The IFIH1/MDA5 rs1990760 Gene Variant (946Thr) Differentiates Early- vs. Late-Onset Skin Disease and Increases the Risk of Arthritis in a Spanish Cohort of Psoriasis
Abstract
1. Introduction
2. Results
3. Discussion
4. Methods
4.1. Study Population and Data Collection
4.2. IFIH1 Variants’ Genotyping
4.3. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Queiro, R.; Tejón, P.; Alonso, S.; Coto, P. Age at disease onset: A key factor for understanding psoriatic disease. Rheumatology 2014, 53, 1178–1185. [Google Scholar] [CrossRef] [PubMed]
- Harden, J.L.; Krueger, J.G.; Bowcock, A.M. The immunogenetics of Psoriasis: A comprehensive review. J. Autoimmun. 2015, 64, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Oka, A.; Mabuchi, T.; Ozawa, A.; Inoko, H. Current understanding of human genetics and genetic analysis of psoriasis. J. Dermatol. 2012, 39, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Nair, R.P.; Stuart, P.E.; Nistor, I.; Hiremagalore, R.; Chia, N.V.C.; Jenisch, S.; Weichenthal, M.; Abecasis, G.R.; Lim, H.W.; Christophers, M.; et al. Sequence and haplotype analysis supports HLA-C as the psoriasis susceptibility 1 gene. Am. J. Hum. Genet. 2006, 78, 827–851. [Google Scholar] [CrossRef]
- Gudjonsson, J.E.; Karason, A.; Runarsdottir, E.H.; Antonsdottir, A.; Hauksson, V.; Jónsson, H.; Gulcher, J.; Stefansson, K.; Valdimarsson, H. Distinct clinical differences between HLA-Cw*0602 positive and negative psoriasis patients—An analysis of 1019 HLA-C- and HLA-B-typed patients. J. Investig. Dermatol. 2006, 126, 740–745. [Google Scholar] [CrossRef] [PubMed]
- Jordan, C.T.; Cao, L.; Roberson, E.D.; Duan, S.; Helms, C.; Nair, R.; Duffin, K.; Stuart, P.E.; Goldgar, D.; Hayashi, G.; et al. Rare and common variants in CARD14, encoding an epidermal regulator of NF-kappaB, in psoriasis. Am. J. Hum. Genet. 2012, 90, 796–808. [Google Scholar] [CrossRef] [PubMed]
- Nair, R.P.; Duffin, K.C.; Helms, C.; Ding, J.; Stuart, P.E.; Goldgar, D.; Gudjonsson, J.E.; Li, Y.; Tejasvi, T.; Feng, B.-J.; et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat. Genet. 2009, 41, 199–204. [Google Scholar] [CrossRef]
- Nikamo, P.; Lysell, J.; Ståhle, M. Association with Genetic Variants in the IL-23 and NF-κB Pathways Discriminates between Mild and Severe Psoriasis Skin Disease. J. Investig. Dermatol. 2015, 135, 1969–1976. [Google Scholar] [CrossRef]
- González-Lara, L.; Coto-Segura, P.; Penedo, A.; Eiris, N.; Díaz, M.; Santos-Juanes, J.; Queiro, R.; Coto, E. SNP rs11652075 in the CARD14 gene as a risk factor for psoriasis (PSORS2) in a Spanish cohort. DNA Cell Biol. 2013, 32, 601–604. [Google Scholar] [CrossRef]
- Dias Junior, A.G.; Sampaio, N.G.; Rehwinkel, J. A Balancing Act: MDA5 in Antiviral Immunity and Autoinflammation. Trends Microbiol. 2019, 27, 75–85. [Google Scholar] [CrossRef]
- Sampaio, N.G.; Chauveau, L.; Hertzog, J.; Bridgeman, A.; Fowler, G.; Moonen, J.P.; Dupont, M.; Russell, R.A.; Noerenberg, M.; Rehwinkel, J. The RNA sensor MDA5 detects SARS-CoV-2 infection. Sci. Rep. 2021, 11, 13638. [Google Scholar] [CrossRef] [PubMed]
