To Break or Not to Break: The Role of TOP2B in Transcription
Abstract
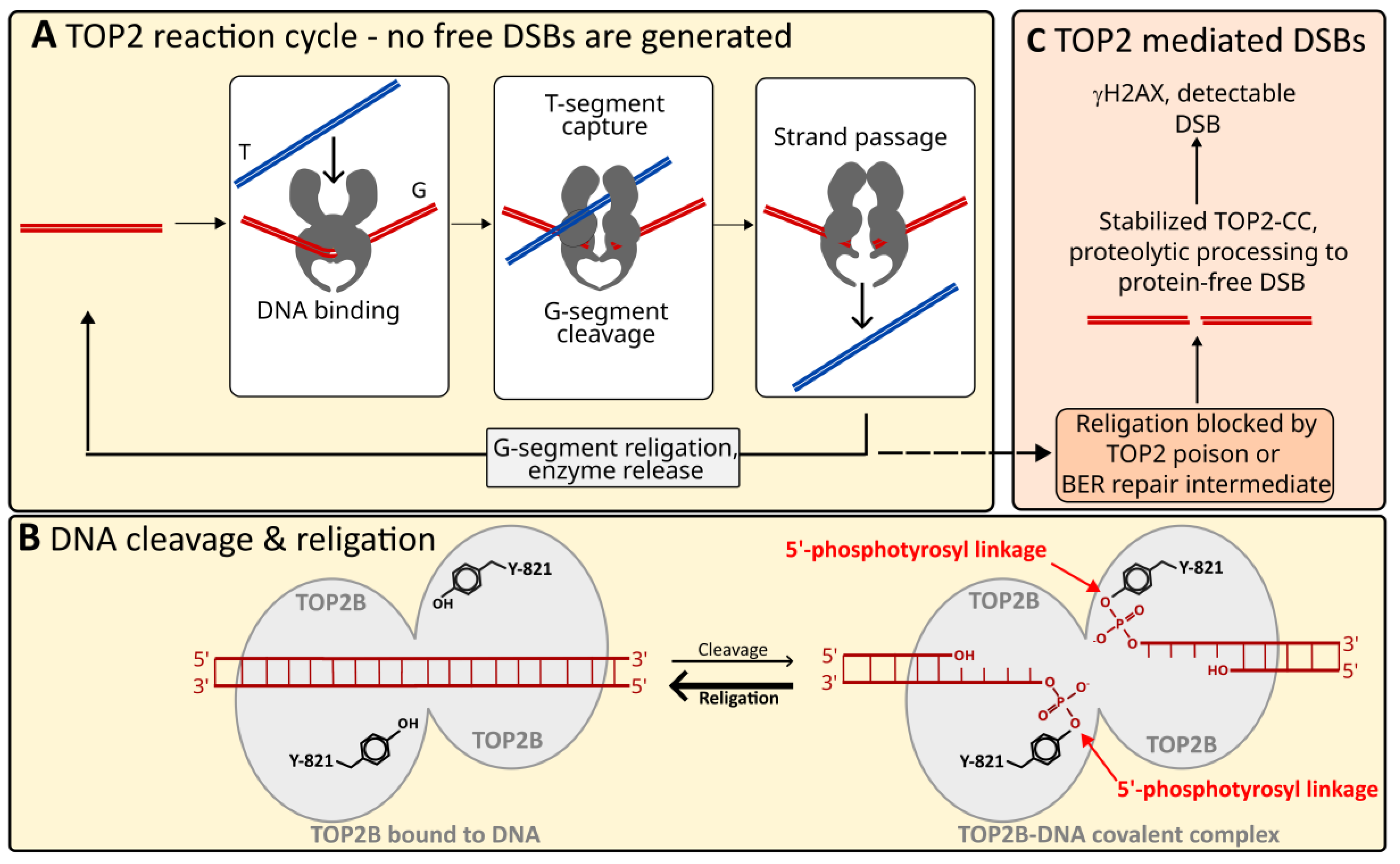
:1. Introduction
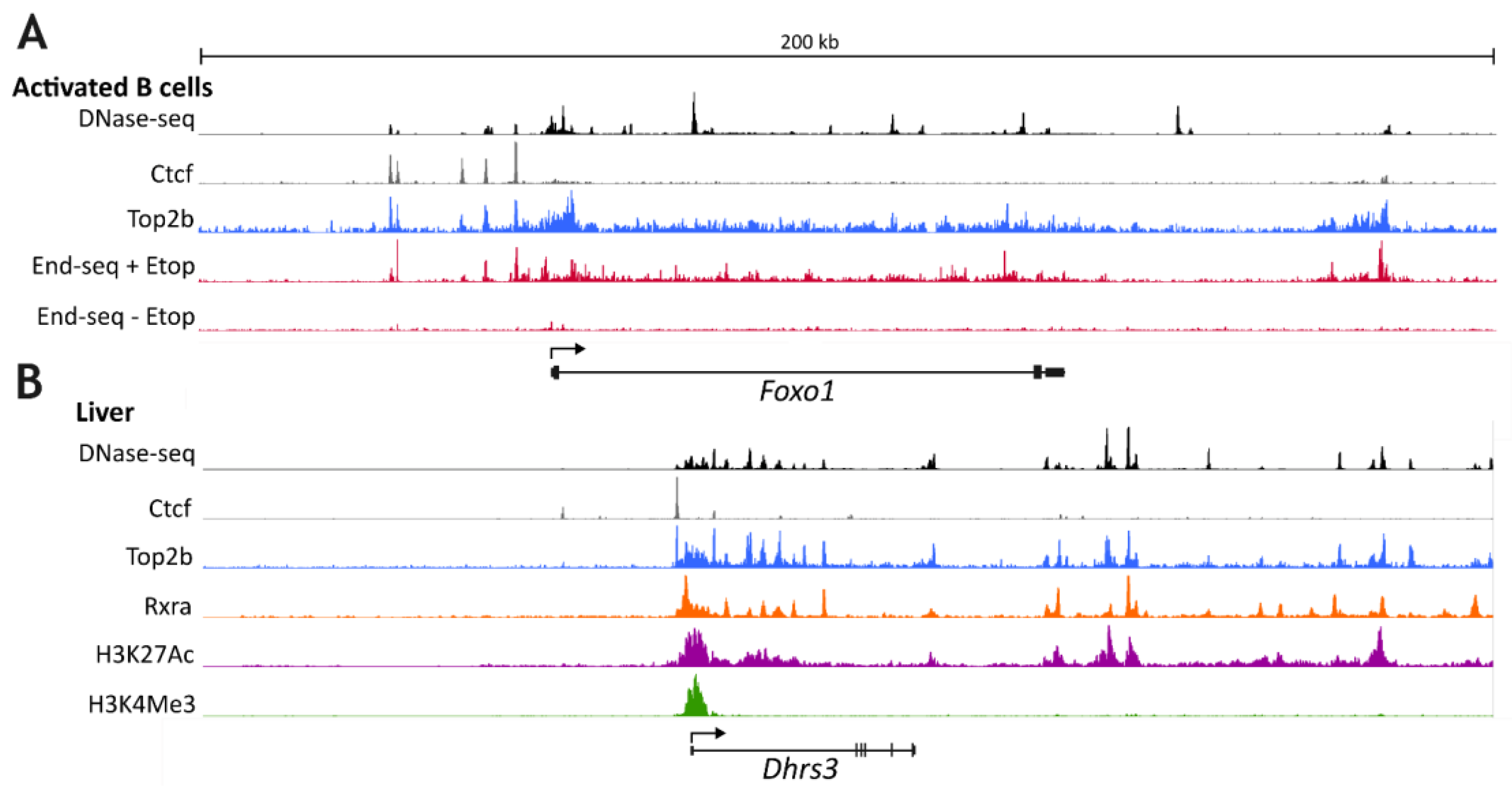
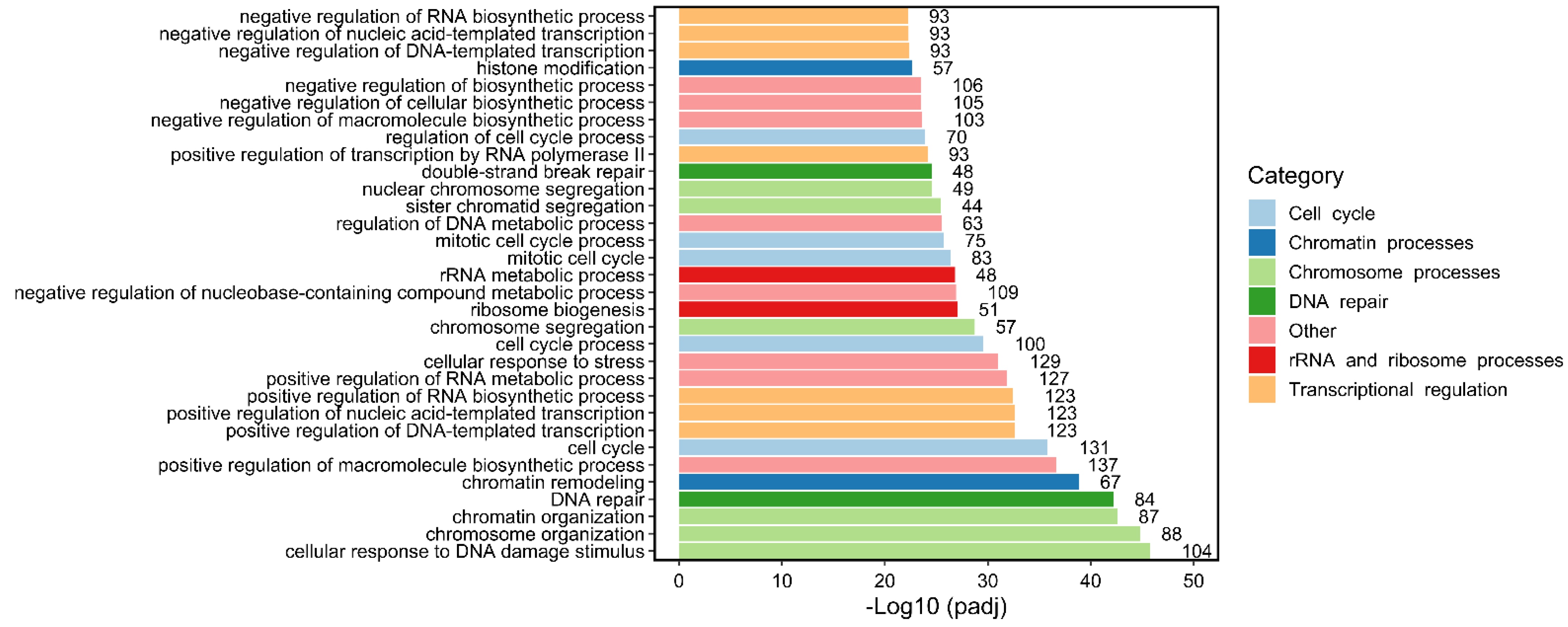
2. Specific Roles for TOP2B in Transcriptional Programs
3. TOP2B and Stimulus-Induced Gene Expression
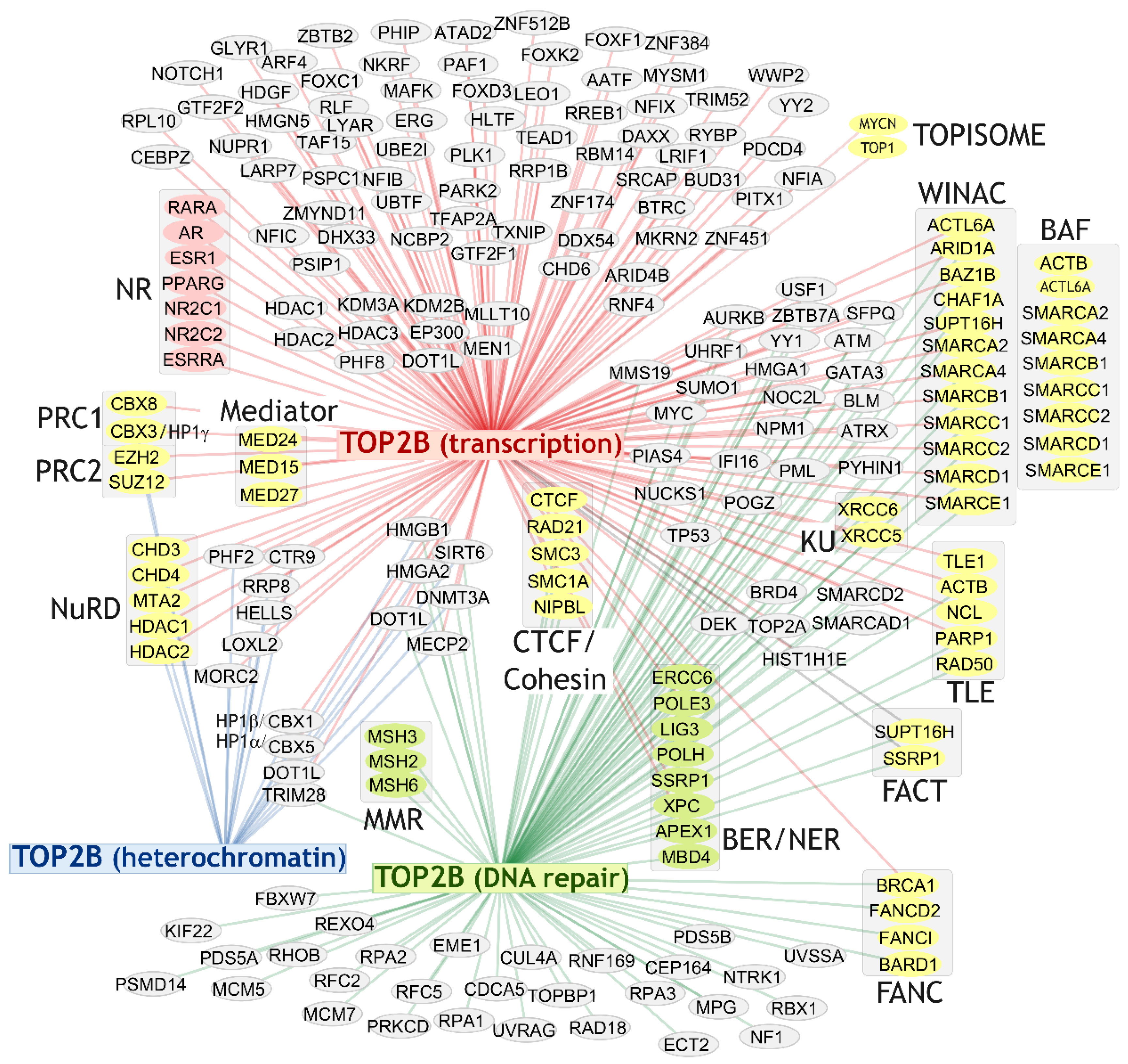
4. TOP2B Protein Interactors
4.1. TOP2B/DNA-PK/PARP Connection
4.2. Broad Range of Protein Interaction Partners
4.3. TOP2B Transcription Factor Associations
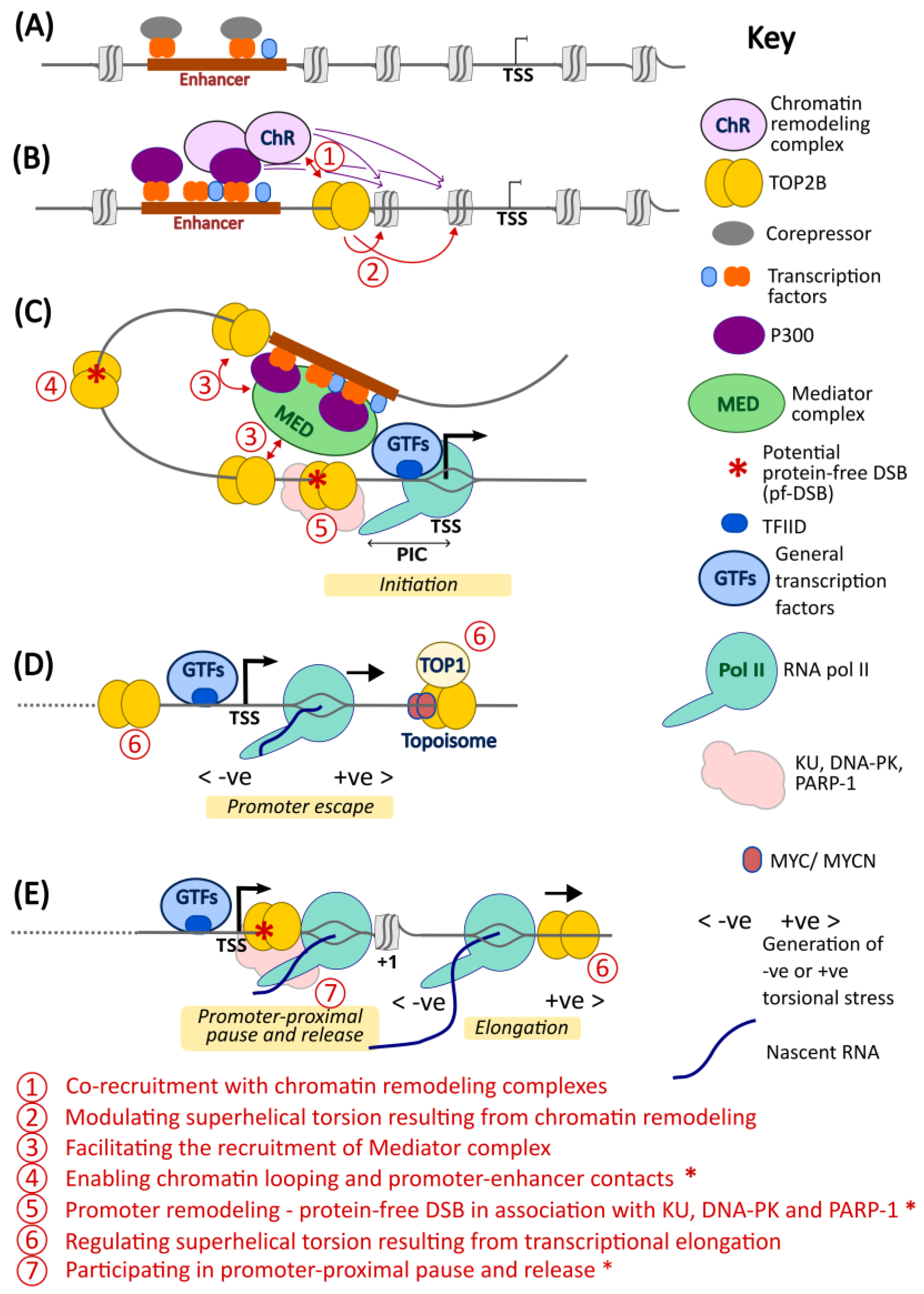
5. A Model for TOP2B Function in Transcription
6. Conclusions and Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pommier, Y.; Nussenzweig, A.; Takeda, S.; Austin, C. Human topoisomerases and their roles in genome stability and organization. Nat. Rev. Mol. Cell Biol. 2022, 23, 407–427. [Google Scholar] [CrossRef] [PubMed]
- Austin, C.A.; Cowell, I.G.; Khazeem, M.M.; Lok, D.; Ng, H.T. TOP2B’s contributions to transcription. Biochem. Soc. Trans. 2021, 49, 2483–2493. [Google Scholar] [CrossRef] [PubMed]
- Austin, C.A.; Lee, K.C.; Swan, R.L.; Khazeem, M.M.; Manville, C.M.; Cridland, P.; Treumann, A.; Porter, A.; Morris, N.J.; Cowell, I.G. TOP2B: The First Thirty Years. Int. J. Mol. Sci. 2018, 19, 2765. [Google Scholar] [CrossRef]
- Wilstermann, A.M.; Osheroff, N. Base excision repair intermediates as topoisomerase II poisons. J. Biol. Chem. 2001, 276, 46290–46296. [Google Scholar] [CrossRef] [PubMed]
- Swan, R.L.; Cowell, I.G.; Austin, C.A. Mechanisms to Repair Stalled Topoisomerase II-DNA Covalent Complexes. Mol. Pharmacol. 2022, 101, 24–32. [Google Scholar] [CrossRef] [PubMed]
- McKie, S.J.; Neuman, K.C.; Maxwell, A. DNA topoisomerases: Advances in understanding of cellular roles and multi-protein complexes via structure-function analysis. Bioessays 2021, 43, e2000286. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.F.; Wang, J.C. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA 1987, 84, 7024–7027. [Google Scholar] [CrossRef]
- Ma, J.; Wang, M.D. DNA supercoiling during transcription. Biophys. Rev. 2016, 8, 75–87. [Google Scholar] [CrossRef]
- Khazeem, M.M.; Casement, J.W.; Schlossmacher, G.; Kenneth, N.S.; Sumbung, N.K.; Chan, J.Y.T.; McGow, J.F.; Cowell, I.G.; Austin, C.A. TOP2B Is Required to Maintain the Adrenergic Neural Phenotype and for ATRA-Induced Differentiation of SH-SY5Y Neuroblastoma Cells. Mol. Neurobiol. 2022, 59, 5987–6008. [Google Scholar] [CrossRef]
