Differential Sensitivity of Germline and Somatic BRCA Variants to PARP Inhibitor in High-Grade Serous Ovarian Cancer
Abstract
:1. Introduction
2. Results
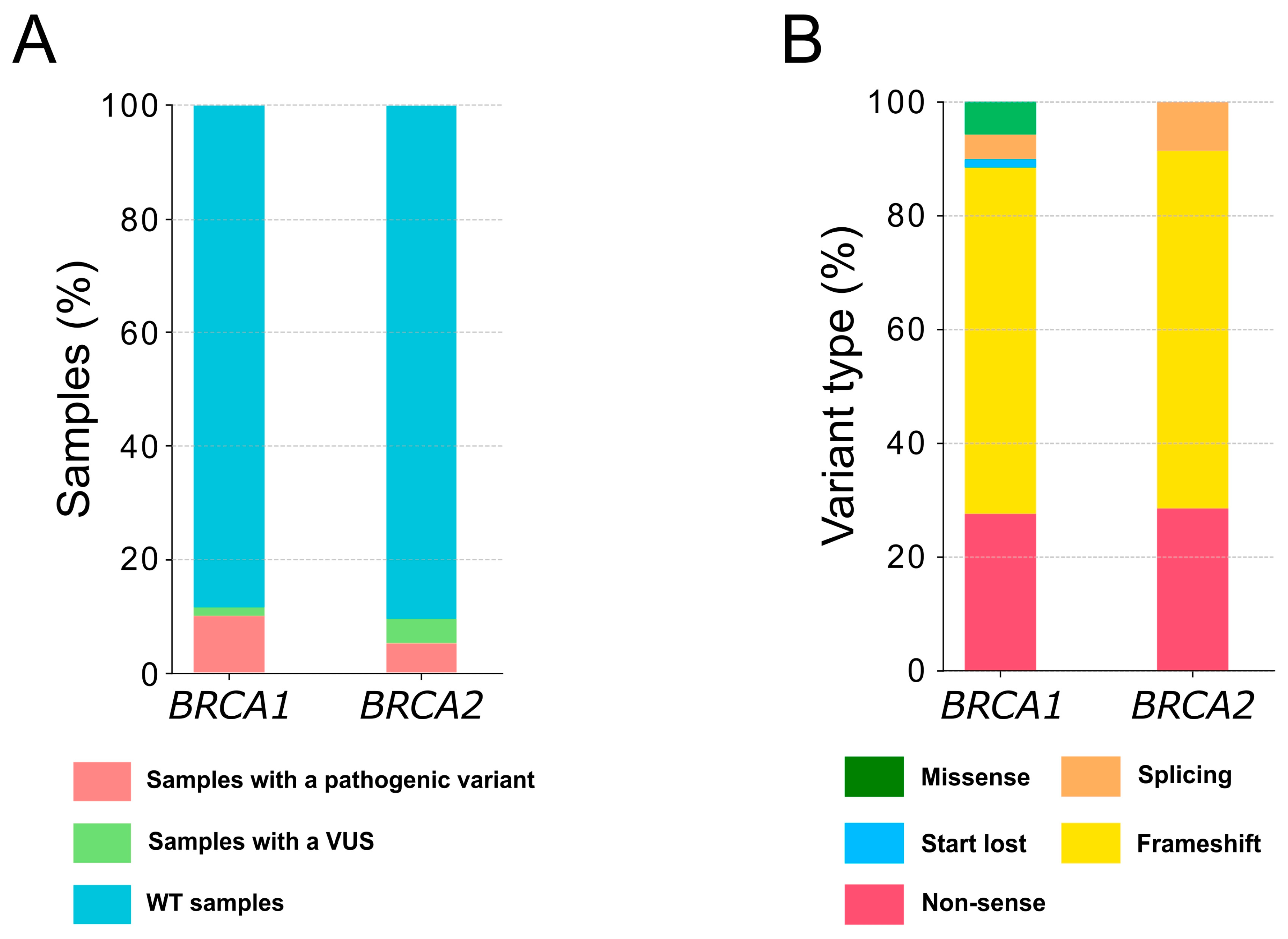
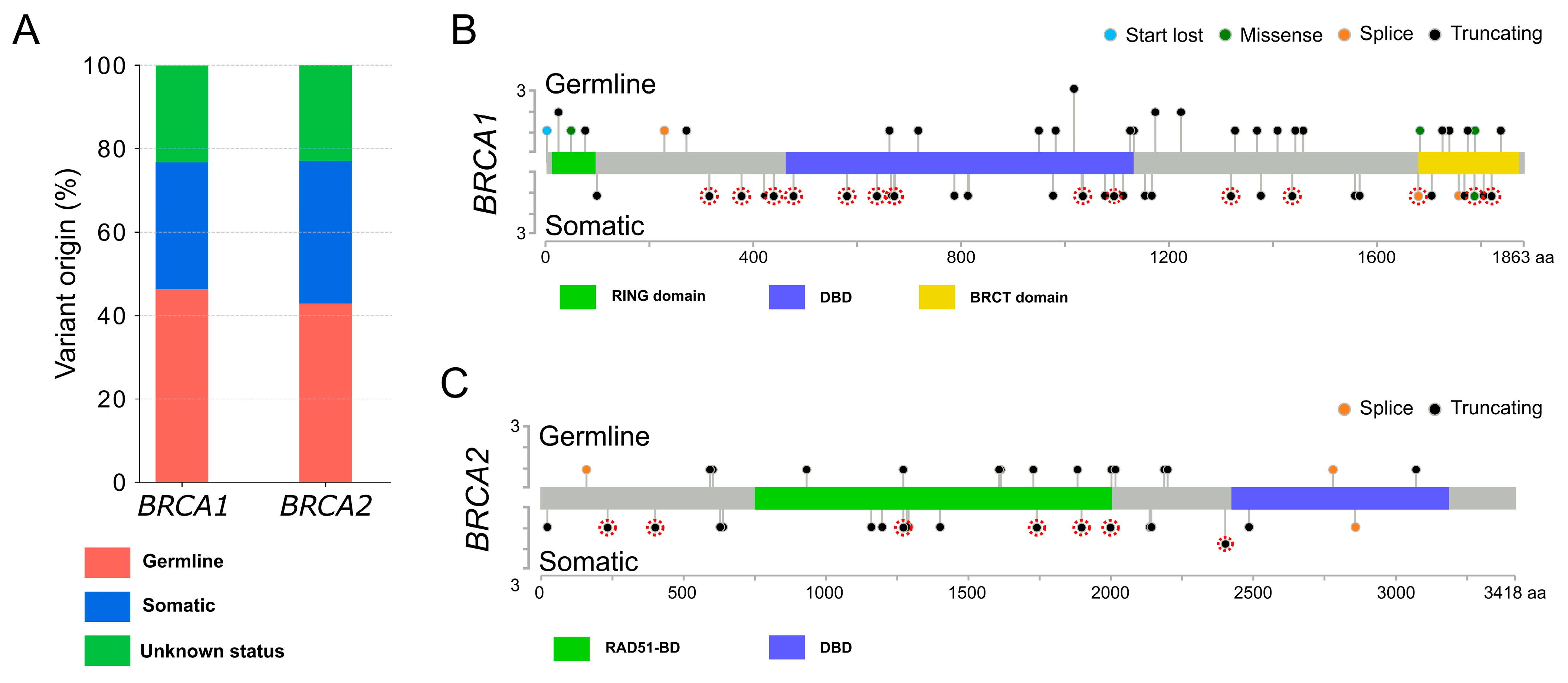
2.1. Patient Cohort and Alteration Descriptions
2.2. Clinicopathological Characteristics of the Patients with a Pathogenic Variant
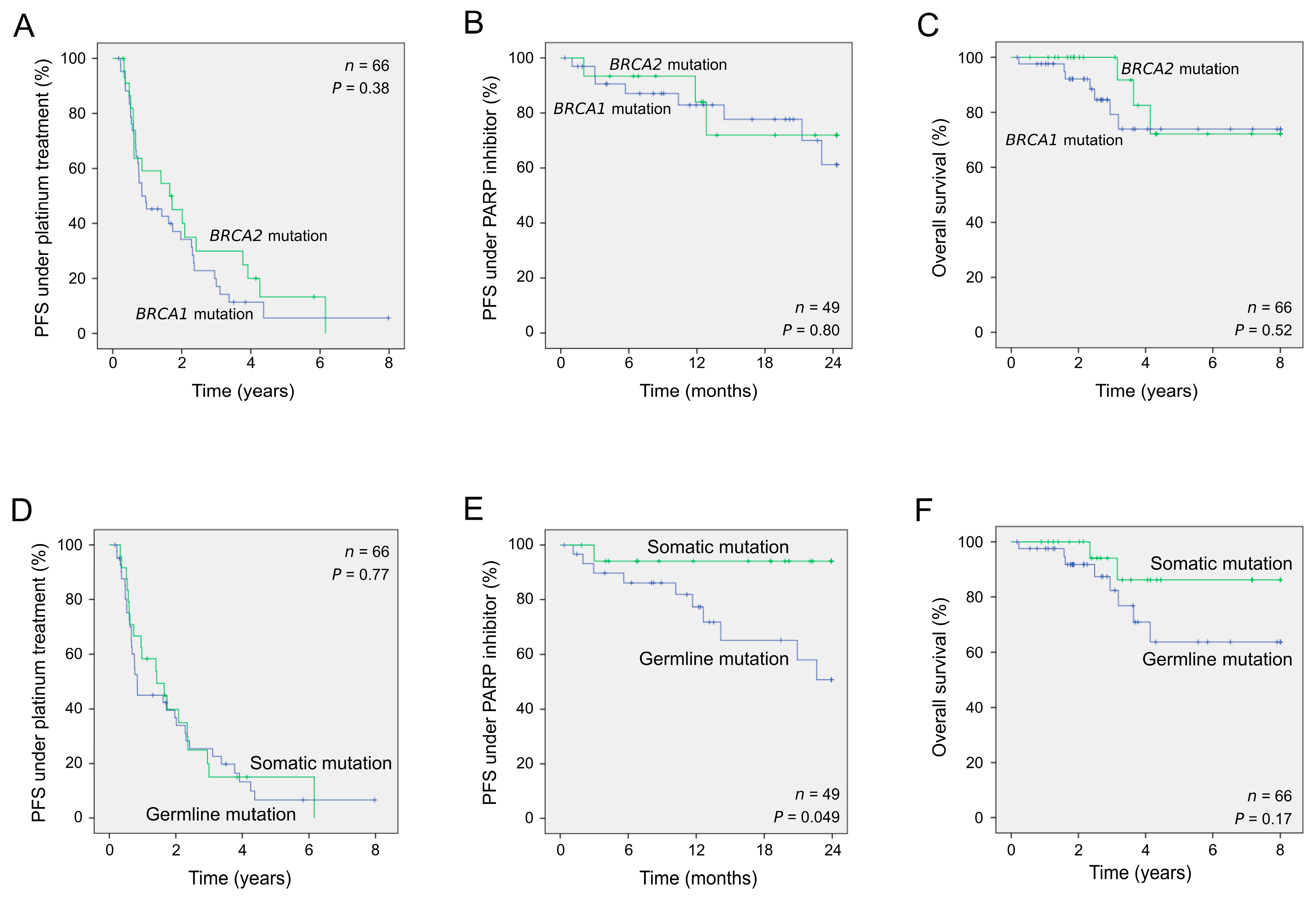
2.3. Clinical Outcome of Patients According to the Presence of Germline or Somatic Pathogenic Alterations
3. Discussion
4. Material and Methods
4.1. Patients and Samples Collection
4.2. DNA Extraction and Qualification
4.3. BRCA1 and BRCA2 Variants Analysis
4.4. Bioinformatics and Data Analysis
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Prat, J. FIGO Committee on Gynecologic Oncology FIGO’s Staging Classification for Cancer of the Ovary, Fallopian Tube, and Peritoneum: Abridged Republication. J. Gynecol. Oncol. 2015, 26, 87–89. [Google Scholar] [CrossRef]
- Moore, K.; Colombo, N.; Scambia, G.; Kim, B.-G.; Oaknin, A.; Friedlander, M.; Lisyanskaya, A.; Floquet, A.; Leary, A.; Sonke, G.S.; et al. Maintenance Olaparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2018, 379, 2495–2505. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Li, N.; Sun, Y.; Li, B.; Xu, L.; Wu, L. Clinical Outcome and Prognostic Factors of Patients with Early-Stage Epithelial Ovarian Cancer. Oncotarget 2017, 8, 23862–23870. [Google Scholar] [CrossRef] [PubMed]
- Lisio, M.-A.; Fu, L.; Goyeneche, A.; Gao, Z.-H.; Telleria, C. High-Grade Serous Ovarian Cancer: Basic Sciences, Clinical and Therapeutic Standpoints. Int. J. Mol. Sci. 2019, 20, 952. [Google Scholar] [CrossRef] [PubMed]