- Thorne, L.G.; Reuschl, A.; Zuliani-Alvarez, L.; Whelan, M.V.X.; Turner, J.; Noursadeghi, M.; Jolly, C.; Towers, G.J. SARS-CoV-2 sensing by RIG-I and MDA5 links epithelial infection to macrophage inflammation. EMBO J. 2021, 40, e107826. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Riva, L.; Pu, Y.; Martin-Sancho, L.; Kanamune, J.; Yamamoto, Y.; Sakai, K.; Gotoh, S.; Miorin, L.; De Jesus, P.D.; et al. MDA5 governs the Innate Immune Response to SARS-CoV-2 in Lung Epithelial Cells. Cell Rep. 2021, 34, 108628. [Google Scholar] [CrossRef]
- Reikine, S.; Nguyen, J.B.; Modis, Y. Pattern Recognition and Signaling Mechanisms of RIG-I and MDA5. Front. Immunol. 2014, 5, 342. [Google Scholar] [CrossRef] [PubMed]
- Vabret, N.; Blander, J.M. Sensing microbial RNA in the cytosol. Front. Immunol. 2013, 4, 468. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Richman, L.; Morehouse, C.; de los Reyes, M.; Higgs, B.W.; Boutrin, A.; White, B.; Coyle, A.; Krueger, J.; Kiener, P.A.; et al. Type I interferon: Potential therapeutic target for psoriasis? PLoS ONE 2008, 3, e2737. [Google Scholar] [CrossRef] [PubMed]
- Van der Fits, L.; van der Wel, L.I.; Laman, J.D.; Prens, E.P.; Verschuren, M.C. In psoriasis lesional skin the type I interferon signaling pathway is activated, whereas interferon-alpha sensitivity is unaltered. J. Investig. Dermatol. 2004, 122, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Schmid, P.; Itin, P.; Cox, D.; McMaster, G.K.; Horisberger, M.A. The type I interferon system is locally activated in psoriatic lesions. J. Interferon Res. 1994, 14, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.-K.; Jain, D.; Sánchez, M.D.; Koziol-White, C.J.; Matthews, K.; Ge, M.Q.; Haczku, A.; Panettieri, R.A.; Frieman, M.B.; López, C.B. Deficiency of melanoma differentiation-associated protein 5 results in exacerbated chronic postviral lung inflammation. Am. J. Respir. Crit. Care Med. 2014, 189, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Asgari, S.; Schlapbach, L.J.; Anchisi, S.; Hammer, C.; Bartha, I.; Junier, T.; Mottet-Osman, G.; Posfay-Barbe, K.M.; Longchamp, D.; Stocker, M.; et al. Severe viral respiratory infections in children with IFIH1 loss-of-function mutations. Proc. Natl. Acad. Sci. USA 2017, 114, 8342–8347. [Google Scholar] [CrossRef]
- Lamborn, I.T.; Jing, H.; Zhang, Y.; Drutman, S.B.; Abbott, J.K.; Munir, S.; Bade, S.; Murdock, H.M.; Santos, C.P.; Brock, L.G.; et al. Recurrent rhinovirus infections in a child with inherited MDA5 deficiency. J. Exp. Med. 2017, 214, 1949–1972. [Google Scholar] [CrossRef] [PubMed]
- Rebendenne, A.; Valadão, A.L.C.; Tauziet, M.; Maarifi, G.; Bonaventure, B.; McKellar, J.; Planès, R.; Nisole, S.; Arnaud-Arnould, M.; Moncorgé, O.; et al. SARS-CoV-2 Triggers an MDA-5-Dependent Interferon Response Which Is Unable to Control Replication in Lung Epithelial Cells. J. Virol. 2021, 95, e02415–e02420. [Google Scholar] [CrossRef] [PubMed]