- Kouzine, F.; Gupta, A.; Baranello, L.; Wojtowicz, D.; Ben-Aissa, K.; Liu, J.; Przytycka, T.M.; Levens, D. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat. Struct. Mol. Biol. 2013, 20, 396–403. [Google Scholar] [CrossRef]
- Joshi, R.S.; Pina, B.; Roca, J. Topoisomerase II is required for the production of long Pol II gene transcripts in yeast. Nucleic Acids Res. 2012, 40, 7907–7915. [Google Scholar] [CrossRef] [PubMed]
- Broderick, L.; Yost, S.; Li, D.; McGeough, M.D.; Booshehri, L.M.; Guaderrama, M.; Brydges, S.D.; Kucharova, K.; Patel, N.C.; Harr, M.; et al. Mutations in topoisomerase IIβ result in a B cell immunodeficiency. Nat. Commun. 2019, 10, 3644. [Google Scholar] [CrossRef] [PubMed]
- Broderick, L.; Clay, G.M.; Blum, R.H.; Liu, Y.; McVicar, R.; Papes, F.; Booshehri, L.M.; Cowell, I.G.; Austin, C.A.; Putnam, C.D.; et al. Disease-associated mutations in topoisomerase IIβ result in defective NK cells. J. Allergy Clin. Immunol. 2022, 149, 2171–2176.e3. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, W.; Prescott, E.D.; Burden, S.J.; Wang, J.C. DNA topoisomerase IIβ and neural development. Science 2000, 287, 131–134. [Google Scholar] [CrossRef] [PubMed]
- Ju, B.G.; Lunyak, V.V.; Perissi, V.; Garcia-Bassets, I.; Rose, D.W.; Glass, C.K.; Rosenfeld, M.G. A topoisomerase IIβ-mediated dsDNA break required for regulated transcription. Science 2006, 312, 1798–1802. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.H.F.; Chang, I.; Hudak, C.S.S.; Hyun, S.; Kwan, H.-Y.; Sul, H.S. A Role of DNA-PK for the Metabolic Gene Regulation in Response to Insulin. Cell 2009, 136, 1056–1072. [Google Scholar] [CrossRef] [PubMed]
- Madabhushi, R.; Kim, T.K. Emerging themes in neuronal activity-dependent gene expression. Mol. Cell. Neurosci. 2018, 87, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Perillo, B.; Ombra, M.N.; Bertoni, A.; Cuozzo, C.; Sacchetti, S.; Sasso, A.; Chiariotti, L.; Malorni, A.; Abbondanza, C.; Avvedimento, E.V. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science 2008, 319, 202–206. [Google Scholar] [CrossRef]
- McNamara, S.; Wang, H.; Hanna, N.; Miller, W.H. Topoisomerase IIβ Negatively Modulates Retinoic Acid Receptor α Function: A Novel Mechanism of Retinoic Acid Resistance. Mol. Cell. Biol. 2008, 28, 2066–2077. [Google Scholar] [CrossRef]
