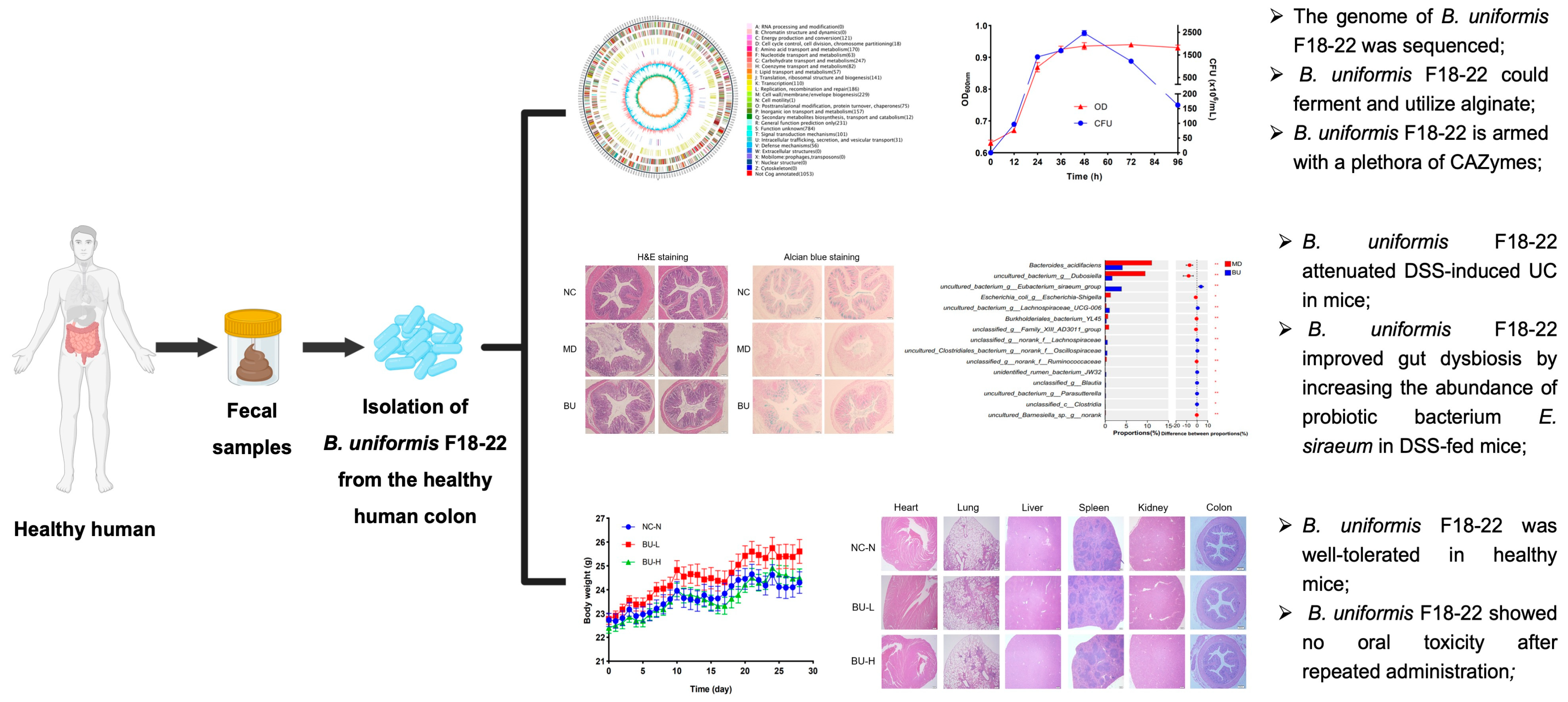
Discovery of Bacteroides uniformis F18-22 as a Safe and Novel Probiotic Bacterium for the Treatment of Ulcerative Colitis from the Healthy Human Colon
Abstract
:1. Introduction
2. Results
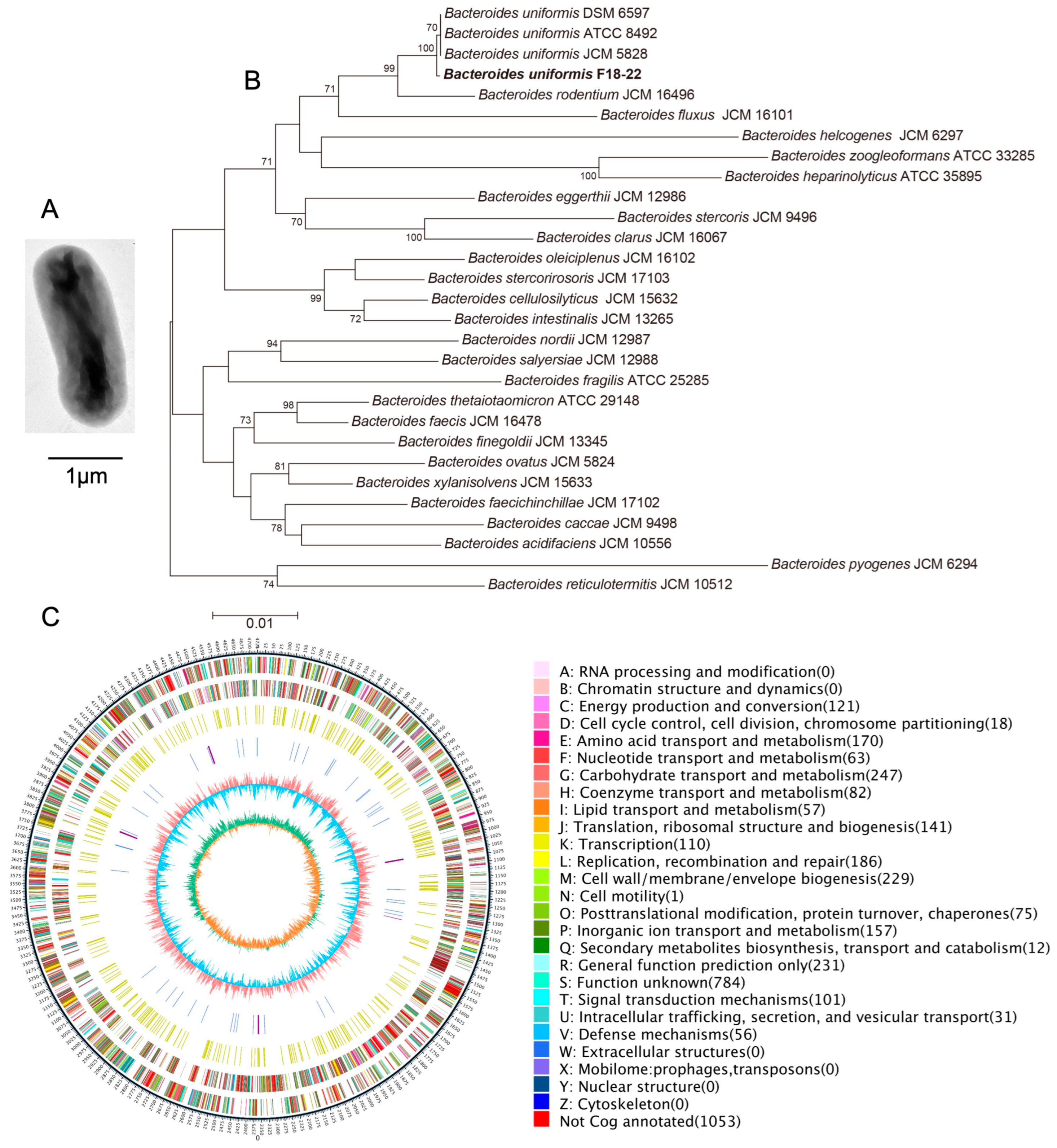
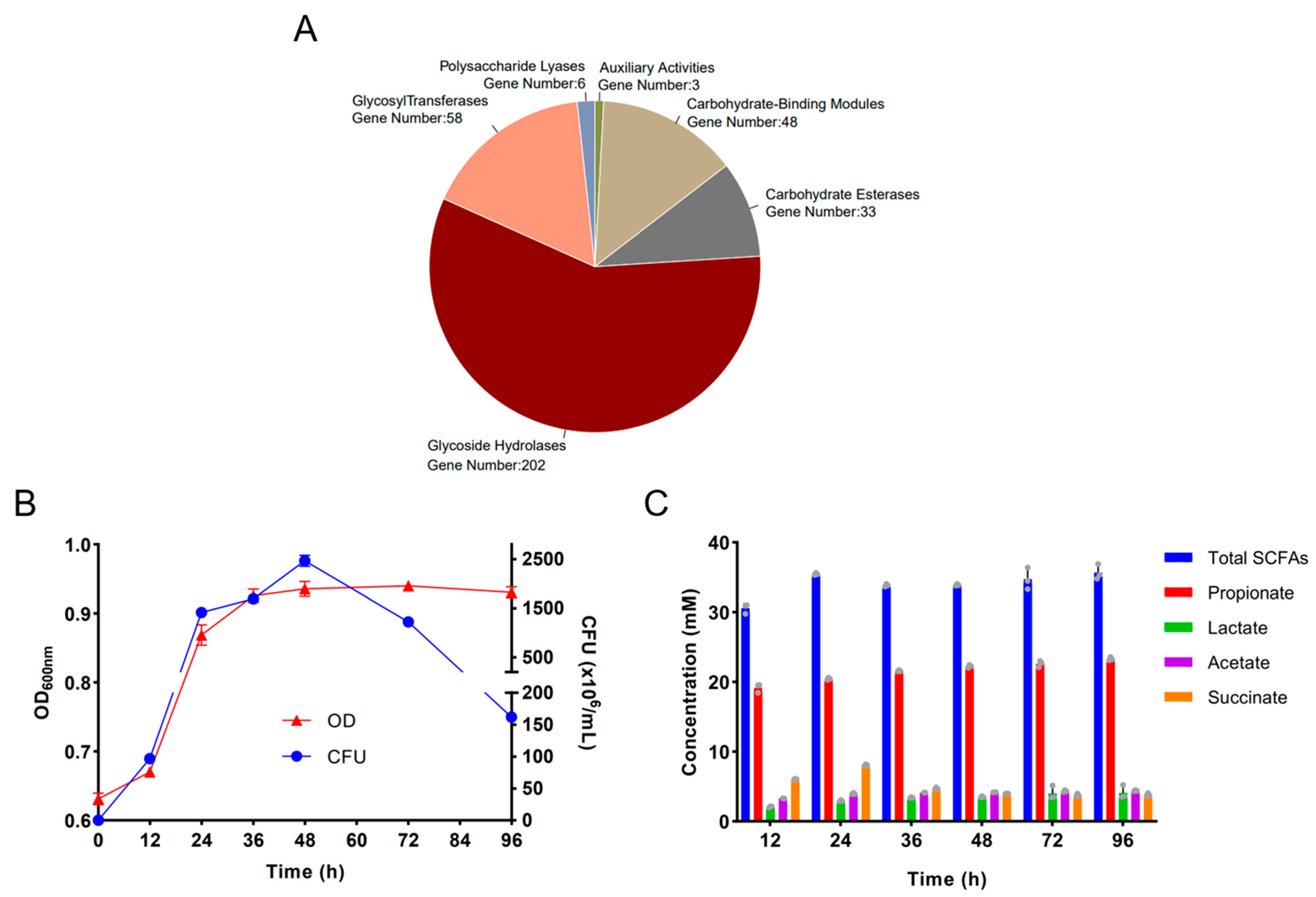
2.1. Genomic Analysis of the Alginate-Fermenting Bacterium, B. uniformis F18-22
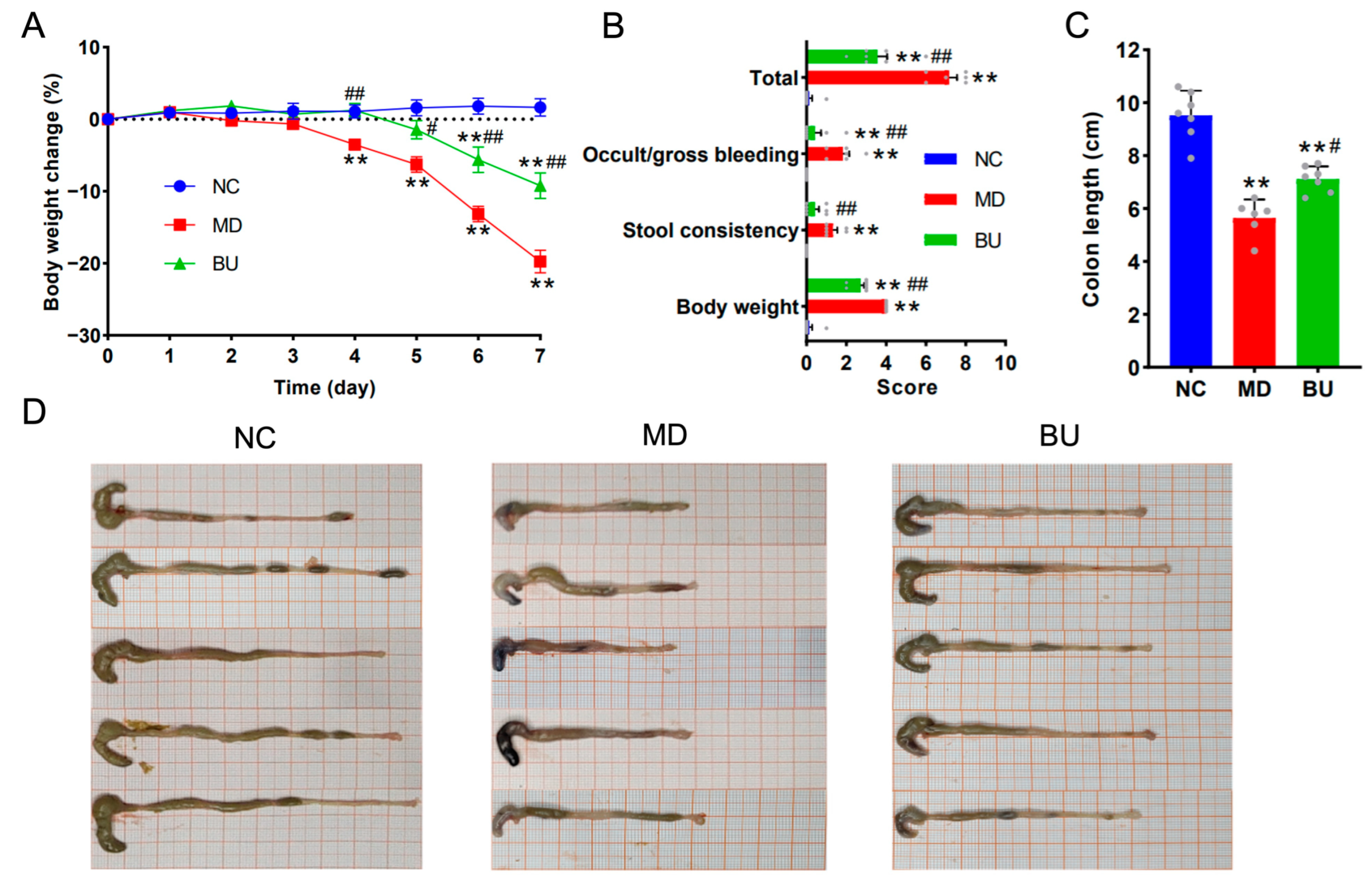
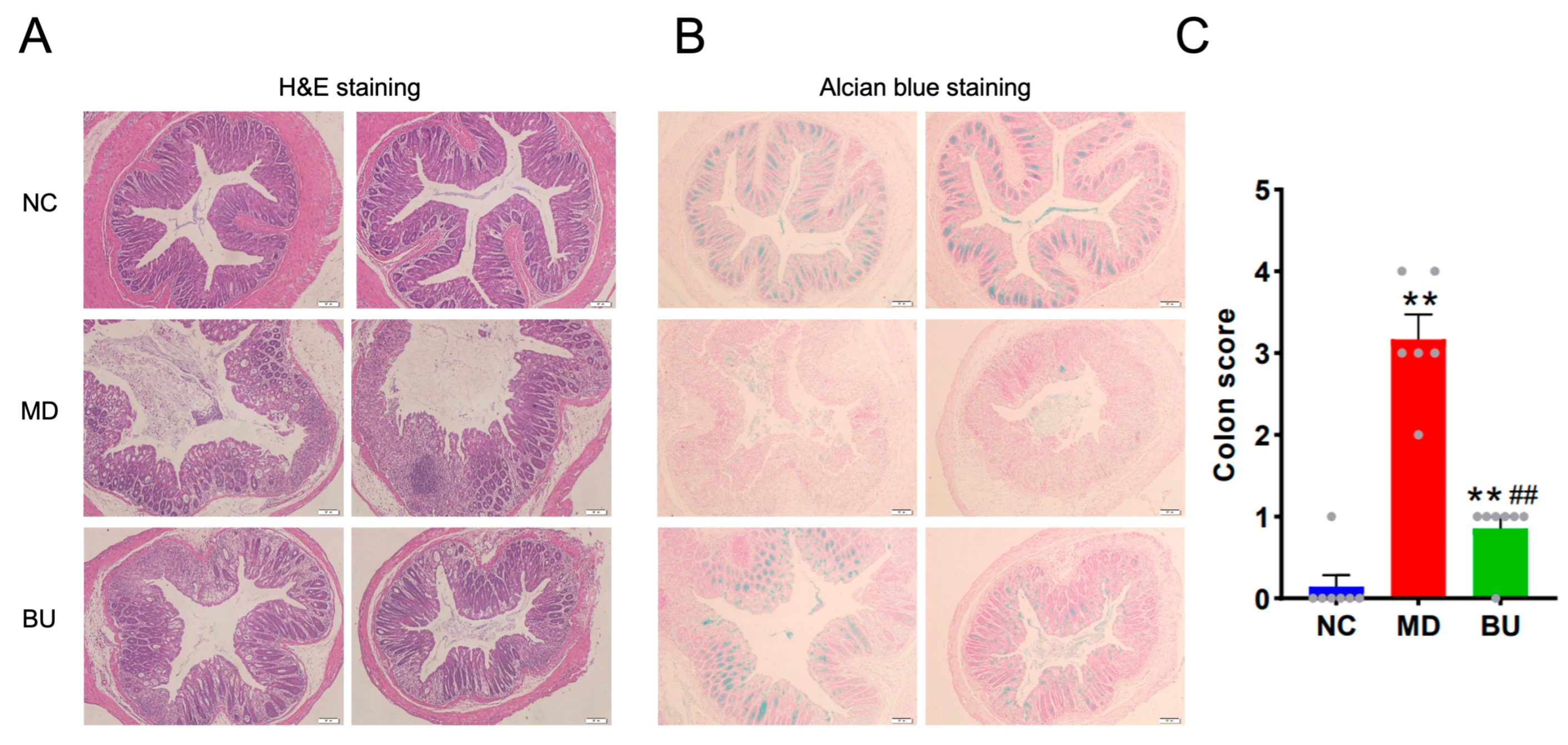
2.2. B. uniformis F18-22 Attenuated DSS-Induced UC in Mice
2.3. B. uniformis F18-22 Improved Gut Dysbiosis by Increasing the Abundance of Eubacterium siraeum and Decreasing the Amount of Escherichia-Shigella spp. in DSS-Fed Mice
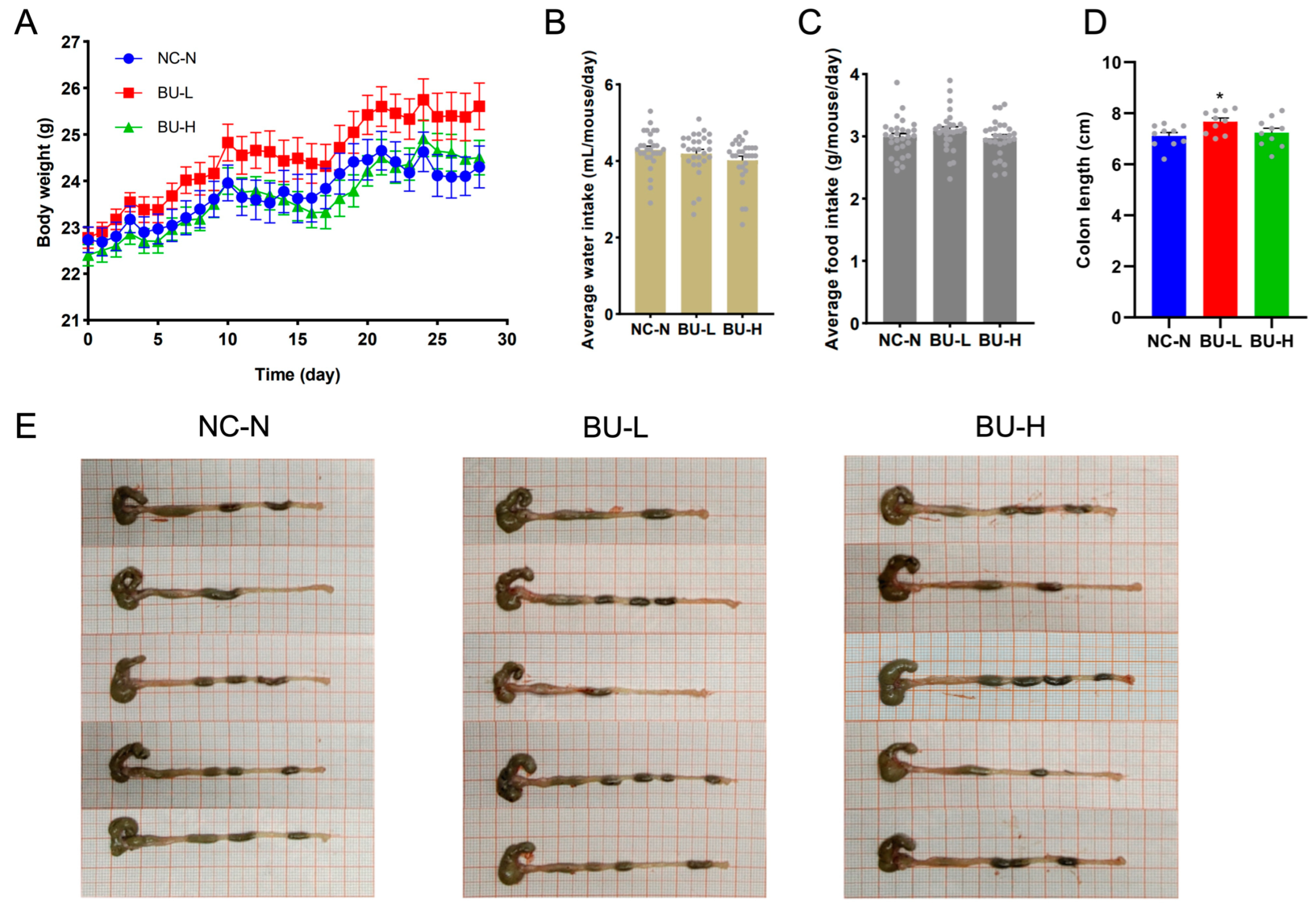
2.4. B. uniformis F18-22 Was Well Tolerated in Mice and Showed No Oral Toxicity after Repeated Administration
3. Discussion
3.1. Potential Applications of B. uniformis F18-22
3.2. Limitations of the Current Study
3.3. Future Directions for the Study of B. uniformis F18-22