- Amado-Rodríguez, L.; del Riego, E.S.; de Ona, J.G.; Alonso, I.L.; Gil-Pena, H.; López-Martínez, C.; Martín-Vicente, P.; Lopez-Vazquez, A.; Lopez, A.G.; Cuesta-Llavona, E.; et al. Effects of IFIH1 Rs1990760 Variants on Systemic Inflammation and Outcome in Critically Ill COVID-19 Patients in an Observational Translational Study. eLife 2022, 11, e73012. [Google Scholar] [CrossRef] [PubMed]
- Rice, G.I.; Duany, Y.d.T.; Jenkinson, E.M.; Forte, G.M.A.; Anderson, B.H.; Ariaudo, G.; Bader-Meunier, B.; Baildam, E.M.; Battini, R.; Beresford, M.W.; et al. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat. Genet. 2014, 46, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Rice, G.I.; Park, S.; Gavazzi, F.; Adang, L.A.; Ayuk, L.A.; Van Eyck, L.; Seabra, L.; Barrea, C.; Battini, R.; Belot, A.; et al. Genetic and phenotypic spectrum associated with IFIH1gain-of-function. Hum. Mutat. 2020, 41, 837–849. [Google Scholar] [CrossRef]
- Tonutti, A.; Motta, F.; Ceribelli, A.; Isailovic, N.; Selmi, C.; De Santis, M. Anti-MDA5Antibody Linking COVID-19, Type I Interferon, and Autoimmunity: A Case Report and Systematic Literature Review. Front. Immunol. 2022, 13, 937667. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, H.; Matsuzaki, Y.; Kimura, K.; Nakano, H.; Imaizumi, T.; Satoh, K.; Hanada, K. Cytokine modulation of retinoic acid-inducible gene-I (RIG-I) expression in human epidermal keratinocytes. J. Dermatol. Sci. 2007, 45, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Prens, E.P.; Kant, M.; van Dijk, G.; van der Wel, L.I.; Mourits, S.; van der Fits, L. IFN-alpha enhances poly-IC responses in human keratinocytes by inducing expression of cytosolic innate RNA receptors: Relevance for psoriasis. J. Investig. Dermatol. 2008, 128, 932–938. [Google Scholar] [CrossRef] [PubMed]
- Rácz, E.; Prens, E.; Kant, M.; Florencia, E.; Jaspers, N.; Laman, J.; de Ridder, D.; van der Fits, L. Narrowband ultraviolet B inhibits innate cytosolic double-stranded RNA receptors in psoriatic skin and keratinocytes. Br. J. Dermatol. 2011, 164, 838–847. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liao, W.; Cargill, M.; Chang, M.; Matsunami, N.; Feng, B.-J.; Poon, A.; Callis-Duffin, K.P.; Catanese, J.J.; Bowcock, A.M.; et al. Carriers of rare missense variants in IFIH1 are protected from psoriasis. J. Investig. Dermatol. 2010, 130, 2768–2772. [Google Scholar] [CrossRef] [PubMed]
- Bijlmakers, M.J.; Kanneganti, S.K.; Barker, J.N.; Trembath, R.C.; Capon, F. Functional analysis of the RNF114 psoriasis susceptibility gene implicates innate immune responses to double-stranded RNA in disease pathogenesis. Hum. Mol. Genet. 2011, 20, 3129–3137. [Google Scholar] [CrossRef]
- Liu, S.; Wang, H.; Jin, Y.; Podolsky, R.; Reddy, M.P.L.; Pedersen, J.; Bode, B.; Reed, J.; Steed, D.; Anderson, S.; et al. IFIH1 Polymorphisms Are Significantly Associated with Type 1 Diabetes and IFIH1 Gene Expression in Peripheral Blood Mononuclear Cells. Hum. Mol. Genet. 2009, 18, 358–365. [Google Scholar] [CrossRef] [PubMed]