- Haffner, M.C.; Aryee, M.J.; Toubaji, A.; Esopi, D.M.; Albadine, R.; Gurel, B.; Isaacs, W.B.; Bova, G.S.; Liu, W.; Xu, J.; et al. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat. Genet. 2010, 42, 668–675. [Google Scholar] [CrossRef]
- Trotter, K.W.; King, H.A.; Archer, T.K. Glucocorticoid Receptor Transcriptional Activation via the BRG1-Dependent Recruitment of TOP2β and Ku70/86. Mol. Cell. Biol. 2015, 35, 2799–2817. [Google Scholar] [CrossRef]
- Uuskula-Reimand, L.; Hou, H.; Samavarchi-Tehrani, P.; Rudan, M.V.; Liang, M.; Medina-Rivera, A.; Mohammed, H.; Schmidt, D.; Schwalie, P.; Young, E.J.; et al. Topoisomerase II beta interacts with cohesin and CTCF at topological domain borders. Genome Biol. 2016, 17, 182. [Google Scholar] [CrossRef] [PubMed]
- Canela, A.; Maman, Y.; Jung, S.; Wong, N.; Callen, E.; Day, A.; Kieffer-Kwon, K.R.; Pekowska, A.; Zhang, H.; Rao, S.S.P.; et al. Genome Organization Drives Chromosome Fragility. Cell 2017, 170, 507–521.e518. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, A.J.; Porter, A.C. Construction, characterization, and complementation of a conditional-lethal DNA topoisomerase IIα mutant human cell line. Mol. Biol. Cell 2004, 15, 5700–5711. [Google Scholar] [CrossRef] [PubMed]
- Akimitsu, N.; Adachi, N.; Hirai, H.; Hossain, M.S.; Hamamoto, H.; Kobayashi, M.; Aratani, Y.; Koyama, H.; Sekimizu, K. Enforced cytokinesis without complete nuclear division in embryonic cells depleting the activity of DNA topoisomerase IIα. Genes Cells 2003, 8, 393–402. [Google Scholar] [CrossRef]
- King, I.F.; Yandava, C.N.; Mabb, A.M.; Hsiao, J.S.; Huang, H.S.; Pearson, B.L.; Calabrese, J.M.; Starmer, J.; Parker, J.S.; Magnuson, T.; et al. Topoisomerases facilitate transcription of long genes linked to autism. Nature 2013, 501, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Nevin, L.M.; Xiao, T.; Staub, W.; Baier, H. Topoisomerase IIβ is required for lamina-specific targeting of retinal ganglion cell axons and dendrites. Development 2011, 138, 2457–2465. [Google Scholar] [CrossRef] [PubMed]
- Tsutsui, K.; Tsutsui, K.; Sano, K.; Kikuchi, A.; Tokunaga, A. Involvement of DNA topoisomerase IIβ in neuronal differentiation. J. Biol. Chem. 2001, 276, 5769–5778. [Google Scholar] [CrossRef]
- Sano, K.; Miyaji-Yamaguchi, M.; Tsutsui, K.M.; Tsutsui, K. Topoisomerase IIβ activates a subset of neuronal genes that are repressed in AT-rich genomic environment. PLoS ONE 2008, 3, e4103. [Google Scholar] [CrossRef]
