Defense Strategies of Rice in Response to the Attack of the Herbivorous Insect, Chilo suppressalis
Abstract
1. Introduction
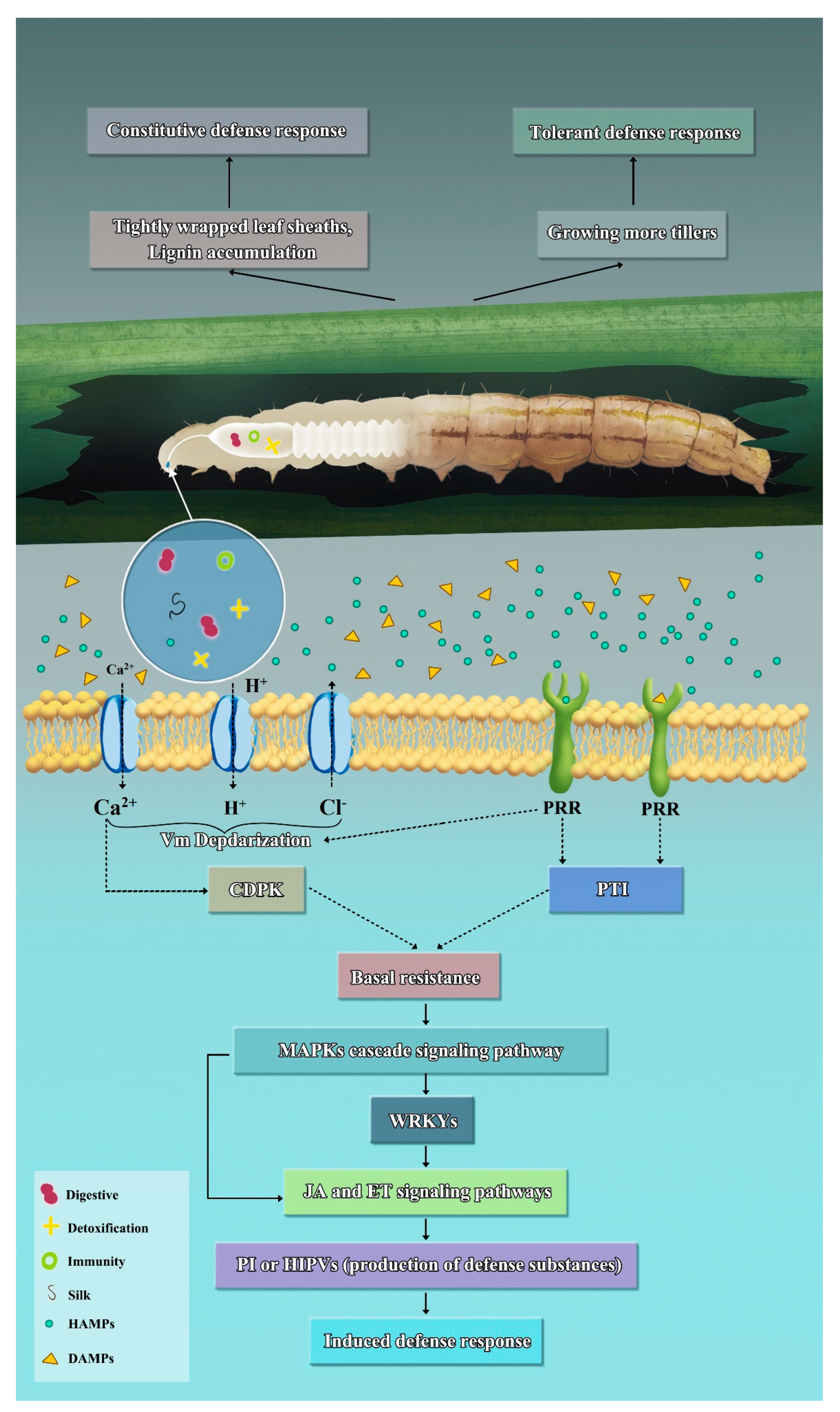
2. Defense Strategies of Rice against the Herbivorous Insect, C. suppressalis
2.1. Constitutive Defense Response
2.2. Tolerant Defense Response
2.3. Induced Defense Response
2.3.1. Herbivore-Associated Molecular Patterns
2.3.2. Activation of Defense Signal Transduction in Rice
2.3.3. Defense Compounds
2.3.4. Applying Omics Techniques to Understand Host Defense
3. Transgenic Strategies
3.1. Signal Transduction Genes
3.2. JA Biosynthesis-Related Genes
3.3. Plant Protease Inhibitor (PPI)
3.4. Host-Induced Gene Silencing (HIGS)
3.5. Bacillus thuringiensis (Bt) Gene
3.6. Other Ways
4. Conclusions and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, H.M.; Li, Y.; Liu, H.F.; Kong, L.G.; Ding, X.H. Research progress on regulatory genes of important agronomic traits and breeding utilization in rice. Biotechnol. Bull. 2020, 36, 155–169. [Google Scholar]
- Saito, K.; Dieng, I.; Toure, A.A.; Somado, E.A.; Wopereis, M.C.S. Rice yield growth analysis for 24 African countries over 1960–2012. Glob. Food Secur. 2015, 5, 62–69. [Google Scholar]
- Douglas, A.E. Strategies for enhanced crop resistance to insect pests. Annu. Rev. Plant Biol. 2018, 69, 637–660. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.J.; Hou, M.L.; Han, L.Z.; Liu, Y.D. Research progress in host selection and underlying mechanisms, and factors affecting population dynamics of Chilo suppressalis. Plant Prot. 2008, 34, 22–28. [Google Scholar]
- Chen, M.; Shelton, A.; Ye, G.Y. Insect-resistant genetically modified rice in China: From research to commercialization. Annu. Rev. Entomol. 2011, 56, 81–101. [Google Scholar] [CrossRef]
- Li, Y.H.; Hallerman, E.M.; Liu, Q.S.; Wu, K.M.; Peng, Y.F. The development and status of Bt rice in China. Plant Biotechnol. J. 2016, 14, 839–848. [Google Scholar]
- Li, Y.H.; Hallerman, E.M.; Wu, K.M.; Peng, Y.F. Insect-resistant genetically engineered crops in China: Development, application, and prospects for use. Annu. Rev. Entomol. 2020, 65, 13.1–13.20. [Google Scholar]
- Yang, M.; Tian, C.H.; Sun, F.Y.; Liu, X.Y.; Yu, F.Q. Study on different morphological indicators used for age division of Chilo suppressalis larvae. Jiangsu Agric. Sci. 2022, 50, 114–118. [Google Scholar]
- Lou, Y.G.; Zhang, G.R.; Zhang, W.Q.; Hu, Y.; Zhang, J. Biological control of rice insect pests in China. Biol. Control 2013, 67, 8–20. [Google Scholar] [CrossRef]
- Chagnon, M.; Kreutzweiser, D.; Mitchell, E.A.D.; Morrissey, C.A.; Noome, D.A.; Van der Sluijs, J.P. Risks of large-scale use of systemic insecticides to ecosystem functioning and services. Environ. Sci. Pollut. Res. 2015, 22, 119–134. [Google Scholar]
- Guo, J.P.; Xu, C.X.; Wu, D.; Zhao, Y.; Qiu, Y.F.; Wang, X.X.; Ouyang, Y.D.; Cai, B.D.; Liu, X.; Jing, S.L.; et al. Bph6 encodes an exocyst-localized protein and confers broad resistance to planthoppers in rice. Nat. Genet. 2018, 50, 297–306. [Google Scholar] [PubMed]
- Guo, J.P.; Wang, H.Y.; Guan, W.; Guo, Q.; Wang, J.; Yang, J.; Peng, Y.X.; Shan, J.H.; Gao, M.Y.; Shi, S.J.; et al. A tripartite rheostat controls self-regulated host plant resistance to insects. Nature 2023, 618, 799–807. [Google Scholar] [PubMed]
- Hu, J.; Zhou, J.B.; Peng, X.X.; Xu, H.H.; Liu, C.X.; Du, B.; Yuan, H.Y.; Zhu, L.L.; He, G.C. The Bphi008a gene interacts with the ethylene pathway and transcriptionally regulates MAPK genes in the response of rice to brown planthopper feeding. Plant Physiol. 2011, 156, 856–872. [Google Scholar] [PubMed]
- Huang, H.J.; Wang, Y.Z.; Li, L.L.; Lu, H.B.; Lu, J.B.; Wang, X.; Ye, Z.X.; Zhang, Z.L.; He, Y.J.; Lu, G.; et al. Planthopper salivary sheath protein LsSP1 contributes to manipulation of rice plant defenses. Nat. Commun. 2023, 14, 737. [Google Scholar]
- Liu, Y.Q.; Wu, H.; Chen, H.; Liu, Y.L.; He, J.; Kang, H.Y.; Sun, Z.G.; Pan, G.; Wang, Q.; Hu, J.L.; et al. A gene cluster encoding lectin receptor kinases confers broad-spectrum and durable insect resistance in rice. Nat. Biotechnol. 2014, 33, 301–305. [Google Scholar]
- Lu, H.P.; Luo, T.; Fu, H.W.; Wang, L.; Tan, Y.Y.; Huang, J.Z.; Wang, Q.; Ye, G.Y.; Gatehouse, A.M.R.; Lou, Y.G.; et al. Resistance of rice to insect pests mediated by suppression of serotonin biosynthesis. Nat. Plants 2018, 4, 338–344. [Google Scholar]
- Howe, G.A.; Jander, G. Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 2008, 59, 41–66. [Google Scholar]
- Chen, M.S. Inducible direct plant defense against insect herbivores: A review. Insect Sci. 2008, 15, 101–114. [Google Scholar]
