Stress and Infection with Potato Virus Y. Front. Microbiol. 2018, 9, 2582. [Google Scholar] [CrossRef] [PubMed]
- Fominykh, T.S.; Ivanova, G.P.; Medvedeva, K.D. Monitoring of Potato Viral Diseases in the Pskov and Astrakhan Regions of Russia. Plant Prot. Vestn. 2017, 4, 29–34. [Google Scholar]
- Della Bartola, M.; Byrne, S.; Mullins, E. Characterization of Potato Virus Y Isolates and Assessment of Nanopore Sequencing to Detect and Genotype Potato Viruses. Viruses 2020, 12, 478. [Google Scholar] [CrossRef]
- Avrahami-Moyal, L.; Tam, Y.; Sela, N.; Prakash, S.; Meller Harel, Y.; Bornstein, M.; Shulchani, R.; Dar, Z.; Gaba, V. Characterization of Potato Virus Y Populations in Potato in Israel. Arch. Virol. 2019, 164, 1691–1695. [Google Scholar] [CrossRef]
- Lukashev, A.N.; Lashkevich, V.A.; Ivanova, O.E.; Koroleva, G.A.; Hinkkanen, A.E.; Ilonen, J. Recombination in Circulating Human Enterovirus B: Independent Evolution of Structural and Non-Structural Genome Regions. J. Gen. Virol. 2005, 86, 3281–3290. [Google Scholar] [CrossRef]
- Moore, J.; Jironkin, A.; Chandler, D.; Burroughs, N.; Evans, D.J.; Ryabov, E.V. Recombinants between Deformed Wing Virus and Varroa Destructor Virus-1 May Prevail in Varroa Destructor-Infested Honeybee Colonies. J. Gen. Virol. 2011, 92, 156–161. [Google Scholar] [CrossRef]
- Ryabov, E.V.; Childers, A.K.; Chen, Y.; Madella, S.; Nessa, A.; vanEngelsdorp, D.; Evans, J.D. Recent Spread of Varroa Destructor Virus-1, a Honey Bee Pathogen, in the United States. Sci. Rep. 2017, 7, 17447. [Google Scholar] [CrossRef] [PubMed]
- Collection of Varieties DokaGene. Available online: https://www.dokagene.ru/pdf/Кoллекция%20сoртoв.pdf (accessed on 17 August 2023).
- Jiang, H.; Lei, R.; Ding, S.-W.; Zhu, S. Skewer: A Fast and Accurate Adapter Trimmer for next-Generation Sequencing Paired-End Reads. BMC Bioinform. 2014, 15, 182. [Google Scholar] [CrossRef] [PubMed]
- Babraham Bioinformatics—FastQC A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 21 August 2023).
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Bushmanova, E.; Antipov, D.; Lapidus, A.; Prjibelski, A.D. RnaSPAdes: A de Novo Transcriptome Assembler and Its Application to RNA-Seq Data. GigaScience 2019, 8, giz100. [Google Scholar] [CrossRef]
- Meleshko, D.; Hajirasouliha, I.; Korobeynikov, A. CoronaSPAdes: From Biosynthetic Gene Clusters to RNA Viral Assemblies. Bioinformatics 2021, 38, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- McGinnis, S.; Madden, T.L. BLAST: At the Core of a Powerful and Diverse Set of Sequence Analysis Tools. Nucleic Acids Res. 2004, 32, W20–W25. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Milne, I.; Bayer, M.; Stephen, G.; Cardle, L.; Marshall, D. Tablet: Visualizing Next-Generation Sequence Assemblies and Mappings. Methods Mol. Biol. Clifton NJ 2016, 1374, 253–268. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, Scalable Generation of High-Quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Castresana, J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Grant, B.J.; Rodrigues, A.P.C.; ElSawy, K.M.; McCammon, J.A.; Caves, L.S.D. Bio3d: An R Package for the Comparative Analysis of Protein Structures. Bioinformatics 2006, 22, 2695–2696. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 1 September 2022).

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samarskaya, V.O.; Ryabov, E.V.; Gryzunov, N.; Spechenkova, N.; Kuznetsova, M.; Ilina, I.; Suprunova, T.; Taliansky, M.E.; Ivanov, P.A.; Kalinina, N.O. The Temporal and Geographical Dynamics of Potato Virus Y Diversity in Russia. Int. J. Mol. Sci. 2023, 24, 14833. https://doi.org/10.3390/ijms241914833
Samarskaya VO, Ryabov EV, Gryzunov N, Spechenkova N, Kuznetsova M, Ilina I, Suprunova T, Taliansky ME, Ivanov PA, Kalinina NO. The Temporal and Geographical Dynamics of Potato Virus Y Diversity in Russia. International Journal of Molecular Sciences. 2023; 24(19):14833. https://doi.org/10.3390/ijms241914833
Chicago/Turabian StyleSamarskaya, Viktoriya O., Eugene V. Ryabov, Nikita Gryzunov, Nadezhda Spechenkova, Maria Kuznetsova, Irina Ilina, Tatiana Suprunova, Michael E. Taliansky, Peter A. Ivanov, and Natalia O. Kalinina. 2023. "The Temporal and Geographical Dynamics of Potato Virus Y Diversity in Russia" International Journal of Molecular Sciences 24, no. 19: 14833. https://doi.org/10.3390/ijms241914833
APA StyleSamarskaya, V. O., Ryabov, E. V., Gryzunov, N., Spechenkova, N., Kuznetsova, M., Ilina, I., Suprunova, T., Taliansky, M. E., Ivanov, P. A., & Kalinina, N. O. (2023). The Temporal and Geographical Dynamics of Potato Virus Y Diversity in Russia. International Journal of Molecular Sciences, 24(19), 14833. https://doi.org/10.3390/ijms241914833