- Gomez, M.K.; Illuzzi, G.; Colomer, C.; Churchman, M.; Hollis, R.L.; O’Connor, M.J.; Gourley, C.; Leo, E.; Melton, D.W. Identifying and Overcoming Mechanisms of PARP Inhibitor Resistance in Homologous Recombination Repair-Deficient and Repair-Proficient High Grade Serous Ovarian Cancer Cells. Cancers 2020, 12, 1503. [Google Scholar] [CrossRef]
- Slade, D. PARP and PARG Inhibitors in Cancer Treatment. Genes Dev. 2020, 34, 360–394. [Google Scholar] [CrossRef] [PubMed]
- Labidi-Galy, S.I.; Rodrigues, M.; Sandoval, J.L.; Kurtz, J.E.; Heitz, F.; Mosconi, A.M.; Romero, I.; Denison, U.; Nagao, S.; Vergote, I.; et al. Association of Location of BRCA1 and BRCA2 Mutations with Benefit from Olaparib and Bevacizumab Maintenance in High-Grade Ovarian Cancer: Phase III PAOLA-1/ENGOT-Ov25 Trial Subgroup Exploratory Analysis. Ann. Oncol. 2023, 34, 152–162. [Google Scholar] [CrossRef]
- Al Hadidi, S.; Aburahma, A.; Badami, S.; Upadhaya, S. PARP (Poly(ADP-Ribose) Polymerase) Inhibitors in Platinum-Sensitive Recurrent Ovarian Cancer: A Meta-Analysis of Randomized Controlled Trials. Oncol. Res. Treat. 2018, 41, 226–235. [Google Scholar] [CrossRef]
- Caruso, G.; Tomao, F.; Parma, G.; Lapresa, M.; Multinu, F.; Palaia, I.; Aletti, G.; Colombo, N. Poly (ADP-Ribose) Polymerase Inhibitors (PARPi) in Ovarian Cancer: Lessons Learned and Future Directions. Int. J. Gynecol. Cancer 2023, 33, 431–443. [Google Scholar] [CrossRef]
- Tomao, F.; Bardhi, E.; Di Pinto, A.; Sassu, C.M.; Biagioli, E.; Petrella, M.C.; Palaia, I.; Muzii, L.; Colombo, N.; Panici, P.B. Parp Inhibitors as Maintenance Treatment in Platinum Sensitive Recurrent Ovarian Cancer: An Updated Meta-Analysis of Randomized Clinical Trials According to BRCA Mutational Status. Cancer Treat. Rev. 2019, 80, 101909. [Google Scholar] [CrossRef]