- Li, Y.; Hao, H.; Tzatzalos, E.; Lin, R.-K.; Doh, S.; Liu, L.F.; Lyu, Y.L.; Cai, L. Topoisomerase IIbeta is required for proper retinal development and survival of postmitotic cells. Biol. Open 2014, 3, 172–184. [Google Scholar] [CrossRef]
- Feng, W.; Kawauchi, D.; Korkel-Qu, H.; Deng, H.; Serger, E.; Sieber, L.; Lieberman, J.A.; Jimeno-Gonzalez, S.; Lambo, S.; Hanna, B.S.; et al. Chd7 is indispensable for mammalian brain development through activation of a neuronal differentiation programme. Nat. Commun. 2017, 8, 14758. [Google Scholar] [CrossRef]
- Lopes, I.; Altab, G.; Raina, P.; de Magalhaes, J.P. Gene Size Matters: An Analysis of Gene Length in the Human Genome. Front. Genet. 2021, 12, 559998. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, V.K.; Burger, L.; Nikoletopoulou, V.; Deogracias, R.; Thakurela, S.; Wirbelauer, C.; Kaut, J.; Terranova, R.; Hoerner, L.; Mielke, C.; et al. Target genes of Topoisomerase IIβ regulate neuronal survival and are defined by their chromatin state. Proc. Natl. Acad. Sci. USA 2012, 109, E934–E943. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Y.L.; Lin, C.P.; Azarova, A.M.; Cai, L.; Wang, J.C.; Liu, L.F. Role of topoisomerase IIβ in the expression of developmentally regulated genes. Mol. Cell Biol. 2006, 26, 7929–7941. [Google Scholar] [CrossRef] [PubMed]
- Madabhushi, R.; Gao, F.; Pfenning, A.R.; Pan, L.; Yamakawa, S.; Seo, J.; Rueda, R.; Phan, T.X.; Yamakawa, H.; Pao, P.C.; et al. Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell 2015, 161, 1592–1605. [Google Scholar] [CrossRef] [PubMed]
- The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Wang, J.; Zhang, T.; Liu, Z.; Liu, L.; Chen, Y.; Li, Z.; Zhao, Y.; Yu, Q.; Liu, T.; et al. Chromatin accessibility dynamics dictate renal tubular epithelial cell response to injury. Nat. Commun. 2022, 13, 7322. [Google Scholar] [CrossRef] [PubMed]
- Bunch, H. DNA Breaks and Damage Response Signaling are Coupled with RNA Polymerase II Promoter-Proximal Pause Release and Required for Effective Transcriptional Elongation. FASEB J. 2016, 30, 589.581. [Google Scholar]
- Bunch, H.; Lawney, B.P.; Lin, Y.-F.; Asaithamby, A.; Murshid, A.; Wang, Y.E.; Chen, B.P.C.; Calderwood, S.K. Transcriptional elongation requires DNA break-induced signalling. Nat. Commun. 2015, 6, 10191. [Google Scholar] [CrossRef]
- Rogakou, E.P.; Pilch, D.R.; Orr, A.H.; Ivanova, V.S.; Bonner, W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998, 273, 5858–5868. [Google Scholar] [CrossRef]
