Elevated E200K Somatic Mutation of the Prion Protein Gene (PRNP) in the Brain Tissues of Patients with Sporadic Creutzfeldt–Jakob Disease (CJD)
Abstract
1. Introduction
2. Results
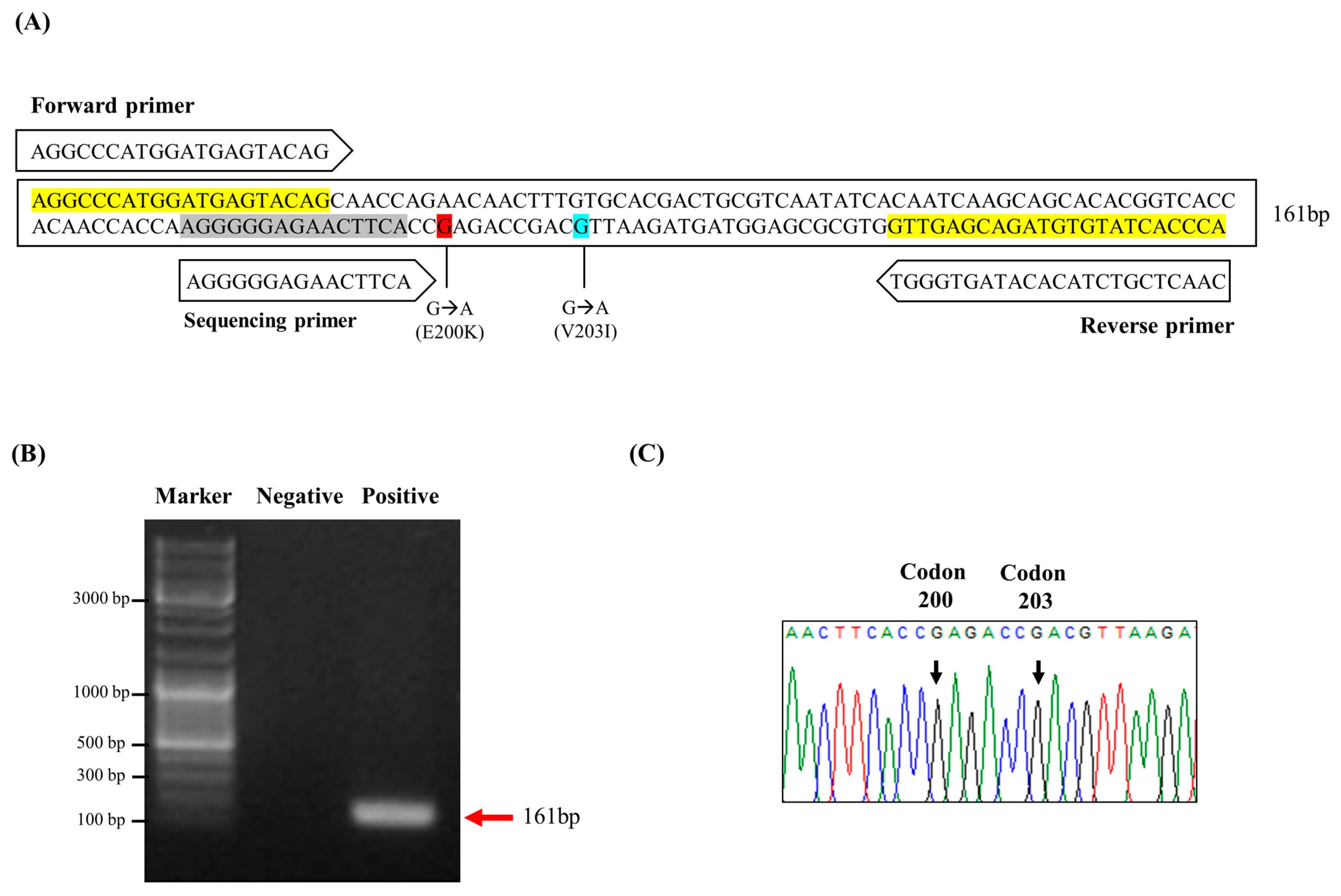
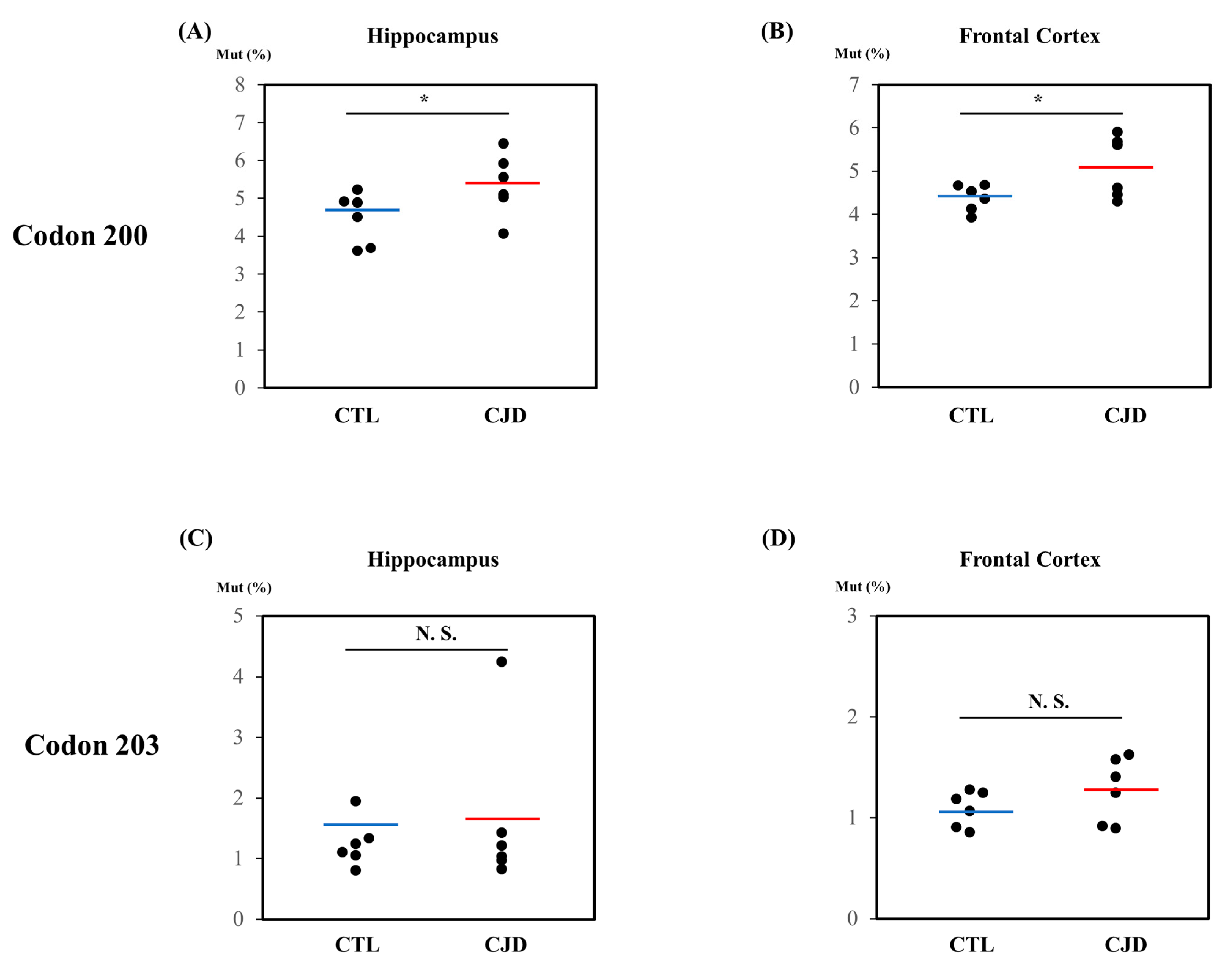
2.1. Investigation of Somatic Mutations of the Human PRNP Gene by Pyrosequencing
2.2. Evaluation of the Effect of Somatic Mutations on the Human PrP Using in Silico Analysis
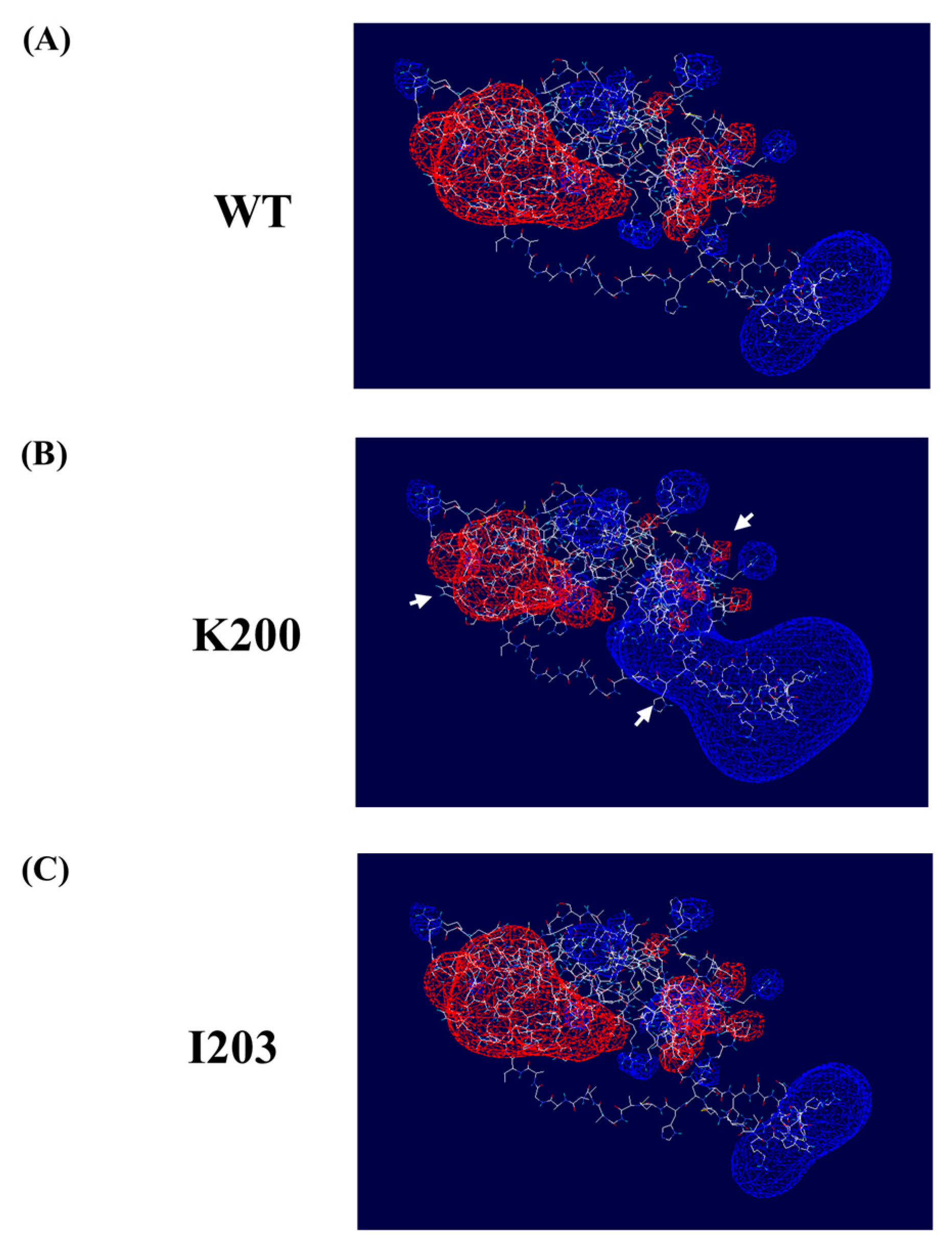
2.3. Evaluation of the Effect of Somatic Mutations on the Electrostatic Potential of the PrP
3. Discussion
4. Materials and Methods
4.1. Subjects
4.2. Ethics Statements
4.3. Genomic DNA Extraction
4.4. Polymerase Chain Reaction (PCR)
4.5. Sanger Sequencing
4.6. Pyrosequencing
4.7. In Silico Estimation of the Impact of E200K and V203I Somatic Mutations on the Human Prion Protein (PrP)
4.8. D Structure and Electrostatic Potential Analyses
4.9. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Prusiner, S.B. Prions. Proc. Natl. Acad. Sci. USA 1998, 95, 13363–13383. [Google Scholar] [CrossRef]
- Prusiner, S.B. The prion diseases. Brain Pathol. 1998, 8, 499–513. [Google Scholar] [CrossRef]
- Kim, Y.C.; Won, S.Y.; Jeong, B.H. Altered expression of glymphatic system-related proteins in prion diseases: Implications for the role of the glymphatic system in prion diseases. Cell Mol. Immunol. 2021, 18, 2281–2283. [Google Scholar] [CrossRef]
- Won, S.Y.; Kim, Y.C.; Jeong, B.H. First Report of the Potential Bovine Spongiform Encephalopathy (BSE)-Related Somatic Mutation E211K of the Prion Protein Gene (PRNP) in Cattle. Int. J. Mol. Sci. 2020, 21, 4246. [Google Scholar] [CrossRef]