- Dicke, M.; Poecke, R.M.P. Signalling in plant-insect interactions: Signal transduction in direct and indirect plant defence. In Plant Signal Transduction; Scheel, D., Wasternack, C., Eds.; Oxford University Press: Oxford, UK, 2002; pp. 289–316. [Google Scholar]
- Gols, R. Direct and indirect chemical defences against insects in a multitrophic framework. Plant Cell Environ. 2014, 37, 1741–1752. [Google Scholar]
- Hulburt, E.M. The four principles of adaptation and their set theory foundation. Ecol. Model. 2004, 180, 253–276. [Google Scholar]
- Valverde, P.L.; Fornoni, J.; NUNez-Farfan, J. Defensive role of leaf trichomes in resistance to herbivorous insects in Datura Stramonium. J. Evolution. Biol. 2001, 14, 424–432. [Google Scholar] [CrossRef]
- He, Y.J.; Ju, D.; Wang, Y.; Yang, X.Q.; Wang, X.Q. Compositive and inductive expression patterns of protease inhibitor genes OsLTPL164 and OsLTPL151 in rice (Oryza sativa). Agric. Sci. China 2018, 51, 2311–2321. [Google Scholar]
- Peumans, W.J.; Van Damme, E.J.M. Lectins as plant defense proteins. Plant Physiol. 1995, 109, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Tian, D.; Peiffer, M.; Shoemaker, E.; Tooker, J.; Haubruge, E.; Francis, F.; Luthe, D.S.; Felton, G.W. Salivary glucose oxidase from caterpillars mediates the induction of rapid and delayed-induced defenses in the tomato plant. PLoS ONE 2012, 7, e36168. [Google Scholar]
- Xu, J.; Lu, M.X.; Huang, D.L.; Du, Y.Z. Molecular cloning, characterization, genomic structure and functional analysis of catalase in Chilo suppressalis. J. Asia-Pac. Entomol. 2017, 20, 331–336. [Google Scholar] [CrossRef]
- Zheng, Y.Q.; Zhang, X.Y.; Liu, X.; Qin, N.N.; Xu, K.F.; Zeng, R.S.; Liu, J.; Song, Y.Y. Nitrogen supply alters rice defense against the striped stem borer Chilo suppressalis. Front. Plant Sci. 2021, 12, 691292. [Google Scholar] [CrossRef]
- Alam, S.N.; Cohen, M.B. Detection and analysis of QTLs for resistance to the brown planthopper, Nilaparvata lugens, in a doubled-haploid rice population. Theor. Appl. Genet. 1998, 97, 1370–1379. [Google Scholar] [CrossRef]
- Frei, A.; Bueno, J.M.; Diaz-Montano, J.; Gu, H.N.; Cardona, C.; Dorn, S. Tolerance as a mechanism of resistance to Thrips palmi in common beans. Entomol. Exp. Appl. 2004, 112, 73–80. [Google Scholar] [CrossRef]
- Li, Y.Q.; Sheng, C.F. Plant overcompensation responses. Plant Physiol. Commun. 1996, 32, 457–464. [Google Scholar]
- Rao, P.R.M.; Prakasa Rao, P.S. Gall midge outbreak on dry season rice in West Godavari District, Andhra Pradesh (AP), India. Int. Rice Res. Newsl. 1989, 14, 28. [Google Scholar]
- Qin, H.G.; Xie, Y.F. A study on the compensation ability for the damage of Chilo suppressalis during rice tillering stage. Plant Prot. 1992, 18, 2–4. [Google Scholar]
- Panda, N.; Heinrichs, E.A. Levels of tolerance and antibiosis in rice varieties having moderate resistance to the brown planthopper, Nilaparvata lugens (Stål) (Hemiptera: Delphacidae). Environ. Entomol. 1983, 12, 1204–1214. [Google Scholar] [CrossRef]
- Qiu, Y.F.; Guo, J.P.; Jing, S.L.; Zhu, L.L.; He, G.C. Development and characterization of japonica rice lines carrying the brown planthopper-resistance genes BPH12 and BPH6. Theor. Appl. Genet. 2012, 124, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Erb, M.; Meldau, S.; Howe, G.A. Role of phytohormones in insect-specific plant reactions. Trends Plant Sci. 2012, 17, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Santamaria, M.E.; Martínez, M.; Cambra, I.; Grbic, V.; Diaz, I. Understanding plant defence responses against herbivore attacks: An essential first step towards the development of sustainable resistance against pests. Transgenic Res. 2013, 22, 697–708. [Google Scholar] [CrossRef]
- Alborn, H.T.; Hansen, T.V.; Jones, T.H.; Bennett, D.C.; Tumlinson, J.H.; Schmelz, E.A.; Teal, P.E.A. Disulfooxy fatty acids from the American bird grasshopper Schistocerca americana, elicitors of plant volatiles. Proc. Natl. Acad. Sci. USA 2007, 104, 12976–12981. [Google Scholar] [CrossRef]
- Aljbory, Z.; Chen, M.S. Indirect plant defense against insect herbivores: A review. Insect Sci. 2018, 25, 2–23. [Google Scholar] [CrossRef]
- Hilker, M.; Meiners, T. How do plants "notice" attack by herbivorous arthropods? Biol. Rev. 2010, 85, 267–280. [Google Scholar] [CrossRef]
- Schuman, M.C.; Baldwin, I.T. The layers of plant responses to insect herbivores. Annu. Rev. Entomol. 2016, 61, 373–394. [Google Scholar] [CrossRef]
- Schuman, M.C.; Barthel, K.; Baldwin, I.T. Herbivory-induced volatiles function as defenses increasing fitness of the native plant Nicotiana attenuata in nature. ELife 2012, 1, e00007. [Google Scholar] [CrossRef]