- Delint-Ramirez, I.; Konada, L.; Heady, L.; Rueda, R.; Jacome, A.S.V.; Marlin, E.; Marchioni, C.; Segev, A.; Kritskiy, O.; Yamakawa, S.; et al. Calcineurin dephosphorylates topoisomerase IIβ and regulates the formation of neuronal-activity-induced DNA breaks. Mol. Cell 2022, 82, 3794–3809.e8. [Google Scholar] [CrossRef] [PubMed]
- Core, L.; Adelman, K. Promoter-proximal pausing of RNA polymerase II: A nexus of gene regulation. Genes Dev. 2019, 33, 960–982. [Google Scholar] [CrossRef]
- Abbasi, S.; Schild-Poulter, C. Mapping the Ku Interactome Using Proximity-Dependent Biotin Identification in Human Cells. J. Proteome Res. 2019, 18, 1064–1077. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Szlachta, K.; Manukyan, A.; Raimer, H.M.; Dinda, M.; Bekiranov, S.; Wang, Y.H. Pausing sites of RNA polymerase II on actively transcribed genes are enriched in DNA double-stranded breaks. J. Biol. Chem. 2020, 295, 3990–4000. [Google Scholar] [CrossRef] [PubMed]
- Ju, B.G.; Solum, D.; Song, E.J.; Lee, K.J.; Rose, D.W.; Glass, C.K.; Rosenfeld, M.G. Activating the PARP-1 sensor component of the groucho/TLE1 corepressor complex mediates a CaMKinase IIδ-dependent neurogenic gene activation pathway. Cell 2004, 119, 815–829. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.B.; Ji, P.; Anish, R.; Jacobson, R.H.; Takada, S. Poly(ADP-ribose) Polymerase 1 Interacts with Nuclear Respiratory Factor 1 (NRF-1) and Plays a Role in NRF-1 Transcriptional Regulation. J. Biol. Chem. 2009, 284, 8621–8632. [Google Scholar] [CrossRef] [PubMed]
- Cho, N.H.; Cheveralls, K.C.; Brunner, A.D.; Kim, K.; Michaelis, A.C.; Raghavan, P.; Kobayashi, H.; Savy, L.; Li, J.Y.; Canaj, H.; et al. OpenCell: Endogenous tagging for the cartography of human cellular organization. Science 2022, 375, eabi6983. [Google Scholar] [CrossRef] [PubMed]
- Havugimana, P.C.; Goel, R.K.; Phanse, S.; Youssef, A.; Padhorny, D.; Kotelnikov, S.; Kozakov, D.; Emili, A. Scalable multiplex co-fractionation/mass spectrometry platform for accelerated protein interactome discovery. Nat. Commun. 2022, 13, 4043. [Google Scholar] [CrossRef]
- Buchel, G.; Carstensen, A.; Mak, K.Y.; Roeschert, I.; Leen, E.; Sumara, O.; Hofstetter, J.; Herold, S.; Kalb, J.; Baluapuri, A.; et al. Association with Aurora-A Controls N-MYC-Dependent Promoter Escape and Pause Release of RNA Polymerase II during the Cell Cycle. Cell Rep. 2017, 21, 3483–3497. [Google Scholar] [CrossRef]