- Kim, Y.C.; Won, S.Y.; Jeong, M.J.; Jeong, B.H. Absence of proteinase K-resistant PrP in Korean Holstein cattle carrying potential bovine spongiform encephalopathy-related E211K somatic mutation. Transbound. Emerg. Dis. 2021, 69, 805–812. [Google Scholar] [CrossRef]
- Kim, Y.C.; Park, K.J.; Hwang, J.Y.; Park, H.C.; Kang, H.E.; Sohn, H.J.; Jeong, B.H. In-depth examination of PrP(Sc) in Holstein cattle carrying the E211K somatic mutation of the bovine prion protein gene (PRNP). Transbound. Emerg. Dis. 2021, 69, e356–e361. [Google Scholar] [CrossRef]
- Benestad, S.L.; Telling, G.C. Chronic wasting disease: An evolving prion disease of cervids. Handb. Clin. Neurol. 2018, 153, 135–151. [Google Scholar] [CrossRef]
- Lloyd, S.; Mead, S.; Collinge, J. Genetics of prion disease. Top. Curr. Chem. 2011, 305, 1–22. [Google Scholar] [CrossRef]
- Jeong, B.H.; Kim, Y.S. Genetic studies in human prion diseases. J. Korean Med. Sci. 2014, 29, 623–632. [Google Scholar] [CrossRef]
- Gambetti, P.; Kong, Q.; Zou, W.; Parchi, P.; Chen, S.G. Sporadic and familial CJD: Classification and characterisation. Br. Med. Bull. 2003, 66, 213–239. [Google Scholar] [CrossRef]
- Atalay, F.O.; Tolunay, S.; Ozgun, G.; Bekar, A.; Zarifoglu, M. Creutzfeldt-Jakob disease: Report of four cases and review of the literature. Turk Patoloji Derg 2015, 31, 148–152. [Google Scholar] [CrossRef][Green Version]
- Collinge, J.; Palmer, M.S.; Dryden, A.J. Genetic predisposition to iatrogenic Creutzfeldt-Jakob disease. Lancet 1991, 337, 1441–1442. [Google Scholar] [CrossRef]
- Thomas, J.G.; Chenoweth, C.E.; Sullivan, S.E. Iatrogenic Creutzfeldt-Jakob disease via surgical instruments. J. Clin. Neurosci. 2013, 20, 1207–1212. [Google Scholar] [CrossRef]
- Mastrianni, J.A. The genetics of prion diseases. Genet. Med. 2010, 12, 187–195. [Google Scholar] [CrossRef]