- Chung, S.H.; Rosa, C.; Scully, E.D.; Peiffer, M.; Tooker, J.F.; Hoover, K.; Luthe, D.S.; Felton, G.W. Herbivore exploits orally secreted bacteria to suppress plant defenses. Proc. Natl. Acad. Sci. USA 2013, 110, 15728–15733. [Google Scholar] [CrossRef]
- Doss, R.P. Treatment of pea pods with Bruchin B results in up-regulation of a gene similar to MtN19. Plant Physiol. Bioch. 2005, 43, 225–231. [Google Scholar] [CrossRef]
- Hilker, M.; Meiners, T. Early herbivore alert: Insect eggs induce plant defense. J. Chem. Ecol. 2006, 32, 1379–1397. [Google Scholar] [CrossRef] [PubMed]
- Hilker, M.; Meiners, T. Plants and insect eggs: How do they affect each other? Phytochemistry 2011, 72, 1612–1623. [Google Scholar] [CrossRef] [PubMed]
- Reymond, P. Perception, signaling and molecular basis of oviposition-mediated plant responses. Planta 2013, 238, 247–258. [Google Scholar] [CrossRef]
- Ray, S.; Basu, S.; Rivera-Vega, L.J.; Acevedo, F.E.; Louis, J.; Felton, G.W.; Luthe, D.S. Lessons from the far end: Caterpillar FRASS-induced defenses in maize, rice, cabbage, and tomato. J. Chem. Ecol. 2016, 42, 1130–1141. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Gaffor, I.; Acevedo, F.E.; Helms, A.; Chuang, W.P.; Tooker, J.; Felton, G.W.; Luthe, D.S. Maize Plants Recognize Herbivore-Associated Cues from Caterpillar Frass. J. Chem. Ecol. 2015, 41, 781–792. [Google Scholar] [CrossRef]
- Zebelo, S.; Piorkowski, J.; Disi, J.; Fadamiro, H. Secretions from the ventral eversible gland of Spodoptera exigua caterpillars activate defense-related genes and induce emission of volatile organic compounds in tomato, Solanum lycopersicum. BMC Plant Biol. 2014, 14, 140. [Google Scholar] [CrossRef]
- Helms, A.M.; De Moraes, C.M.; Tooker, J.F.; Mescher, M.C. Exposure of Solidago altissima plants to volatile emissions of an insect antagonist (Eurosta solidaginis) deters subsequent herbivory. Proc. Natl. Acad. Sci. USA 2013, 110, 199–204. [Google Scholar] [CrossRef]
- Helms, A.M.; De Moraes, C.M.; Tröger, A.; Alborn, H.T.; Francke, W.; Tooker, J.F.; Mescher, M.C. Identification of an insect-produced olfactory cue that primes plant defenses. Nat. Commun. 2017, 8, 337. [Google Scholar] [CrossRef]
- Hu, L.F.; Ye, M.; Kuai, P.; Ye, M.F.; Erb, M.; Lou, Y.G. OsLRR-RLK1, an early responsive leucine-rich repeat receptor-like kinase, initiates rice defense responses against a chewing herbivore. New Phytol. 2018, 219, 1097–1111. [Google Scholar] [CrossRef]
- Berens, M.L.; Berry, H.M.; Mine, A.; Argueso, C.T.; Tsuda, K. Evolution of hormone signaling networks in plant defense. Annu. Rev. Phytopathol. 2017, 55, 401–425. [Google Scholar] [CrossRef]
- Maffei, M.E.; Mithofer, A.; Arimura, G.I.; Uchtenhagen, H.; Bossi, S.; Bertea, C.M.; Cucuzza, L.S.; Novero, M.; Quadro, S.; Boland, W. Effects of feeding Spodoptera littoralis on lima bean leaves. III. membrane depolarization and involvement of hydrogen peroxide. Plant Physiol. 2006, 140, 1022–1035. [Google Scholar] [CrossRef] [PubMed]
- Zebelo, S.A.; Maffei, M.E. Role of early signalling events in plant-insect interactions. J. Exp. Bot. 2014, 66, 435–448. [Google Scholar] [CrossRef] [PubMed]
- Batistič, O.; Kudla, J. Plant calcineurin B-like proteins and their interacting protein kinases. BBA Mol. Cell Res. 2009, 1793, 985–992. [Google Scholar] [CrossRef] [PubMed]
- Edel, K.H.; Marchadier, E.; Brownlee, C.; Kudla, J.; Hetherington, A.M. The evolution of calcium-based signalling in plants. Curr. Biol. 2017, 27, R667–R679. [Google Scholar] [CrossRef]
- Yuan, P.G.; Jauregui, E.; Du, L.Q.; Tanaka, K.; Poovaiah, B. Calcium signatures and signaling events orchestrate plant-microbe interactions. Curr. Opin. Plant Biol. 2017, 38, 173–183. [Google Scholar] [CrossRef]
- Doke, N. Involvement of superoxide anion generation in the hypersensitive response of potato tuber tissues to infection with an incompatible race of Phytophthora infestans and to the hyphal wall components. Physiol. Plant Pathol. 1983, 23, 345–357. [Google Scholar] [CrossRef]
- Chakradhar, T.; Mahanty, S.; Reddy, R.A.; Divya, K.; Reddy, P.S.; Reddy, M.K. Biotechnological perspective of reactive oxygen species (ROS)-mediated stress tolerance in plants. In Reactive Oxygen Species and Antioxidant Systems in Plants: Role and Regulation Under Abiotic Stress; Springer: Berlin/Heidelberg, Germany, 2017; pp. 53–87. [Google Scholar]
- Guo, H.M.; Li, H.C.; Zhou, S.R.; Xue, H.W.; Miao, X.X. Deficiency of mitochondrial outer membrane protein 64 confers rice resistance to both piercing-sucking and chewing insects in rice. J. Integ. Plant Biol. 2020, 12, 1967–1982. [Google Scholar] [CrossRef]
- Zhou, G.X.; Qi, J.F.; Ren, N.; Cheng, J.A.; Erb, M.; Mao, B.Z.; Lou, Y.G. Silencing OsHI-LOX makes rice more susceptible to chewing herbivores, but enhances resistance to a phloem feeder. Plant J. 2009, 60, 638–648. [Google Scholar] [CrossRef]