- Cen, H.; Wang, W.; Leng, R.-X.; Wang, T.-Y.; Pan, H.-F.; Fan, Y.-G.; Wang, B.; Ye, D.-Q. Association of IFIH1 Rs1990760 Polymorphism with Susceptibility to Autoimmune Diseases: A Meta-Analysis. Autoimmunity 2013, 46, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Gorman, J.A.; Hundhausen, C.; Errett, J.S.; Stone, A.E.; Allenspach, E.J.; Ge, Y.; Arkatkar, T.; Clough, C.; Dai, X.; Khim, S.; et al. The A946T Variant of the RNA Sensor IFIH1 Mediates an Interferon Program That Limits Viral Infection but Increases the Risk for Autoimmunity. Nat. Immunol. 2017, 18, 744–752. [Google Scholar] [CrossRef]
- Borysewicz-Sańczyk, H.; Sawicka, B.; Wawrusiewicz-Kurylonek, N.; Głowińska-Olszewska, B.; Kadłubiska, A.; Gościk, J.; Szadkowska, A.; Łosiewicz, A.; Młynarski, W.; Kretowski, A.; et al. Genetic Association Study of IL2RA, IFIH1, and CTLA-4 Polymorphisms with Autoimmune Thyroid Diseases and Type 1 Diabetes. Front. Pediatr. 2020, 8, 481. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, M.R.; Thirawatananond, P.; Peters, L.; Sharp, R.C.; Ogundare, S.; Posgai, A.L.; Perry, D.J.; Brusko, T.M. De-Coding Genetic Risk Variants in Type 1 Diabetes. Immunol. Cell Biol. 2021, 99, 496–508. [Google Scholar] [CrossRef]
- Funabiki, M.; Kato, H.; Miyachi, Y.; Toki, H.; Motegi, H.; Inoue, M.; Minowa, O.; Yoshida, A.; Deguchi, K.; Sato, H.; et al. Autoimmune disorders associated with gain of function of the intracellular sensor MDA5. Immunity 2014, 40, 199–212. [Google Scholar] [CrossRef]
- Bamming, D.; Horvath, C.M. Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I, and LGP2. J. Biol. Chem. 2009, 284, 9700–9712. [Google Scholar] [CrossRef] [PubMed]
- Chistiakov, D.A.; Voronova, N.V.; Savost’Anov, K.V.; Turakulov, R.I. Loss-of-Function Mutations E6 27X and I923V of IFIH1 Are Associated with Lower Poly(I: C)–Induced Interferon-β Production in Peripheral Blood Mononuclear Cells of Type 1 Diabetes Patients. Hum. Immunol. 2010, 71, 1128–1134. [Google Scholar] [CrossRef] [PubMed]
- Downes, K.; Pekalski, M.; Angus, K.L.; Hardy, M.; Nutland, S.; Smyth, D.J.; Walker, N.M.; Wallace, C.; Todd, J.A. Reduced expression of IFIH1 is protective for type 1 diabetes. PLoS ONE 2010, 5, e12646. [Google Scholar] [CrossRef] [PubMed]
- Forbester JL and Humphreys, I.R. Genetic Influences on Viral-Induced Cytokine Responses in the Lung. Mucosal Immunol. 2021, 14, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Nejentsev, S.; Walker, N.; Riches, D.; Egholm, M.; Todd, J.A. Rare Variants of IFIH1, a Gene Implicated in Antiviral Responses, Protect against Type 1 Diabetes. Science 2009, 324, 387–389. [Google Scholar] [CrossRef] [PubMed]
- Mine, K.; Yoshikai, Y.; Takahashi, H.; Mori, H.; Anzai, K.; Nagafuchi, S. Genetic Susceptibility of the Host in Virus-Induced Diabetes. Microorganisms 2020, 8, 1133. [Google Scholar] [CrossRef] [PubMed]