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. In Vitro Fermentation
4.3. Animals and Treatment
4.4. High-Throughput Sequencing and Bioinformatic Analyses
4.5. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, M.; Song, Y.; Xu, Y.; Xu, H. Manipulating Microbiota in Inflammatory Bowel Disease Treatment: Clinical and Natural Product Interventions Explored. Int. J. Mol. Sci. 2023, 24, 11004. [Google Scholar] [PubMed]
- Hu, Y.; Chen, Z.; Xu, C.; Kan, S.; Chen, D. Disturbances of the gut microbiota and microbiota-derived metabolites in inflammatory bowel disease. Nutrients 2022, 14, 5140. [Google Scholar] [CrossRef]
- Oliveira, E.C.S.D.; Quaglio, A.E.V.; Magro, D.O.; Di Stasi, L.C.; Sassaki, L.Y. Intestinal Microbiota and miRNA in IBD: A Narrative Review about Discoveries and Perspectives for the Future. Int. J. Mol. Sci. 2023, 24, 7176. [Google Scholar]
- Manichanh, C.; Borruel, N.; Casellas, F.; Guarner, F. The gut microbiota in IBD. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 599–608. [Google Scholar] [PubMed]
- O’Toole, P.W.; Marchesi, J.R.; Hill, C. Next-generation probiotics: The spectrum from probiotics to live biotherapeutics. Nat. Microbiol. 2017, 2, 17057. [Google Scholar] [CrossRef] [PubMed]
- Lavelle, A.; Sokol, H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 223–237. [Google Scholar] [PubMed]
- Noor, S.O.; Ridgway, K.; Scovell, L.; Kemsley, E.K.; Lund, E.K.; Jamieson, C.; Johnson, I.T.; Narbad, A. Ulcerative colitis and irritable bowel patients exhibit distinct abnormalities of the gut microbiota. BMC Gastroenterol. 2010, 10, 134. [Google Scholar] [CrossRef]
- Nomura, K.; Ishikawa, D.; Okahara, K.; Ito, S.; Haga, K.; Takahashi, M.; Arakawa, A.; Shibuya, T.; Osada, T.; Kuwahara-Arai, K.; et al. Bacteroidetes species are correlated with disease activity in ulcerative colitis. J. Clin. Med. 2021, 10, 1749. [Google Scholar] [CrossRef]
- Morita, H.; Kano, C.; Ishii, C.; Kagata, N.; Ishikawa, T.; Hirayama, A.; Uchiyama, Y.; Hara, S.; Nakamura, T.; Fukuda, S. Bacteroides uniformis and its preferred substrate, α-cyclodextrin, enhance endurance exercise performance in mice and human males. Sci. Adv. 2023, 9, eadd2120. [Google Scholar] [CrossRef]