- Raja, V.; Majeed, U.; Kang, H.; Andrabi, K.I.; John, R. Abiotic stress: Interplay between ROS, hormones and MAPKs. Environ. Exp. Bot. 2017, 137, 142–157. [Google Scholar] [CrossRef]
- Hettenhausen, C.; Schuman, M.C.; Wu, J.Q. MAPK signaling: A key element in plant defense response to insects. Insect Sci. 2015, 22, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, J.C.; Hu, L.F.; Zhang, T.F.; Zhang, G.R.; Lou, Y.G. OsMPK3 positively regulates the JA signaling pathway and plant resistance to a chewing herbivore in rice. Plant Cell Rep. 2013, 32, 1075–1084. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Ju, H.P.; Zhou, G.X.; Zhu, C.S.; Erb, M.; Wang, X.P.; Wang, P.; Lou, Y.G. An EAR-motif-containing ERF transcription factor affects herbivore-induced signaling, defense and resistance in rice. Plant J. 2011, 68, 583–596. [Google Scholar] [CrossRef]
- Hu, L.F.; Ye, M.; Li, R.; Zhang, T.F.; Zhou, G.X.; Wang, Q.; Lu, J. The rice transcription factor WRKY53 suppresses herbivore-induced defenses by acting as a negative feedback modulator of map kinase activity. Plant Physiol. 2015, 169, 2907–2921. [Google Scholar] [CrossRef]
- Lyons, R.; Manners, J.M.; Kazan, K. Jasmonate biosynthesis and signaling in monocots: A comparative overview. Plant Cell Rep. 2013, 32, 815–827. [Google Scholar] [CrossRef]
- Schaller, F. Enzymes of the biosynthesis of octadecanoid-derived signalling molecules. J. Exp. Bot. 2001, 52, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.G.; Ellis, C.; Devoto, A. The jasmonate signal pathway. Plant Cell 2002, 14, S153–S164. [Google Scholar] [CrossRef]
- Wasternack, C. Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 2007, 100, 681–697. [Google Scholar] [CrossRef]
- Browse, J. Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 2009, 60, 183–205. [Google Scholar] [CrossRef]
- Ray, R.; Li, D.P.; Halitschke, R.; Baldwin, I.T. Using natural variation to achieve a whole-plant functional understanding of the responses mediated by jasmonate signaling. Plant J. 2019, 99, 414–425. [Google Scholar] [CrossRef] [PubMed]
- Goossens, J.; Fernández-Calvo, P.; Schweizer, F.; Goossens, A. Jasmonates: Signal transduction components and their roles in environmental stress responses. Plant Mol. Biol. 2016, 91, 673–689. [Google Scholar] [CrossRef]
- Zhou, G.X.; Wang, X.; Yan, F.; Wang, X.; Li, R.; Cheng, J.A.; Lou, Y.G. Genome-wide transcriptional changes and defence-related chemical profiling of rice in response to infestation by the rice striped stem borer Chilo suppressalis. Physiol. Plant. 2011, 143, 21–40. [Google Scholar] [CrossRef]
- Liu, Q.S.; Hu, X.Y.; Su, S.L.; Ning, Y.S.; Peng, Y.F.; Ye, G.Y.; Lou, Y.G.; Turlings, T.C.J.; Li, Y.H. Cooperative herbivory between two important pests of rice. Nat. Commun. 2021, 12, 6772. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.X.; Ren, N.; Qi, J.F.; Lu, J.; Xiang, C.Y.; Ju, H.P.; Cheng, J.A.; Lou, Y.G. The 9-lipoxygenase Osr9-LOX1 interacts with the 13-lipoxygenase-mediated pathway to regulate resistance to chewing and piercing-sucking herbivores in rice. Physiol. Plant. 2014, 152, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.M.; Li, H.C.; Zhou, S.R.; Xue, H.W.; Miao, X.X. Cis-12-oxo-phytodienoic acid stimulates rice defense response to a piercing-sucking insect. Mol. Plant 2014, 7, 1683–1692. [Google Scholar] [CrossRef]
- Yao, C.C.; Du, L.X.; Liu, Q.S.; Hu, X.Y.; Ye, W.F.; Turlings, T.C.J.; Li, Y.H. Stemborer-induced rice plant volatiles boost direct and indirect resistance in neighboring plants. New Phytol. 2023, 237, 2375–2387. [Google Scholar] [CrossRef]
- Tong, X.H.; Qi, J.F.; Zhu, X.D.; Mao, B.Z.; Zeng, L.J.; Wang, B.H.; Li, Q.; Zhou, G.X.; Xu, X.J.; Lou, Y.G.; et al. The rice hydroperoxide lyase OsHPL3 functions in defense responses by modulating the oxylipin pathway. Plant J. 2012, 71, 763–775. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, Y.J.; Cao, G.C.; Gu, S.H.; Wu, K.M.; Gao, X.W.; Guo, Y.Y. Rice gene expression profiles responding to larval feeding of the striped stem borer at the 1st to 2nd instar stage. Insect Sci. 2010, 18, 273–281. [Google Scholar] [CrossRef]
- Zeng, J.M.; Zhang, T.F.; Huangfu, J.Y.; Li, R.; Lou, Y.G. Both allene oxide synthases genes are involved in the biosynthesis of herbivore-induced jasmonic acid and herbivore resistance in rice. Plants 2021, 10, 442. [Google Scholar] [CrossRef]
- Erb, M.; Reymond, P. Molecular interactions between plants and insect herbivores. Annu. Rev. Plant Biol. 2019, 70, 4.1–4.31. [Google Scholar] [CrossRef] [PubMed]