- Uttley, L.; Carroll, C.; Wong, R.; Hilton, D.A.; Stevenson, M. Creutzfeldt-Jakob disease: A systematic review of global incidence, prevalence, infectivity, and incubation. Lancet Infect Dis. 2020, 20, e2–e10. [Google Scholar] [CrossRef]
- Park, J.S.; Lee, J.; Jung, E.S.; Kim, M.H.; Kim, I.B.; Son, H.; Kim, S.; Kim, S.; Park, Y.M.; Mook-Jung, I.; et al. Brain somatic mutations observed in Alzheimer’s disease associated with aging and dysregulation of tau phosphorylation. Nat. Commun. 2019, 10, 3090. [Google Scholar] [CrossRef]
- Miller, M.B.; Huang, A.Y.; Kim, J.; Zhou, Z.; Kirkham, S.L.; Maury, E.A.; Ziegenfuss, J.S.; Reed, H.C.; Neil, J.E.; Rento, L.; et al. Somatic genomic changes in single Alzheimer’s disease neurons. Nature 2022, 604, 714–722. [Google Scholar] [CrossRef]
- Alzualde, A.; Moreno, F.; Martinez-Lage, P.; Ferrer, I.; Gorostidi, A.; Otaegui, D.; Blazquez, L.; Atares, B.; Cardoso, S.; Martinez de Pancorbo, M.; et al. Somatic mosaicism in a case of apparently sporadic Creutzfeldt-Jakob disease carrying a de novo D178N mutation in the PRNP gene. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2010, 153B, 1283–1291. [Google Scholar] [CrossRef]
- Lobon, I.; Solís-Moruno, M.; Juan, D.; Muhaisen, A.; Abascal, F.; Esteller-Cucala, P.; García-Pérez, R.; Martí, M.J.; Tolosa, E.; Ávila, J.; et al. Somatic Mutations Detected in Parkinson Disease Could Affect Genes With a Role in Synaptic and Neuronal Processes. Front. Aging 2022, 3, 851039. [Google Scholar] [CrossRef]
- Leija-Salazar, M.; Piette, C.; Proukakis, C. Review: Somatic mutations in neurodegeneration. Neuropathol. Appl. Neurobiol. 2018, 44, 267–285. [Google Scholar] [CrossRef]
- Beck, J.A.; Poulter, M.; Campbell, T.A.; Uphill, J.B.; Adamson, G.; Geddes, J.F.; Revesz, T.; Davis, M.B.; Wood, N.W.; Collinge, J.; et al. Somatic and germline mosaicism in sporadic early-onset Alzheimer’s disease. Hum. Mol. Genet. 2004, 13, 1219–1224. [Google Scholar] [CrossRef]