- Das, S.K.; Kuzin, V.; Cameron, D.P.; Sanford, S.; Jha, R.K.; Nie, Z.; Rosello, M.T.; Holewinski, R.; Andresson, T.; Wisniewski, J.; et al. MYC assembles and stimulates topoisomerases 1 and 2 in a “topoisome”. Mol. Cell 2021, 82, 140–158.e12. [Google Scholar] [CrossRef]
- Sears, R.M.; May, D.G.; Roux, K.J. BioID as a Tool for Protein-Proximity Labeling in Living Cells. Methods Mol. Biol. 2019, 2012, 299–313. [Google Scholar] [CrossRef] [PubMed]
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.J.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021, 30, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Tsitsiridis, G.; Steinkamp, R.; Giurgiu, M.; Brauner, B.; Fobo, G.; Frishman, G.; Montrone, C.; Ruepp, A. CORUM: The comprehensive resource of mammalian protein complexes—2022. Nucleic Acids Res. 2022, 51, D539–D545. [Google Scholar] [CrossRef] [PubMed]
- Malik, S.; Roeder, R.G. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 2010, 11, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Wan, C.; Borgeson, B.; Phanse, S.; Tu, F.; Drew, K.; Clark, G.; Xiong, X.; Kagan, O.; Kwan, J.; Bezginov, A.; et al. Panorama of ancient metazoan macromolecular complexes. Nature 2015, 525, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Hoffmeister, H.; Fuchs, A.; Erdel, F.; Pinz, S.; Gröbner-Ferreira, R.; Bruckmann, A.; Deutzmann, R.; Schwartz, U.; Maldonado, R.; Huber, C.; et al. CHD3 and CHD4 form distinct NuRD complexes with different yet overlapping functionality. Nucleic Acids Res. 2017, 45, 10534–10554. [Google Scholar] [CrossRef]
- Tsai, S.C.; Valkov, N.; Yang, W.M.; Gump, J.; Sullivan, D.; Seto, E. Histone deacetylase interacts directly with DNA topoisomerase II. Nat. Genet. 2000, 26, 349–353. [Google Scholar] [CrossRef]
- Johnson, C.A.; Padget, K.; Austin, C.A.; Turner, B.M. Deacetylase activity associates with topoisomerase II and is necessary for etoposide-induced apoptosis. J. Biol. Chem. 2001, 276, 4539–4542. [Google Scholar] [CrossRef]
- Mathelier, A.; Zhao, X.; Zhang, A.W.; Parcy, F.; Worsley-Hunt, R.; Arenillas, D.J.; Buchman, S.; Chen, C.-y.; Chou, A.; Ienasescu, H.; et al. JASPAR 2014: An extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2013, 42, D142–D147. [Google Scholar] [CrossRef]
- Manville, C.M.; Smith, K.; Sondka, Z.; Rance, H.; Cockell, S.; Cowell, I.G.; Lee, K.C.; Morris, N.J.; Padget, K.; Jackson, G.H.; et al. Genome-wide ChIP-seq analysis of human TOP2B occupancy in MCF7 breast cancer epithelial cells. Biol. Open 2015, 4, 1436–1447. [Google Scholar] [CrossRef]