- Dempsey, D.A.; Vlot, A.C.; Wildermuth, M.C.; Klessig, D.F. Salicylic acid biosynthesis and metabolism. Arab. Book 2011, 9, e0156. [Google Scholar] [CrossRef] [PubMed]
- An, C.F.; Mou, Z.L. Salicylic acid and its function in plant immunity. J. Integr. Plant Biol. 2011, 53, 412–428. [Google Scholar] [CrossRef] [PubMed]
- Koo, Y.M.; Heo, A.Y.; Choi, H.W. Salicylic acid as a safe plant protector and growth regulator. Plant Pathol. J. 2020, 36, 1–10. [Google Scholar] [CrossRef]
- Bari, R.; Jones, J.D.G. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef]
- Chen, H.; Wilkerson, C.G.; Kuchar, J.A.; Phinney, B.S.; Howe, G.A. Jasmonate-inducible plant enzymes degrade essential amino acids in the herbivore midgut. Proc. Natl. Acad. Sci. USA 2005, 102, 19237–19242. [Google Scholar] [CrossRef]
- Walling, L.L. The myriad plant responses to herbivores. J. Plant Growth Regul. 2000, 19, 195–216. [Google Scholar] [CrossRef]
- Von Dahl, C.C.; Winz, R.A.; Halitschke, R.; Kühnemann, F.; Gase, K.; Baldwin, I.T. Tuning the herbivore-induced ethylene burst: The role of transcript accumulation and ethylene perception in Nicotiana attenuata. Plant J. 2007, 51, 293–307. [Google Scholar] [CrossRef]
- Alonso, J.M.; Stepanova, A.N. The ethylene signaling pathway. Science 2004, 306, 1513–1515. [Google Scholar] [CrossRef]
- Guo, H.W.; Ecker, J.R. Plant responses to ethylene gas are mediated by SCFEBF1/EBF2-dependent proteolysis of EIN3 transcription factor. Cell 2003, 115, 667–677. [Google Scholar] [CrossRef]
- Lu, J.; Li, J.C.; Ju, H.P.; Liu, X.L.; Erb, M.; Wang, X.; Lou, Y.G. Contrasting effects of ethylene biosynthesis on induced plant resistance against a chewing and a piercing-sucking herbivore in rice. Mol. Plant 2014, 7, 1670–1682. [Google Scholar] [CrossRef] [PubMed]
- Mithöfer, A.; Boland, W. Plant defense against herbivores: Chemical aspects. Annu. Rev. Plant Biol. 2012, 63, 431–450. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.J.; Sheng, L.P.; Zhang, H.R.; Du, X.P.; An, C.; Xia, X.L.; Chen, F.D.; Jiang, J.F.; Chen, S.M. CmMYB19 over-expression improves aphid tolerance in chrysanthemum by promoting lignin synthesis. Int. J. Mol. Sci. 2017, 18, 619. [Google Scholar] [CrossRef] [PubMed]
- Lalthazuali; Mathew, N. Mosquito repellent activity of volatile oils from selected aromatic plants. Parasitol. Res. 2016, 116, 821–825. [Google Scholar] [CrossRef]
- Hincapié, L.C.A.; Monsalve, F.Z.; Parada, K.; Lamilla, C.; Alarcón, J.; Céspedes, A.C.L.; Seigler, D. Insect growth regulatory activity of Blechnum chilense. Nat. Prod. Commun. 2011, 6, 1085–1088. [Google Scholar]
- Shan, L.H.; Chen, L.; Gao, F.; Zhou, X.L. Diterpenoid alkaloids from Delphinium naviculare var. lasiocarpum with their antifeedant activity on Spodoptera exigua. Nat. Prod. Res. 2019, 33, 3254–3259. [Google Scholar] [CrossRef]
- Vandenborre, G.; Smagghe, G.; Van Damme, E.J.M. Plant lectins as defense proteins against phytophagous insects. Phytochemistry 2011, 72, 1538–1550. [Google Scholar] [CrossRef]
- Chen, L.; Cao, T.T.; Zhang, J.; Lou, Y.G. Overexpression of OsGID1 enhances the resistance of rice to the brown planthopper Nilaparvata lugens. Int. J. Mol. Sci. 2018, 19, 2744. [Google Scholar] [CrossRef]
- Quilis, J.; López-García, B.; Meynard, D.; Guiderdoni, E.; San Segundo, B. Inducible expression of a fusion gene encoding two proteinase inhibitors leads to insect and pathogen resistance in transgenic rice. Plant Biotechnol. J. 2014, 12, 367–377. [Google Scholar] [CrossRef]
- Lu, J.; Robert, C.A.M.; Riemann, M.; Cosme, M.; Mène-Saffrané, L.; Massana, J.; Stout, M.J.; Lou, Y.G.; Gershenzon, J.; Erb, M. Induced jasmonate signaling leads to contrasting effects on root damage and herbivore performance. Plant Physiol. 2015, 167, 1100–1116. [Google Scholar] [CrossRef]
- Robert, C.A.M.; Ferrieri, R.A.; Schirmer, S.; Babst, B.A.; Schueller, M.J.; Machado, R.A.R.; Arce, C.C.M.; Hibbard, B.E.; Gershenzon, J.; Turlings, T.C.J.; et al. Induced carbon reallocation and compensatory growth as root herbivore tolerance mechanisms. Plant Cell Environ. 2014, 37, 2613–2622. [Google Scholar] [CrossRef]
- Wang, Y.; Ju, D.; Yang, X.Q.; Ma, D.R.; Wang, X.Q. Comparative transcriptome analysis between resistant and susceptible rice cultivars responding to striped stem borer (SSB), Chilo suppressalis (Walker) infestation. Front. Physiol. 2018, 9, 1717. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, X.Y.; Tzin, V.; Romeis, J.; Peng, Y.F.; Li, Y.H. Combined transcriptome and metabolome analyses to understand the dynamic responses of rice plants to attack by the rice stem borer Chilo suppressalis (Lepidoptera: Crambidae). BMC Plant Biol. 2016, 16, 259. [Google Scholar] [CrossRef]