- Hébert, H.L.; Bowes, J.; Smith, R.L.; Flynn, E.; Parslew, R.; Alsharqi, A.; McHugh, N.J.; Barker, J.N.; Griffiths, C.E.; Barton, A.; et al. Identification of loci associated with late-onset psoriasis using dense genotyping of immune-related regions. Br. J. Dermatol. 2015, 172, 933–939. [Google Scholar] [CrossRef] [PubMed]
- Stuart, P.E.; Nair, R.P.; Tsoi, L.C.; Tejasvi, T.; Das, S.; Kang, H.M.; Ellinghaus, E.; Chandran, V.; Callis-Duffin, K.; Ike, R.; et al. Genome-wide Association Analysis of Psoriatic Arthritis and Cutaneous Psoriasis Reveals Differences in Their Genetic Architecture. Am. J. Hum. Genet. 2015, 97, 816–836. [Google Scholar] [CrossRef] [PubMed]
- Tsoi, L.C.; Spain, S.L.; Knight, J.; Ellinghaus, E.; Stuart, P.E.; Capon, F.; Ding, J.; Li, Y.; Tejasvi, T.; Gudjonsson, J.E.; et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat. Genet. 2012, 44, 1341–1348. [Google Scholar] [CrossRef] [PubMed]
- Strange, A.; Capon, F.; Spencer, C.C.; Knight, J.; Weale, M.E.; Allen, M.H. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat. Genet. 2010, 42, 985–990. [Google Scholar] [CrossRef] [PubMed]
- Patrick, M.T.; Stuart, P.E.; Raja, K.; Gudjonsson, J.E.; Tejasvi, T.; Yang, J.; Chandran, V.; Das, S.; Callis-Duffin, K.; Ellinghaus, E.; et al. Genetic signature to provide robust risk assessment of psoriatic arthritis development in psoriasis patients. Nat. Commun. 2018, 9, 4178. [Google Scholar] [CrossRef]
- Pang, L.; Gong, X.; Liu, N.; Xie, G.; Gao, W.; Kong, G.; Li, X.; Zhang, J.; Jin, Y.; Duan, Z. A polymorphism in melanoma differentiation-associated gene 5 may be a risk factor for enterovirus 71 infection. Clin. Microbiol. Infect. 2014, 20, O711–O717. [Google Scholar] [CrossRef]
- Yao, Y.; Shen, Y.; Shao, H.; Liu, Y.; Ji, Y.; Du, G.; Ye, X.; Huang, P.; Chen, H. Polymorphisms of RIG-I-like receptor influence HBV clearance in Chinese Han population. J. Med. Virol. 2021, 93, 4957–4965. [Google Scholar] [CrossRef]
- Domsgen, E.; Lind, K.; Kong, L.; Hühn, M.H.; Rasool, O.; van Kuppeveld, F.; Korsgren, O.; Lahesmaa, R.; Flodström-Tullberg, M. An IFIH1 gene polymorphism associated with risk for autoimmunity regulates canonical antiviral defence pathways in Coxsackievirus infected human pancreatic islets. Sci. Rep. 2016, 6, 39378. [Google Scholar] [CrossRef] [PubMed]
- Dou, Y.; Yim, H.C.; Kirkwood, C.D.; Williams, B.R.; Sadler, A.J. The innate immune receptor MDA5 limits rotavirus infection but promotes cell death and pancreatic inflammation. EMBO J. 2017, 36, 2742–2757. [Google Scholar] [CrossRef] [PubMed]
- Dieter, C.; de Almeida Brondani, L.; Lemos, N.E.; Schaeffer, A.F.; Zanotto, C.; Ramos, D.T.; Girardi, E.; Pellenz, F.M.; Camargo, J.L.; Moresco, K.S.; et al. Polymorphismsin ACE1, TMPRSS2, IFIH1, IFNAR2, and TYK2 Genes Are Associated with Worse Clinical Outcomes in COVID-19. Genes 2022, 14, 29. [Google Scholar] [CrossRef] [PubMed]