- Canela, A.; Maman, Y.; Huang, S.N.; Wutz, G.; Tang, W.; Zagnoli-Vieira, G.; Callen, E.; Wong, N.; Day, A.; Peters, J.M.; et al. Topoisomerase II-Induced Chromosome Breakage and Translocation Is Determined by Chromosome Architecture and Transcriptional Activity. Mol. Cell 2019, 75, 252–266.e8. [Google Scholar] [CrossRef] [PubMed]
- Castro-Mondragon, J.A.; Riudavets-Puig, R.; Rauluseviciute, I.; Berhanu Lemma, R.; Turchi, L.; Blanc-Mathieu, R.; Lucas, J.; Boddie, P.; Khan, A.; Manosalva Pérez, N.; et al. JASPAR 2022: The 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2021, 50, D165–D173. [Google Scholar] [CrossRef] [PubMed]
- Gavin, I.; Horn, P.J.; Peterson, C.L. SWI/SNF chromatin remodeling requires changes in DNA topology. Mol. Cell 2001, 7, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Jha, R.K.; Levens, D.; Kouzine, F. Mechanical determinants of chromatin topology and gene expression. Nucleus 2022, 13, 94–115. [Google Scholar] [CrossRef] [PubMed]
- Kagey, M.H.; Newman, J.J.; Bilodeau, S.; Zhan, Y.; Orlando, D.A.; van Berkum, N.L.; Ebmeier, C.C.; Goossens, J.; Rahl, P.B.; Levine, S.S.; et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 2010, 467, 430–435. [Google Scholar] [CrossRef] [PubMed]
- Dellino, G.I.; Palluzzi, F.; Chiariello, A.M.; Piccioni, R.; Bianco, S.; Furia, L.; Conti, G.D.; Bouwman, B.A.M.; Melloni, G.; Guido, D.; et al. Release of paused RNA polymerase II at specific loci favors DNA double-strand-break formation and promotes cancer translocations. Nat. Genet. 2019, 51, 1011–1023. [Google Scholar] [CrossRef] [PubMed]
- Iacovoni, J.S.; Caron, P.; Lassadi, I.; Nicolas, E.; Massip, L.; Trouche, D.; Legube, G. High-resolution profiling of γH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010, 29, 1446–1457. [Google Scholar] [CrossRef]
- Caron, P.; van der Linden, J.; van Attikum, H. Bon voyage: A transcriptional journey around DNA breaks. DNA Repair 2019, 82, 102686. [Google Scholar] [CrossRef]
- Sengupta, S.; Wang, H.; Yang, C.; Szczesny, B.; Hegde, M.L.; Mitra, S. Ligand-induced gene activation is associated with oxidative genome damage whose repair is required for transcription. Proc. Natl. Acad. Sci. USA 2020, 117, 22183–22192. [Google Scholar] [CrossRef]
- Pankotai, T.; Bonhomme, C.; Chen, D.; Soutoglou, E. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat. Struct. Mol. Biol. 2012, 19, 276–282. [Google Scholar] [CrossRef]