- Mochizuki, A.; Nishizawa, Y.; Onodera, H.; Tabei, Y.; Toki, S.; Habu, Y.; Ugaki, M.; Ohashi, Y. Transgenic rice plants expressing a trypsin inhibitor are resistant against rice stem borers, Chilo suppressalis. Entomol. Exp. Appl. 1999, 93, 173–178. [Google Scholar] [CrossRef]
- Huang, J.Q.; Wei, Z.M.; An, H.L.; Zhu, Y.X. Agrobacterium tumefaciens-mediated transformation of rice with the spider insecticidal gene conferring resistance to leaffolder and striped stem borer. Cell Res. 2001, 11, 149–155. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vila, L.; Quilis, J.; Meynard, D.; Breitler, J.C.; Marfà, V.; Murillo, I.; Vassal, J.M.; Messeguer, J.; Guiderdoni, E.; Segundo, B.S. Expression of the maize proteinase inhibitor (mpi) gene in rice plants enhances resistance against the striped stem borer (Chilo suppressalis): Effects on larval growth and insect gut proteinases. Plant Biotechnol. J. 2005, 3, 187–202. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Chen, H.; Tang, W.; Hua, H.X.; Lin, Y.H. Development and characterisation of transgenic rice expressing two Bacillus thuringiensis genes. Pest. Manag. Sci. 2011, 67, 414–422. [Google Scholar] [CrossRef]
- Jiang, S.; Wu, H.; Liu, H.J.; Zheng, J.; Lin, Y.J.; Chen, H. The overexpression of insect endogenous small RNAs in transgenic rice inhibits growth and delays pupation of striped stem borer (Chilo suppressalis). Pest. Manag. Sci. 2017, 73, 1453–1461. [Google Scholar] [CrossRef]
- Xu, C.; Cheng, J.H.; Lin, H.Y.; Lin, C.Y.; Gao, J.H.; Shen, Z.C. Characterization of transgenic rice expressing fusion protein Cry1Ab/Vip3A for insect resistance. Sci. Rep. 2018, 8, 15788. [Google Scholar] [CrossRef]
- Jiao, Y.Y.; Hu, X.Y.; Peng, Y.F.; Wu, K.M.; Romeis, J.; Li, Y.H. Bt rice plants may protect neighbouring non-Bt rice plants against the striped stem borer, Chilo suppressalis. Proc. R. Soc. B Biol. Sci. 2018, 285, 20181283. [Google Scholar] [CrossRef]
- He, K.; Xiao, H.M.; Sun, Y.; Ding, S.M.; Situ, G.M.; Li, F. Transgenic microRNA-14 rice shows high resistance to rice stem borer. Plant Biotechnol. J. 2019, 17, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Wen, N.; Chen, J.J.; Chen, G.; Du, L.X.; Chen, H.; Li, Y.H.; Peng, Y.F.; Yang, X.W.; Han, L.Z. The overexpression of insect endogenous microRNA in transgenic rice inhibits the pupation of Chilo suppressalis and Cnaphalocrocis medinalis. Pest. Manag. Sci. 2021, 77, 3990–3999. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.J.; Shen, E.L.; Wu, H.; Ma, W.H.; Chen, H.; Lin, Y.J. Trans-kingdom expression of an insect endogenous microRNA in rice enhances resistance to striped stem borer Chilo suppressalis. Pest. Manag. Sci. 2022, 78, 770–777. [Google Scholar] [CrossRef]
- Mao, C.; Zhu, X.P.; Wang, P.P.; Sun, Y.J.; Huang, R.L.; Zhao, M.C.; Hull, J.J.; Lin, Y.J.; Zhou, F.; Chen, H.; et al. Transgenic double-stranded RNA rice, a potential strategy for controlling striped stem borer (Chilo suppressalis). Pest. Manag. Sci. 2022, 78, 785–792. [Google Scholar] [CrossRef]
- Sun, Y.J.; Gong, Y.W.; He, Q.Z.; Kuang, S.J.; Gao, Q.; Ding, W.B.; He, H.L.; Xue, J.; Li, Y.Z.; Qiu, L. FAR knockout significantly inhibits Chilo suppressalis survival and transgene expression of double-stranded FAR in rice exhibits strong pest resistance. Plant Biotechnol. J. 2022, 20, 2272–2283. [Google Scholar] [CrossRef]
- Bostock, R.M. Signal crosstalk and induced resistance: Straddling the line between cost and benefit. Annu. Rev. Phytopathol. 2005, 43, 545–580. [Google Scholar] [CrossRef] [PubMed]
- Browse, J.; Howe, G.A. New weapons and a rapid response against insect attack. Plant Physiol. 2008, 146, 832–838. [Google Scholar] [CrossRef]
- Chung, H.S.; Koo, A.J.K.; Gao, X.L.; Jayanty, S.; Thines, B.; Jones, A.D.; Howe, G.A. Regulation and function of arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 2008, 146, 952–964. [Google Scholar] [CrossRef]
- Stork, W.; Diezel, C.; Halitschke, R.; Gális, I.; Baldwin, I.T. An ecological analysis of the herbivory-elicited JA burst and its metabolism: Plant memory processes and predictions of the moving target model. PLoS ONE 2009, 4, e4697. [Google Scholar] [CrossRef]
- Hartl, M.; Giri, A.P.; Kaur, H.; Baldwin, I.T. Serine protease inhibitors specifically defend Solanum nigrum against generalist herbivores but do not influence plant growth and development. Plant Cell 2010, 22, 4158–4175. [Google Scholar] [CrossRef]
- Hartl, M.; Giri, A.P.; Kaur, H.; Baldwin, I.T. The multiple functions of plant serine protease inhibitors. Plant Signal. Behav. 2011, 6, 1009–1011. [Google Scholar] [CrossRef] [PubMed]
- Baum, J.A.; Bogaert, T.; Clinton, W.; Heck, G.R.; Feldmann, P.; Ilagan, O.; Johnson, S.; Plaetinck, G.; Munyikwa, T.; Pleau, M.; et al. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007, 25, 1322–1326. [Google Scholar] [CrossRef]