- Nicolas, G.; Acuna-Hidalgo, R.; Keogh, M.J.; Quenez, O.; Steehouwer, M.; Lelieveld, S.; Rousseau, S.; Richard, A.C.; Oud, M.S.; Marguet, F.; et al. Somatic variants in autosomal dominant genes are a rare cause of sporadic Alzheimer’s disease. Alzheimer’s Dement. 2018, 14, 1632–1639. [Google Scholar] [CrossRef]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013, 76, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Thomas, P.D. PANTHER-PSEP: Predicting disease-causing genetic variants using position-specific evolutionary preservation. Bioinformatics 2016, 32, 2230–2232. [Google Scholar] [CrossRef]
- Choi, Y.; Chan, A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015, 31, 2745–2747. [Google Scholar] [CrossRef]
- Lodato, M.A.; Rodin, R.E.; Bohrson, C.L.; Coulter, M.E.; Barton, A.R.; Kwon, M.; Sherman, M.A.; Vitzthum, C.M.; Luquette, L.J.; Yandava, C.N.; et al. Aging and neurodegeneration are associated with increased mutations in single human neurons. Science 2018, 359, 555–559. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, G.G.; Puopolo, M.; Ladogana, A.; Pocchiari, M.; Budka, H.; van Duijn, C.; Collins, S.J.; Boyd, A.; Giulivi, A.; Coulthart, M.; et al. Genetic prion disease: The EUROCJD experience. Hum. Genet. 2005, 118, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Chen, C.; Xiao, K.; Zhou, W.; Gao, L.P.; Chen, D.D.; Wu, Y.Z.; Wang, Y.; Hu, C.; Gao, C.; et al. Genetic Prion Disease: Insight from the Features and Experience of China National Surveillance for Creutzfeldt-Jakob Disease. Neurosci. Bull. 2021, 37, 1570–1582. [Google Scholar] [CrossRef]
| Somatic Mutations | Methods | Score | Prediction |