| WINAC/WSTF/ISWI Complex (CORUM:1230)—ATP Dependent Chromatin Remodeling | ||
| Component | Synonyms | Reference |
| ACTL6A | BAF53A, INO80K, Arp4, ACTL6 | [48] |
| ARID1A | BAF250A, SMARCF1, | [48] |
| BAZ1B | WSTF (n.b. H2AX Tyr142-kinase) | [48] |
| CHAF1A | CAF1P150 | [22,48] |
| SMARCA2 | BRM, SNF2L2, STH1P, BAF190, SNF2 | [48] |
| SMARCA4 | BRG1, BAF190A, SNF2L4 | [21,48] |
| SMARCB1 | INI1, BAF47, SNF5L1, SWNTS1 | [48] |
| SMARCC1 | Rsc8, CRACC1, SWI3, BAF155, SRG3 | [48] |
| SMARCC2 | CRACC2, BAF170 | [48] |
| SMARCD1 | Rsc6p, BAF60A, CRACD1 | [48] |
| SMARCE1 | BAF57, CSS5 | [48] |
| SUPT16H | CDC68, SPT16, FACTP140 | [22,56] |
| BAF Complex (CORUM:18)—ATP Dependent Chromatin Remodeling | ||
| ACTB | β-actin | |
| ACTL6A | BAF53A, INO80K, Arp4 | |
| ARID1A | BAF250A, SMARCF1, | [48] |
| SMARCA2 | BRM, SNF2L2, STH1P, BAF190, SNF2 | [48] |
| SMARCA4 | BRG1, BAF190A, SNF2L4 | [21,48] |
| SMARCB1 | INI1, BAF47, SNF5L1, SWNTS1 | [48] |
| SMARCC1 | Rsc8, CRACC1, SWI3, BAF155, SRG3 | [48] |
| SMARCC2 | CRACC2, BAF170 | [48] |
| SMARCD1 | Rsc6p, BAF60A, CRACD1 | [48] |
| SMARCE1 | BAF57, CSS5 | [48] |
| FACT Complex (CORUM:936)—Histone Chaperone | ||
| SUPT16H | CDC68, SPT16, FACTP140 | [22,56] |
| SSRP1 | FACT, FACT80, T160 | [22,47] |
| NuRD Complex (CORUM: 587)—Chromatin Remodeling and Histone Deacetylation | ||
| CHD3 | MI-2A | [57] |
| CHD4 | MI-2B | [57] |
| MTA2 | PID | [58] |
| HDAC1 | [58,59] | |
| HDAC2 | [58,59] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cowell, I.G.; Casement, J.W.; Austin, C.A. To Break or Not to Break: The Role of TOP2B in Transcription. Int. J. Mol. Sci. 2023, 24, 14806. https://doi.org/10.3390/ijms241914806
Cowell IG, Casement JW, Austin CA. To Break or Not to Break: The Role of TOP2B in Transcription. International Journal of Molecular Sciences. 2023; 24(19):14806. https://doi.org/10.3390/ijms241914806
Chicago/Turabian StyleCowell, Ian G., John W. Casement, and Caroline A. Austin. 2023. "To Break or Not to Break: The Role of TOP2B in Transcription" International Journal of Molecular Sciences 24, no. 19: 14806. https://doi.org/10.3390/ijms241914806
APA StyleCowell, I. G., Casement, J. W., & Austin, C. A. (2023). To Break or Not to Break: The Role of TOP2B in Transcription. International Journal of Molecular Sciences, 24(19), 14806. https://doi.org/10.3390/ijms241914806