- Gauffin Cano, P.; Santacruz, A.; Moya, Á.; Sanz, Y. Bacteroides uniformis CECT 7771 ameliorates metabolic and immunological dysfunction in mice with high-fat-diet induced obesity. PLoS ONE 2012, 7, e41079. [Google Scholar] [CrossRef]
- López-Almela, I.; Romaní-Pérez, M.; Bullich-Vilarrubias, C.; Benítez-Páez, A.; Gómez Del Pulgar, E.M.; Francés, R.; Liebisch, G.; Sanz, Y. Bacteroides uniformis combined with fiber amplifies metabolic and immune benefits in obese mice. Gut Microbes 2021, 13, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Fu, T.; Wang, Y.; Ma, M.; Dai, W.; Pan, L.; Shang, Q.; Yu, G. Isolation of alginate-degrading bacteria from the human gut microbiota and discovery of Bacteroides xylanisolvens AY11-1 as a novel anti-colitis probiotic bacterium. Nutrients 2023, 15, 1352. [Google Scholar] [CrossRef] [PubMed]
- Fu, T.; Pan, L.; Shang, Q.; Yu, G. Fermentation of alginate and its derivatives by different enterotypes of human gut microbiota: Towards personalized nutrition using enterotype-specific dietary fibers. Int. J. Biol. Macromol. 2021, 183, 1649–1659. [Google Scholar] [CrossRef]
- Pan, L.; Fu, T.; Cheng, H.; Mi, J.; Shang, Q.; Yu, G. Polysaccharide from edible alga Gloiopeltis furcata attenuates intestinal mucosal damage by therapeutically remodeling the interactions between gut microbiota and mucin O-glycans. Carbohydr. Polym. 2022, 278, 118921. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.S.; Liao, Y.C.; Huang, C.T.; Lin, C.M.; Cheung, C.H.Y.; Ruan, J.W.; Yu, W.H.; Tsai, Y.T.; Lin, I.J.; Huang, C.H.; et al. Identification of a gut microbiota member that ameliorates DSS-induced colitis in intestinal barrier enhanced Dusp6-deficient mice. Cell Rep. 2021, 37, 110016. [Google Scholar] [CrossRef]
- Liu, L. Investigation of Optimal Culture Conditions and Development of a Protective Delivery Method of Eubacterium siraeum as a Potential Probiotic. Master’s Thesis, Wayne State University, Detroit, MI, USA, 2017. [Google Scholar]