- Zheng, X.X.; Weng, Z.J.; Li, H.; Kong, Z.C.; Zhou, Z.H.; Li, F.; Ma, W.H.; Lin, Y.J.; Chen, H. Transgenic rice overexpressing insect endogenous microRNA csu-novel-260 is resistant to striped stem borer under field conditions. Plant Biotechnol. J. 2021, 19, 421–423. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.S.; Romeis, J.; Yu, H.L.; Zhang, Y.J.; Li, Y.H.; Peng, Y.F. Bt rice does not disrupt the host-searching behavior of the parasitoid Cotesia chilonis. Sci. Rep. 2015, 5, 15295. [Google Scholar] [PubMed]
- Li, Z.Y.; Sui, H.; Xu, Y.B.; Han, L.Z.; Chen, F.J. Effects of insect-resistant transgenic Bt rice with a fused Cry1Ab+Cry1Ac gene on population dynamics of the stem borers, Chilo suppressalis and Sesamia inferens, occurring in paddy field. Acta Ecol. Sin. 2012, 32, 1783–1789. [Google Scholar]
- Wang, Y.N.; Ke, K.Q.; Li, Y.H.; Han, L.Z.; Liu, Y.M.; Hua, H.X.; Peng, Y.F. Comparison of three transgenic Bt rice lines for insecticidal protein expression and resistance against a target pest, Chilo suppressalis (Lepidoptera: Crambidae). Insect Sci. 2016, 23, 78–87. [Google Scholar]
- Li, H.P.; Wang, Z.J.; Han, K.H.; Guo, M.J.; Zou, Y.L.; Zhang, W.; Ma, W.H.; Hua, H.X. Cloning and functional identification of a Chilo suppressalis-inducible promoter of rice gene, OsHPL2. Pest. Manag. Sci. 2020, 76, 3177–3187. [Google Scholar] [CrossRef]
- Shangguan, X.X.; Zhang, J.; Liu, B.F.; Zhao, Y.; Wang, H.Y.; Wang, Z.Z.; Guo, J.P.; Rao, W.W.; Jing, S.L.; Guan, W. A mucin-like protein of planthopper is required for feeding and induces immunity response in plants. Plant Physiol. 2017, 176, 552–565. [Google Scholar] [CrossRef]
- Li, R.; Zhang, J.; Li, J.C.; Zhou, G.X.; Wang, Q.; Bian, W.B.; Erb, M.; Lou, Y.G. Prioritizing plant defence over growth through WRKY regulation facilitates infestation by non-target herbivores. eLife 2015, 4, e04805. [Google Scholar]
- Hu, L.F.; Ye, M.; Li, R.; Lou, Y.G. OsWRKY53, a versatile switch in regulating herbivore-induced defense responses in rice. Plant Signal. Behav. 2016, 11, e1169357. [Google Scholar]
- Vincent, T.R.; Avramova, M.; Canham, J.; Higgins, P.; Bilkey, N.; Mugford, S.T.; Pitino, M.; Toyota, M.; Gilroy, S.; Miller, A.J.; et al. Interplay of plasma membrane and vacuolar ion channels, together with BAK1, elicits rapid cytosolic calcium elevations in Arabidopsis during aphid feeding. Plant Cell 2017, 29, 1460–1479. [Google Scholar]
- Bricchi, I.; Bertea, C.M.; Occhipinti, A.; Paponov, I.A.; Maffei, M.E. Dynamics of membrane potential variation and gene expression induced by Spodoptera littoralis, Myzus persicae, and Pseudomonas syringae in Arabidopsis. PLoS ONE 2012, 7, e46673. [Google Scholar]
- Drerup, M.M.; Schlücking, K.; Hashimoto, K.; Manishankar, P.; Steinhorst, L.; Kuchitsu, K.; Kudla, J. The calcineurin B-like calcium sensors CBL1 and CBL9 together with their interacting protein kinase CIPK26 regulate the Arabidopsis NADPH oxidase RBOHF. Mol. Plant 2013, 6, 559–569. [Google Scholar] [PubMed]
- Kimura, S.; Kawarazaki, T.; Nibori, H.; Michikawa, M.; Imai, A.; Kaya, H.; Kuchitsu, K. The CBL-interacting protein kinase CIPK26 is a novel interactor of Arabidopsis NADPH oxidase AtRbohF that negatively modulates its ROS-producing activity in a heterologous expression system. J. Biochem. 2012, 153, 191–195. [Google Scholar] [PubMed]
- Moctezuma, C.; Hammerbacher, A.; Heil, M.; Gershenzon, J.; Méndez-Alonzo, R.; Oyama, K. Specific polyphenols and tannins are associated with defense against insect herbivores in the tropical oak Quercus oleoides. J. Chem. Ecol. 2014, 40, 458–467. [Google Scholar]
- Vasconcelos, I.M.; Oliveira, J.T.A. Antinutritional properties of plant lectins. Toxicon 2004, 44, 385–403. [Google Scholar]
- Mahanil, S.; Attajarusit, J.; Stout, M.J.; Thipyapong, P. Overexpression of tomato polyphenol oxidase increases resistance to common cutworm. Plant Sci. 2008, 174, 456–466. [Google Scholar]
- Amirhusin, B.; Shade, R.E.; Koiwa, H.; Hasegawa, P.M.; Bressan, R.A.; Murdock, L.L.; Zhu-Salzman, K. Soyacystatin N inhibits proteolysis of wheat α-amylase inhibitor and potentiates toxicity against cowpea weevil. J. Econ. Entomol. 2004, 97, 2095–2100. [Google Scholar] [CrossRef]
- Chen, X.Y.; Duan, Y.H.; Qiao, F.G.; Liu, H.; Huang, J.B.; Luo, C.X.; Chen, X.L.; Li, G.T.; Xie, K.B.; Hsiang, T.; et al. A secreted fungal effector suppresses rice immunity through host histone hypoacetylation. New Phytol. 2022, 235, 1977–1994. [Google Scholar]
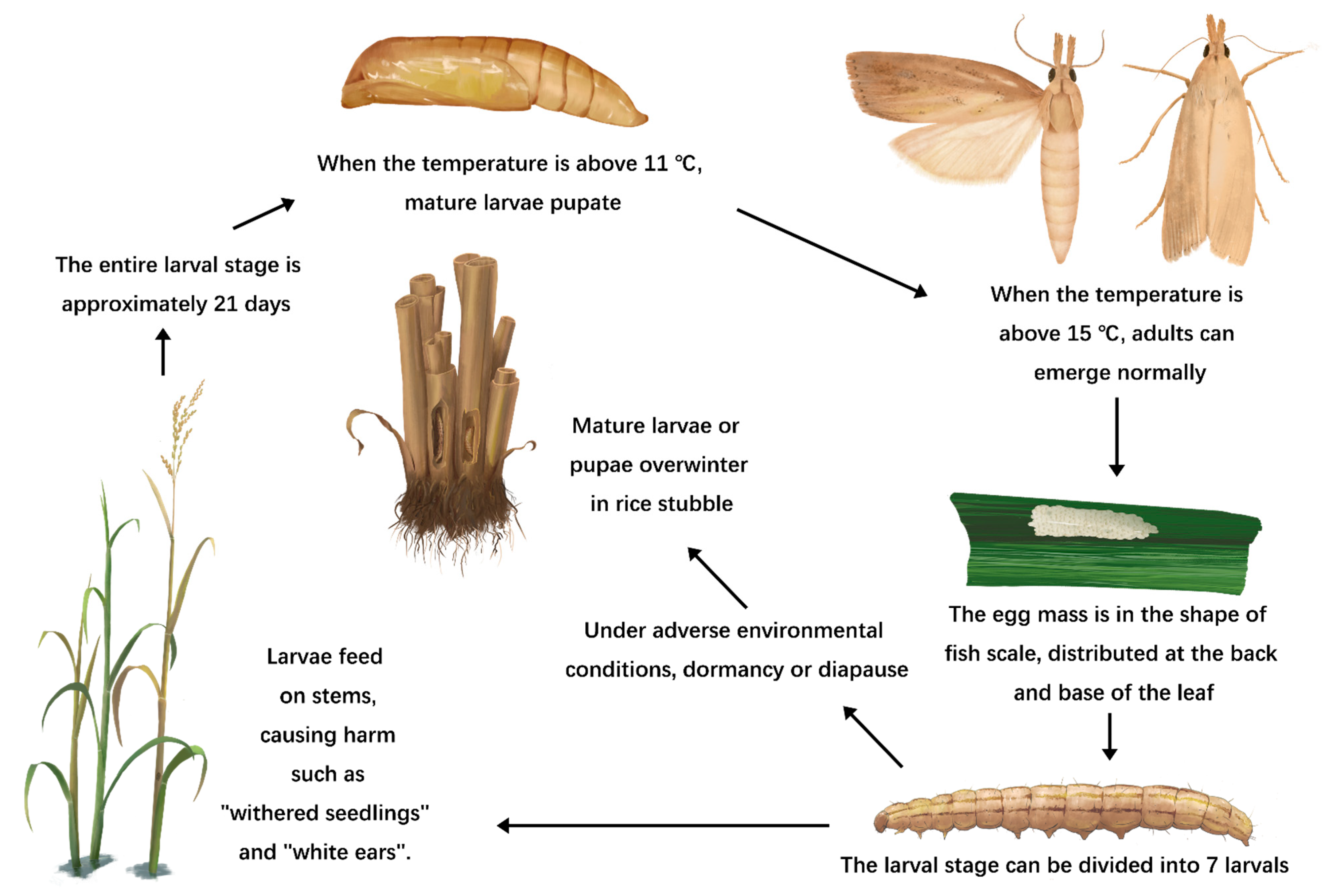
| Rice Cultivar | Transformation Method | Type | Gene | Origin | Comments | References |
|---|---|---|---|---|---|---|