- Nln, I.; Fernandez-Ruiz, R.; Muskardin, T.L.W.; Paredes, J.L.; Blazer, A.D.; Tuminello, S.; Attur, M.; Iturrate, E.; Petrilli, C.M.; Abramson, S.B.; et al. Interferon pathway lupus risk alleles modulate risk of death from acute COVID-19. Transl. Res. 2022, 244, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Muñiz-Banciella, M.G.; Albaiceta, G.M.; Amado-Rodríguez, L.; Del Riego, E.S.; Alonso, I.L.; López-Martínez, C.; Martín-Vicente, P.; García-Clemente, M.; Hermida-Valverde, T.; Enríquez-Rodriguez, A.I.; et al. Age-dependent effect of the IFIH1/MDA5 gene variants on the risk of critical COVID-19. Immunogenetics 2023, 75, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Zurawek, M.; Fichna, M.; Fichna, P.; Skowronska, B.; Dzikiewicz-Krawczyk, A.; Januszkiewicz, D.; Nowak, J. Cumulative effect of IFIH1 variants and increased gene expression associated with type 1 diabetes. Diabetes Res. Clin. Pract. 2015, 107, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, H.M.; Pallas, K.; King, V.; Jamieson, T.; McKimmie, C.S.; Nibbs, R.J.B.; Carballido, J.M.; Jaritz, M.; Rot, A.; Graham, G.J. Microarray analyses demonstrate the involvement of type I interferons in psoriasiform pathology development in D6-deficient mice. J. Biol. Chem. 2013, 288, 36473–36483. [Google Scholar] [CrossRef]
- Nestle, F.O.; Conrad, C.; Tun-Kyi, A.; Homey, B.; Gombert, M.; Boyman, O.; Burg, G.; Liu, Y.-J.; Gilliet, M. Plasmacytoid predendritic cells initiate psoriasis through interferon-alpha production. J. Exp. Med. 2005, 202, 135–143. [Google Scholar] [CrossRef]
- Gregorio, J.; Meller, S.; Conrad, C.; Di Nardo, A.; Homey, B.; Lauerma, A.; Arai, N.; Gallo, R.L.; DiGiovanni, J.; Gilliet, M. Plasmacytoid dendritic cells sense skin injury and promote wound healing through type I interferons. J. Exp. Med. 2010, 207, 2921–2930. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Kane, S.; Chen, H.; Leon, A.; Levin, E.; Nguyen, T.; Debbaneh, M.; Millsop, J.W.; Gupta, R.; Huynh, M.; et al. The role of 39 psoriasis risk variants on age of psoriasis onset. ISRN Dermatol. 2013, 2013, 203941. [Google Scholar] [CrossRef]
- Reich, K.; Mössner, R.; König, I.R.; Westphal, G.; Ziegler, A.; Neumann, C. Promoter polymorphisms of the genes encoding tumor necrosis factor-alpha and interleukin-1 beta are associated with different subtypes of psoriasis characterized by early and late disease onset. J. Investig. Dermatol. 2002, 118, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Qin, Q.; Li, Y.; Zheng, X.; Chen, W.; Zhen, Q.; Li, B.; Wang, W.; Sun, L. Multifactor dimensionality reduction reveals the effect of interaction between ERAP1 and IFIH1 polymorphisms in psoriasis susceptibility genes. Front. Genet. 2022, 13, 1009589. [Google Scholar] [CrossRef] [PubMed]
- Budu-Aggrey, A.; Bowes, J.; Stuart, P.E.; Zawistowski, M.; Tsoi, L.C.; Nair, R.; Jadon, D.R.; McHugh, N.; Korendowych, E.; Elder, J.T.; et al. A rare coding allele in IFIH1 is protective for psoriatic arthritis. Ann. Rheum. Dis. 2017, 76, 1321–1324. [Google Scholar] [CrossRef] [PubMed]
- Bowes, J.; Budu-Aggrey, A.; Huffmeier, U.; Uebe, S.; Steel, K.; Hebert, H.L.; Wallace, C.; Massey, J.; Bruce, I.N.; Bluett, J.; et al. Dense genotyping of immune-related susceptibility loci reveals new insights into the genetics of psoriatic arthritis. Nat. Commun. 2015, 6, 6046. [Google Scholar] [CrossRef] [PubMed]