- Abecassis, I.; Sedgewick, A.J.; Romkes, M.; Buch, S.; Nukui, T.; Kapetanaki, M.G.; Vogt, A.; Kirkwood, J.M.; Benos, P.V.; Tawbi, H. PARP1 Rs1805407 Increases Sensitivity to PARP1 Inhibitors in Cancer Cells Suggesting an Improved Therapeutic Strategy. Sci. Rep. 2019, 9, 3309. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, Z.-Y.; Wu, N.; Chen, Y.-C.; Cheng, Q.; Wang, J. PARP Inhibitor Resistance: The Underlying Mechanisms and Clinical Implications. Mol. Cancer 2020, 19, 107. [Google Scholar] [CrossRef] [PubMed]
- Ragupathi, A.; Singh, M.; Perez, A.M.; Zhang, D. Targeting the BRCA1/2 Deficient Cancer with PARP Inhibitors: Clinical Outcomes and Mechanistic Insights. Front. Cell Dev. Biol. 2023, 11, 1133472. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, C.; Betella, I.; Rappa, A.; di Giminiani, M.; Gaiano, M.; De Vitis, L.A.; Zambetti, B.; Vacirca, D.; Multinu, F.; Venetis, K.; et al. Tumor BRCA Testing in Epithelial Ovarian Cancers: Past and Future-Five-Years’ Single-Institution Experience of 762 Consecutive Patients. Cancers 2022, 14, 1638. [Google Scholar] [CrossRef] [PubMed]
- Palacios, J.; de la Hoya, M.; Bellosillo, B.; de Juan, I.; Matías-Guiu, X.; Lázaro, C.; Palanca, S.; Osorio, A.; Rojo, F.; Rosa-Rosa, J.M.; et al. Mutational Screening of BRCA1/2 Genes as a Predictive Factor for Therapeutic Response in Epithelial Ovarian Cancer: A Consensus Guide from the Spanish Society of Pathology (SEAP-IAP) and the Spanish Society of Human Genetics (AEGH). Virchows Arch. 2020, 476, 195–207. [Google Scholar] [CrossRef]
- Pinto, C.; Bella, M.A.; Capoluongo, E.; Carrera, P.; Clemente, C.; Colombo, N.; Cortesi, L.; De Rosa, G.; Fenizia, F.; Genuardi, M.; et al. Recommendations for the Implementation of BRCA Testing in the Care and Treatment Pathways of Ovarian Cancer Patients. Future Oncol. 2016, 12, 2071–2075. [Google Scholar] [CrossRef]