| Nipponbare | Agrobacterium mediated | Overexpression | mwti1b | Winged bean | Trypsin inhibitor | Mochizuki et al. (1999) [106] |
| Xiushui 11, Chunjiang 11 | Agrobacterium mediated | Overexpression | SpI | Spider | Spider insect toxin | Huang et al. (2001) [107] |
| Senia, Ariete | particle bombardment and Agrobacterium mediated | Overexpression | mpi | Maize | Proteinase inhibitor | Vila et al. (2005) [108] |
| Xiushui 11 | Agrobacterium mediated | Antisense expression | OsHI-LOX | Rice | Type 2 13-lipoxygenase | Zhou et al. (2009) [62] |
| Xiushui 110 | Agrobacterium mediated | Overexpression | OsERF3 | Rice | Ethylene-responsive factors | Lu et al. (2011) [66] |
| Minghui 63 | Agrobacterium mediated | Overexpression | Cry1Ab, Cry1AC, Cry1C, Cry2A | Bt strains | Insecticidal crystal protein | Yang et al. (2011) [109] |
| Zhonghua 11 | γ-rays | Mutation | OsHPL3 | Rice | A hydroperoxide lyase | Tong et al. (2012) [80] |
| Xiushui 11 | Agrobacterium mediated | RNAi | OsMPK3 | Rice | Mitogen-activated protein kinases | Wang et al. (2013) [65] |
| Zhonghua 11 | Agrobacterium mediated | Overexpression | OsAOC | Rice | Aladiene oxide cyclase | Guo et al. (2014) [78] |
| Zhonghua 11 | Agrobacterium mediated | Overexpression | OsOPR3 | Rice | Cis-12-oxo-phytodienoic acid reductase 3 | Guo et al. (2014) [78] |
| Rice cultivar | Transformation method | Type | Gene | Origin | Comments | References |
| Xiushui 11 | Agrobacterium mediated | Antisense expression | Osr9-LOX1 | Rice | 9-lipoxygenase | Zhou et al. (2014) [77] |
| Ariete | Agrobacterium mediated | Overexpression | mpi, pci | Maize, Potato | Maize proteinase inhibitor, potato carboxypeptidase inhibitor | Quilis et al. (2014) [101] |
| Xiushui 11 | Agrobacterium mediated | RNAi | OsWRKY53 | Rice | Transcription factor | Hu et al. (2015) [67] |
| Zhonghua 11 | Agrobacterium mediated | HIGS | csu-novel-miR15 | SSB | Insect endogenous small RNAs | Jiang et al. (2016) [110] |
| Xiushui 134 | Agrobacterium mediated | Overexpression | Cry1Ab, Vip3A | Bt strains | Insecticidal crystal protein | Xu et al. (2018) [111] |
| Minghui 63 | Agrobacterium mediated | Overexpression | Cry1C | Bt strains | Insecticidal crystal protein | Jiao et al. (2018) [112] |
| Xidao NO1 | Lipofectin transfection | knocking out | CYP71A1 | Rice | Tryptamine 5-hydroxylase | Lu et al. (2018) [16] |
| Zhonghua 11 | Agrobacterium mediated | HIGS | miR-14 | SSB | Insect endogenous small RNAs | He et al. (2019) [113] |
| Zhonghua 11 | Agrobacterium mediated | Overexpression | OsHPL2 promoter and Cry1C | Rice and Bt strains | SSB-inducible promoter of rice gene, OsHPL2 | Li et al. (2020) [7] |
| Zhonghua 11 | Agrobacterium mediated | T-DNA insertion | om64 | Rice | rice mitochondrial outer membrane protein 64 | Guo et al. (2020) [61] |
| Rice cultivar | Transformation method | Type | Gene | Origin | Comments | References |
| Nipponbare | Agrobacterium mediated | Overexpression | APIP4-OX-16-2 | Rice | the Bowman–Birk inhibitor AvrPiz-t interacting protein 4 | Liu et al. (2021) [76] |
| Zhonghua 11 | Agrobacterium mediated | HIGS | csu-novel-260 | SSB | Insect endogenous small RNAs | Zheng et al. (2021) [27] Wen et al. (2021) [114] |
| Zhonghua 11 | Agrobacterium mediated | HIGS | csu-novel-miR53 | SSB | Insect endogenous small RNAs | Liu et al. (2022) [115] |
| Zhonghua 11 | Agrobacterium mediated | HIGS | CssHsp | SSB | Small heat shock protein | Mao et al. (2022) [116] |
| Zhonghua 11 | Agrobacterium mediated | HIGS | CsFAR | SSB | Fatty acyl-CoA reductase | Sun et al. (2022) [117] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiang, X.; Liu, S.; Li, H.; Danso Ofori, A.; Yi, X.; Zheng, A. Defense Strategies of Rice in Response to the Attack of the Herbivorous Insect, Chilo suppressalis. Int. J. Mol. Sci. 2023, 24, 14361. https://doi.org/10.3390/ijms241814361
Xiang X, Liu S, Li H, Danso Ofori A, Yi X, Zheng A. Defense Strategies of Rice in Response to the Attack of the Herbivorous Insect, Chilo suppressalis. International Journal of Molecular Sciences. 2023; 24(18):14361. https://doi.org/10.3390/ijms241814361
Chicago/Turabian StyleXiang, Xing, Shuhua Liu, Hongjian Li, Andrews Danso Ofori, Xiaoqun Yi, and Aiping Zheng. 2023. "Defense Strategies of Rice in Response to the Attack of the Herbivorous Insect, Chilo suppressalis" International Journal of Molecular Sciences 24, no. 18: 14361. https://doi.org/10.3390/ijms241814361
APA StyleXiang, X., Liu, S., Li, H., Danso Ofori, A., Yi, X., & Zheng, A. (2023). Defense Strategies of Rice in Response to the Attack of the Herbivorous Insect, Chilo suppressalis. International Journal of Molecular Sciences, 24(18), 14361. https://doi.org/10.3390/ijms241814361