- Julià, A.; Tortosa, R.; Hernanz, J.M.; Cañete, J.D.; Fonseca, E.; Ferrándiz, C.; Unamuno, P.; Puig, L.; Fernández-Sueiro, J.L.; Sanmartí, R.; et al. Risk variants for psoriasis vulgaris in a large case-control collection and association with clinical subphenotypes. Hum. Mol. Genet. 2012, 21, 4549–4557. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Lou, F.; Yin, Q.; Gao, Y.; Sun, Y.; Bai, J.; Xu, Z.; Liu, Z.; Cai, W.; Ke, F.; et al. RIG-I antiviral signaling drives interleukin-23 production and psoriasis-like skin disease. EMBO Mol. Med. 2017, 9, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Yao, Z. Roles of Infection in Psoriasis. Int. J. Mol. Sci. 2022, 23, 6955. [Google Scholar] [CrossRef] [PubMed]
- Gladman, D.D.; Antoni, C.; Mease, P.; Clegg, D.O.; Nash, P. Discussion: Clinical features, epidemiology, classification criteria, and quality of life in psoriasis and psoriatic arthritis. Ann. Rheum. Dis. 2005, 64, ii24–ii25. [Google Scholar] [CrossRef]
- Ahnini, R.T.; Camp, N.J.; Cork, M.J.; Mee, J.B.; Keohane, S.G.; Duff, G.W.; di Giovine, F.S. Novel genetic association between the corneodesmosin (MHC S) gene and susceptibility to psoriasis. Hum. Mol. Genet. 1999, 8, 1135–1140. [Google Scholar] [CrossRef]
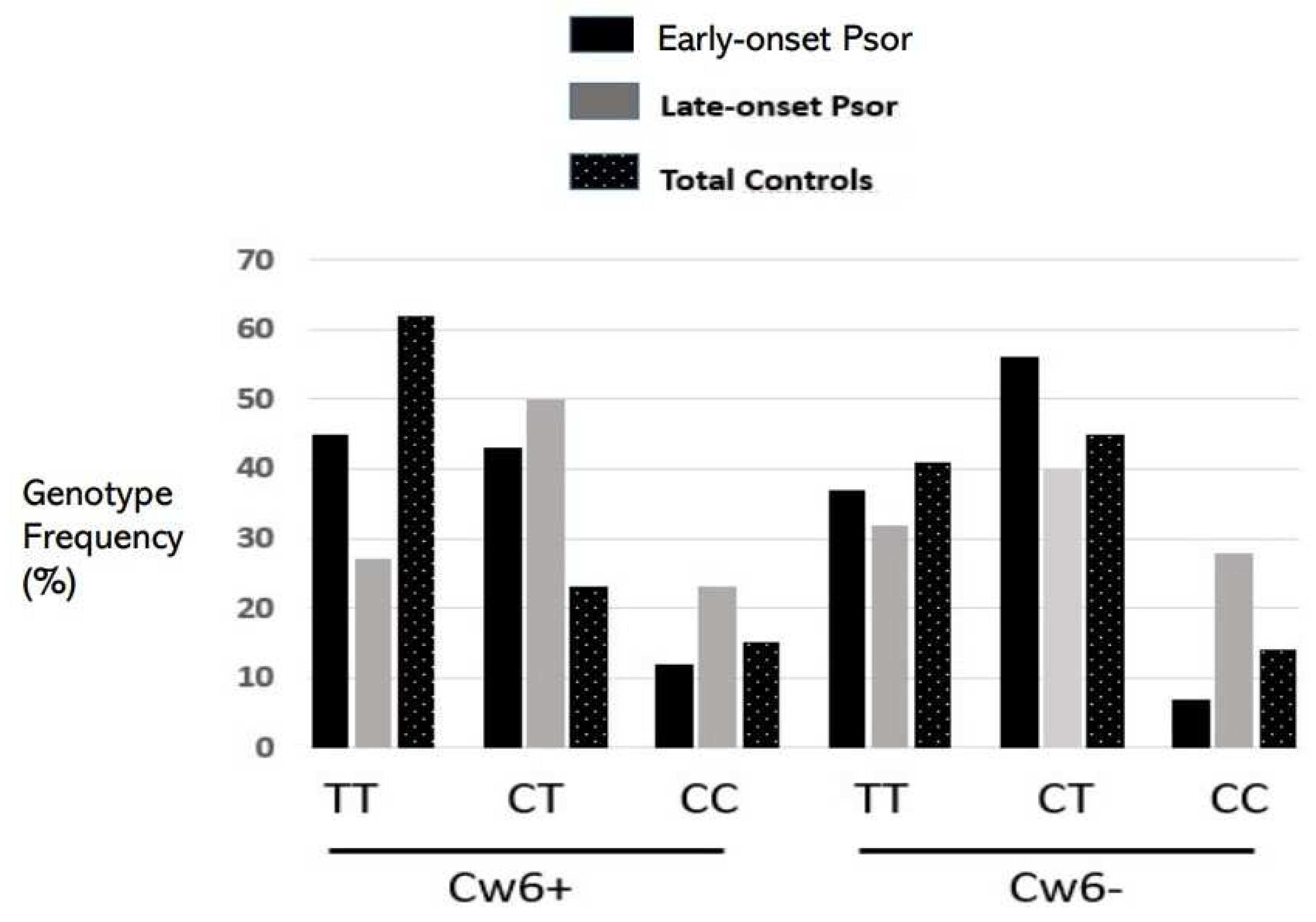
| Early Onset N = 440 | Late Onset N = 132 | p-Value Univariate | OR (95%CI) Multivariate | |
|---|---|---|---|---|
| Male | 232 (53%) | 77 (58%) | 0.26 | |
| Female | 208 (47%) | 55 (42%) | 1.24 (0.81–1.90) | |
| Onset age range | 18–40 | 41–78 | ||
| Median PASI (range) | 11 (1–75) | 6.1 (4.1–7.2) | ||
| Severe (PASI > 10) | 248 (56%) | 46 (35%) | <0.001 | 2.35 (1.52–3.66) |
| Arthritis yes | 139 (32%) | 32 (24%) | 0.11 | 1.22 (0.76–2.01) |
| Cw6*0602 | 211 (48%) | 30 (23%) | <0.001 | 3.25 (2.05–5.29) |
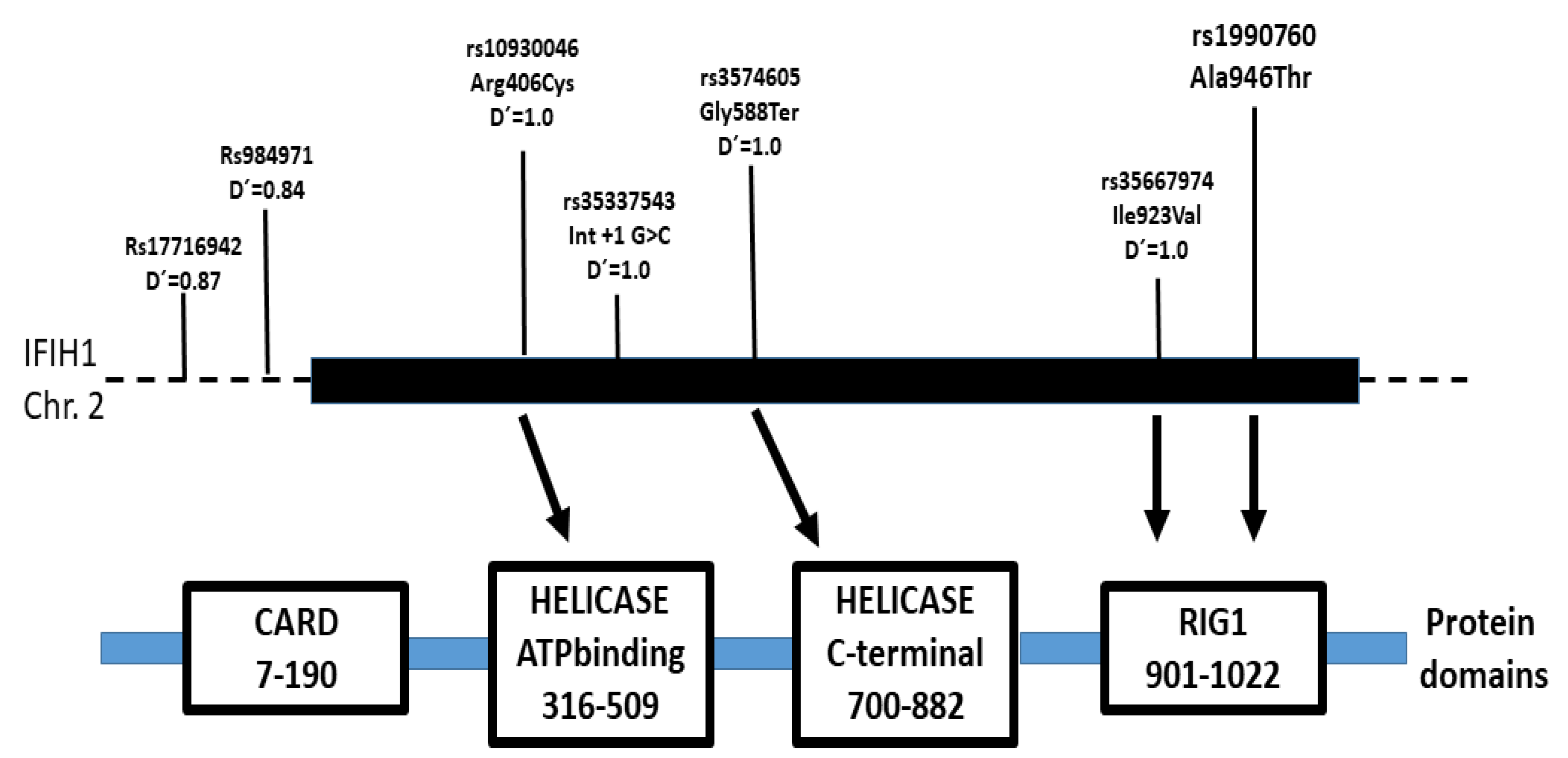
| rs1990760 C>T * (Ala946Thr) | ||||
| TT | 179 (41%) | 40 (30%) | <0.001 | 4.07 (2.37–7.04) |
| CT | 220 (50%) | 56 (42%) | ||
| CC | 41 (9%) | 36 (27%) | ||
| ALLELE T (946Thr) | 0.66 | 0.52 | ||
| rs35337543 int8 +1G>C | ||||
| GG | 430 (98%) | 128 (97%) | 0.62 | 1.12 (0.29–3.65) |
| GC | 10 (2%) | 4 (3%) | ||
| ALLELE G | 0.99 | 0.98 | ||
| rs35744605 C>A (Glu627Stop) | ||||
| CC | 432 (98%) | 129 (98%) | 0.74 | 1.11 (0.23–4.10) |
| CA | 8 (2%) | 3 (2%) | ||
| ALLELE C | 0.99 | 0.99 |
| Early-Onset Psoriasis N = 440 | Late-Onset Psoriasis N = 132 | |||||
|---|---|---|---|---|---|---|
| PsA Yes N = 139 | PsA No N = 301 | p-Value | PsA Yes N = 32 | PsA No N = 100 | p-Value | |
| Male | 65 (47%) | 167 (55%) | 0.09 | 16 (50%) | 61 (61%) | 0.27 |
| Female | 74 (53%) | 134 (45%) | 16 (50%) | 39 (39%) | ||
| Severe psoriasis | 95 (68%) | 153 (51%) | 0.001 | 15 (47%) | 31 (31%) | 0.08 |
| Cw6*0602 | 61 (44%) | 150 (50%) | 0.14 | 15 (47%) | 31 (31%) | 0.08 |
| rs1990760 | ||||||
| TT | 64 (46%) | 115 (38%) | 0.12 | 16 (50%) | 24 (24%) | 0.005 |
| CT | 61 (44%) | 159 (53%) | 12 (38%) | 44 (44%) | ||