|---|---|---|---|
| c.598G > A (E200K) | PolyPhen-2 | 0.987 | Probably damaging |
| PANTHER | 361 | Possibly damaging | |
| PROVEAN | −1.478 | Neutral | |
| c.607G > A (V203I) | PolyPhen-2 | 0.882 | Possibly damaging |
| PANTHER | 176 | Probably benign | |
| PROVEAN | −0.004 | Neutral |
| Sample No. | Type | Sex | Age | Average | Brain Tissue | K200 Somatic Mutation Rates (%) | I203 Somatic Mutation Rates (%) | Codon 129 Genotype |
|---|---|---|---|---|---|---|---|---|
| Sample 1 | Control 1 | Male | 84 | 64.6 ± 9.2 | Frontal cortex | 4.13 | 1.19 | MM |
| Sample 2 | Hippocampus | 4.92 | 1.25 | |||||
| Sample 3 | Control 2 | Male | 67 | Frontal cortex | 4.68 | 1.25 | MM | |
| Sample 4 | Hippocampus | 4.51 | 0.81 | |||||
| Sample 5 | Control 3 | Male | 73 | Frontal cortex | 4.67 | 1.28 | VV | |
| Sample 6 | Hippocampus | 5.23 | 1.95 | |||||
| Sample 7 | Control 4 | Female | 57 | Frontal cortex | 4.36 | 0.91 | MV | |
| Sample 8 | Hippocampus | 3.62 | 1.06 | |||||
| Sample 9 | Control 5 | Female | 73 | Frontal cortex | 3.93 | 0.86 | MV | |
| Sample 10 | Hippocampus | 3.69 | 1.11 | |||||
| Sample 11 | Control 6 | Female | 53 | Frontal cortex | 4.53 | 1.07 | MV | |
| Sample 12 | Hippocampus | 4.89 | 1.34 | |||||
| Sample 13 | Case 1 | Male | 86 | 70.7 ± 13.3 | Frontal cortex | 5.68 | 0.92 | MM |
| Sample 14 | Hippocampus | 4.07 | 4.25 | |||||
| Sample 15 | Case 2 | Male | 66 | Frontal cortex | 4.61 | 1.41 | MV | |
| Sample 16 | Hippocampus | 5.03 | 0.98 | |||||
| Sample 17 | Case 3 | Male | 86 | Frontal cortex | 4.30 | 1.63 | MV | |
| Sample 18 | Hippocampus | 6.45 | 1.43 | |||||
| Sample 19 | Case 4 | Female | 60 | Frontal cortex | 5.61 | 1.25 | MM | |
| Sample 20 | Hippocampus | 5.10 | 1.04 | |||||
| Sample 21 | Case 5 | Female | 72 | Frontal cortex | 4.46 | 1.58 | MM | |