- Moore, W.C.; Johnson, J.L.; Holdeman, L.V. Emendation of Bacteroidaceae and Butyrivibrio and descriptions of Desulfomonas gen. nov. and ten new species in the genera Desulfomonas, Butyrivibrio, Eubacterium, Clostridium, and Ruminococcus. Int. J. Syst. Evol. Microbiol. 1976, 26, 238–252. [Google Scholar] [CrossRef]
- Gómez del Pulgar, E.M.; Benítez-Páez, A.; Sanz, Y. Safety assessment of Bacteroides uniformis CECT 7771, a symbiont of the gut microbiota in infants. Nutrients 2020, 12, 551. [Google Scholar] [CrossRef]
- El Kaoutari, A.; Armougom, F.; Gordon, J.I.; Raoult, D.; Henrissat, B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 2013, 11, 497–504. [Google Scholar] [CrossRef]
- Hehemann, J.H.; Kelly, A.G.; Pudlo, N.A.; Martens, E.C.; Boraston, A.B. Bacteria of the human gut microbiome catabolize red seaweed glycans with carbohydrate-active enzyme updates from extrinsic microbes. Proc. Natl. Acad. Sci. USA 2012, 109, 19786–19791. [Google Scholar] [CrossRef]
- Hehemann, J.H.; Correc, G.; Barbeyron, T.; Helbert, W.; Czjzek, M.; Michel, G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 2010, 464, 908–912. [Google Scholar] [CrossRef]