| CC | 14 (10%) | 27 (9%) | 4 (12%) | 32 (32%) | ||
| Allele T | 0.68 | 0.65 | 0.69 | 0.46 | ||
| rs35337543 | ||||||
| GG | 139 (100%) | 291 (97%) | 0.02 | 30 (94%) | 98 (98%) | 0.24 |
| GC | 0 | 10 (3%) | 2 (6%) | 2 (2%) | ||
| Allele G | 1.0 | 0.99 | 0.88 | 0.99 | ||
| rs35744605 | ||||||
| CC | 137 (98%) | 295 (98%) | 0.38 | 31 (97%) | 98 (98%) | 0.57 |
| CA | 2 (2%) | 6 (2%) | 1 (3%) | 2 (2%) | ||
| Allele C | 0.99 | 0.99 | 0.98 | 0.99 | ||
| Patients ≤40 Years N = 440 | Controls ≤40 Years N = 100 | p-Value | Patients >40 Years N = 132 | Controls >40 Years N = 100 | p-Value | |
|---|---|---|---|---|---|---|
| Male | 232 (53%) | 55 (55%) | n.s. | 77 (58%) | 57 (57%) | n.s. |
| Female | 208 (47%) | 45 (45%) | 55 (42%) | 43 (43%) | ||
| Cw6*0602 | 211 (48%) | 7 (7%) | <0.001 | 30 (23%) | 6 (6%) | 0.001 |
| rs1990760 | ||||||
| TT | 179 (41%) | 42 (42%) | 40 (30%) | 43 (43%) | ||
| CT | 220 (50%) | 43 (43%) | 56 (42%) | 44 (44%) | ||
| CC | 41 (9%) | 15 (15%) | 36 (27%) | 13 (13%) | ||
| MAF C Eurs: 0.36–0.40 | 0.34 | 0.37 | n.s. | 0.48 | 0.35 | 0.005 * |
| rs35337543 | ||||||
| GG | 430 (98%) | 98 (98%) | 128 (97%) | 98 (98%) | ||
| GC | 10 (2%) | 2 (2%) | 4 (3%) | 2 (2%) | ||
| MAF C Eurs: 0.02 | 0.01 | 0.01 | n.s. | 0.02 | 0.01 | n.s. |
| rs35744605 | ||||||
| CC | 432 (98%) | 99 (99%) | 129 (98%) | 98 (98%) | ||
| CA | 8 (2%) | 1 (1%) | 3 (2%) | 2 (2%) | ||
| MAF A Eurs: 0.01 | 0.01 | 0.005 | n.s. | 0.01 | 0.01 | n.s. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coto-Segura, P.; Vázquez-Coto, D.; Velázquez-Cuervo, L.; García-Lago, C.; Coto, E.; Queiro, R. The IFIH1/MDA5 rs1990760 Gene Variant (946Thr) Differentiates Early- vs. Late-Onset Skin Disease and Increases the Risk of Arthritis in a Spanish Cohort of Psoriasis. Int. J. Mol. Sci. 2023, 24, 14803. https://doi.org/10.3390/ijms241914803
Coto-Segura P, Vázquez-Coto D, Velázquez-Cuervo L, García-Lago C, Coto E, Queiro R. The IFIH1/MDA5 rs1990760 Gene Variant (946Thr) Differentiates Early- vs. Late-Onset Skin Disease and Increases the Risk of Arthritis in a Spanish Cohort of Psoriasis. International Journal of Molecular Sciences. 2023; 24(19):14803. https://doi.org/10.3390/ijms241914803
Chicago/Turabian StyleCoto-Segura, Pablo, Daniel Vázquez-Coto, Lucinda Velázquez-Cuervo, Claudia García-Lago, Eliecer Coto, and Rubén Queiro. 2023. "The IFIH1/MDA5 rs1990760 Gene Variant (946Thr) Differentiates Early- vs. Late-Onset Skin Disease and Increases the Risk of Arthritis in a Spanish Cohort of Psoriasis" International Journal of Molecular Sciences 24, no. 19: 14803. https://doi.org/10.3390/ijms241914803
APA StyleCoto-Segura, P., Vázquez-Coto, D., Velázquez-Cuervo, L., García-Lago, C., Coto, E., & Queiro, R. (2023). The IFIH1/MDA5 rs1990760 Gene Variant (946Thr) Differentiates Early- vs. Late-Onset Skin Disease and Increases the Risk of Arthritis in a Spanish Cohort of Psoriasis. International Journal of Molecular Sciences, 24(19), 14803. https://doi.org/10.3390/ijms241914803