- Konstantinopoulos, P.A.; Ceccaldi, R.; Shapiro, G.I.; D’Andrea, A.D. Homologous Recombination Deficiency: Exploiting the Fundamental Vulnerability of Ovarian Cancer. Cancer Discov. 2015, 5, 1137–1154. [Google Scholar] [CrossRef]
- Coleman, R.L.; Oza, A.M.; Lorusso, D.; Aghajanian, C.; Oaknin, A.; Dean, A.; Colombo, N.; Weberpals, J.I.; Clamp, A.; Scambia, G.; et al. Rucaparib Maintenance Treatment for Recurrent Ovarian Carcinoma after Response to Platinum Therapy (ARIEL3): A Randomised, Double-Blind, Placebo-Controlled, Phase 3 Trial. Lancet 2017, 390, 1949–1961. [Google Scholar] [CrossRef]
- Mirza, M.R.; Monk, B.J.; Herrstedt, J.; Oza, A.M.; Mahner, S.; Redondo, A.; Fabbro, M.; Ledermann, J.A.; Lorusso, D.; Vergote, I.; et al. Niraparib Maintenance Therapy in Platinum-Sensitive, Recurrent Ovarian Cancer. N. Engl. J. Med. 2016, 375, 2154–2164. [Google Scholar] [CrossRef]
- Ray-Coquard, I.; Pautier, P.; Pignata, S.; Pérol, D.; González-Martín, A.; Berger, R.; Fujiwara, K.; Vergote, I.; Colombo, N.; Mäenpää, J.; et al. Olaparib plus Bevacizumab as First-Line Maintenance in Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2416–2428. [Google Scholar] [CrossRef]
- Ray-Coquard, I.; Leary, A.; Pignata, S.; Cropet, C.; González-Martín, A.; Marth, C.; Nagao, S.; Vergote, I.; Colombo, N.; Mäenpää, J.; et al. Olaparib plus Bevacizumab First-Line Maintenance in Ovarian Cancer: Final Overall Survival Results from the PAOLA-1/ENGOT-Ov25 Trial. Ann. Oncol. 