- Wardman, J.F.; Bains, R.K.; Rahfeld, P.; Withers, S.G. Carbohydrate-active enzymes (CAZymes) in the gut microbiome. Nat. Rev. Microbiol. 2022, 20, 542–556. [Google Scholar] [CrossRef] [PubMed]
- National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals. Guide for the Care and Use of Laboratory Animals, 8th ed.; National Academies Press: Washington, DC, USA, 2011. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Yu, G.; Shi, C.P.; Liu, L.M.; Guo, Q.; Han, C.; Zhang, D.; Zhang, L.; Liu, B.; Gao, H.; et al. Majorbio Cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses. iMeta 2022, 1, e12. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, W.; Zhang, J.; Chen, L.; Yu, J.; Zhang, J.; Yin, H.; Shang, Q.; Yu, G. Discovery of Bacteroides uniformis F18-22 as a Safe and Novel Probiotic Bacterium for the Treatment of Ulcerative Colitis from the Healthy Human Colon. Int. J. Mol. Sci. 2023, 24, 14669. https://doi.org/10.3390/ijms241914669
Dai W, Zhang J, Chen L, Yu J, Zhang J, Yin H, Shang Q, Yu G. Discovery of Bacteroides uniformis F18-22 as a Safe and Novel Probiotic Bacterium for the Treatment of Ulcerative Colitis from the Healthy Human Colon. International Journal of Molecular Sciences. 2023; 24(19):14669. https://doi.org/10.3390/ijms241914669
Chicago/Turabian StyleDai, Wei, Jiaxue Zhang, Lu Chen, Junhong Yu, Junyi Zhang, Hua Yin, Qingsen Shang, and Guangli Yu. 2023. "Discovery of Bacteroides uniformis F18-22 as a Safe and Novel Probiotic Bacterium for the Treatment of Ulcerative Colitis from the Healthy Human Colon" International Journal of Molecular Sciences 24, no. 19: 14669. https://doi.org/10.3390/ijms241914669
APA StyleDai, W., Zhang, J., Chen, L., Yu, J., Zhang, J., Yin, H., Shang, Q., & Yu, G. (2023). Discovery of Bacteroides uniformis F18-22 as a Safe and Novel Probiotic Bacterium for the Treatment of Ulcerative Colitis from the Healthy Human Colon. International Journal of Molecular Sciences, 24(19), 14669. https://doi.org/10.3390/ijms241914669