| Sample 22 | Hippocampus | 5.92 | 1.22 | |||||
| Sample 23 | Case 6 | Female | 54 | Frontal cortex | 5.91 | 0.90 | MV | |
| Sample 24 | Hippocampus | 5.56 | 0.83 |
| Purpose | Name | Sequence | Size | Annealing Temperature | |
|---|---|---|---|---|---|
| Sanger sequencing | Amplification | F | CAACCGCTACCCACCTCAG | 564 bp | 60 °C |
| R | AGGACCATGCTCGATCCTCT | ||||
| Pyrosequencing | Amplification | PF | AGGCCCATGGATGAGTACAG | 161 bp | 60 °C |
| Biotin_PR | TGGGTGATACACATCTGCTCAAC | ||||
| Sequencing | Seq | AGGGGGAGAACTTCA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Won, S.-Y.; Kim, Y.-C.; Jeong, B.-H. Elevated E200K Somatic Mutation of the Prion Protein Gene (PRNP) in the Brain Tissues of Patients with Sporadic Creutzfeldt–Jakob Disease (CJD). Int. J. Mol. Sci. 2023, 24, 14831. https://doi.org/10.3390/ijms241914831
Won S-Y, Kim Y-C, Jeong B-H. Elevated E200K Somatic Mutation of the Prion Protein Gene (PRNP) in the Brain Tissues of Patients with Sporadic Creutzfeldt–Jakob Disease (CJD). International Journal of Molecular Sciences. 2023; 24(19):14831. https://doi.org/10.3390/ijms241914831
Chicago/Turabian StyleWon, Sae-Young, Yong-Chan Kim, and Byung-Hoon Jeong. 2023. "Elevated E200K Somatic Mutation of the Prion Protein Gene (PRNP) in the Brain Tissues of Patients with Sporadic Creutzfeldt–Jakob Disease (CJD)" International Journal of Molecular Sciences 24, no. 19: 14831. https://doi.org/10.3390/ijms241914831
APA StyleWon, S.-Y., Kim, Y.-C., & Jeong, B.-H. (2023). Elevated E200K Somatic Mutation of the Prion Protein Gene (PRNP) in the Brain Tissues of Patients with Sporadic Creutzfeldt–Jakob Disease (CJD). International Journal of Molecular Sciences, 24(19), 14831. https://doi.org/10.3390/ijms241914831