2023, 34, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Musella, A.; Vertechy, L.; Romito, A.; Marchetti, C.; Giannini, A.; Sciuga, V.; Bracchi, C.; Tomao, F.; Di Donato, V.; De Felice, F.; et al. Bevacizumab in Ovarian Cancer: State of the Art and Unanswered Questions. Chemotherapy 2017, 62, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Monk, B.J.; Parkinson, C.; Lim, M.C.; O’Malley, D.M.; Oaknin, A.; Wilson, M.K.; Coleman, R.L.; Lorusso, D.; Bessette, P.; Ghamande, S.; et al. A Randomized, Phase III Trial to Evaluate Rucaparib Monotherapy as Maintenance Treatment in Patients with Newly Diagnosed Ovarian Cancer (ATHENA-MONO/GOG-3020/ENGOT-Ov45). J. Clin. Oncol. 2022, 40, 3952–3964. [Google Scholar] [CrossRef] [PubMed]
- González-Martín, A.; Pothuri, B.; Vergote, I.; DePont Christensen, R.; Graybill, W.; Mirza, M.R.; McCormick, C.; Lorusso, D.; Hoskins, P.; Freyer, G.; et al. Niraparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2391–2402. [Google Scholar] [CrossRef]
- Pennington, K.P.; Walsh, T.; Harrell, M.I.; Lee, M.K.; Pennil, C.C.; Rendi, M.H.; Thornton, A.; Norquist, B.M.; Casadei, S.; Nord, A.S.; et al. Germline and Somatic Mutations in Homologous Recombination Genes Predict Platinum Response and Survival in Ovarian, Fallopian Tube, and Peritoneal Carcinomas. Clin. Cancer Res. 2014, 20, 764–775. [Google Scholar] [CrossRef]
- Callens, C.; Vaur, D.; Soubeyran, I.; Rouleau, E.; Just, P.-A.; Guillerm, E.; Golmard, L.; Goardon, N.; Sevenet, N.; Cabaret, O.; et al. Concordance Between Tumor and Germline BRCA Status in High-Grade Ovarian Carcinoma Patients in the Phase III PAOLA-1/ENGOT-Ov25 Trial. J. Natl. Cancer Inst. 2021, 113, 917–923. [Google Scholar] [CrossRef]
- Hennessy, B.T.J.; Timms, K.M.; Carey, M.S.; Gutin, A.; Meyer, L.A.; Flake, D.D.; Abkevich, V.; Potter, J.; Pruss, D.; Glenn, P.; et al. Somatic Mutations in BRCA1 and BRCA2 Could Expand the Number of Patients That Benefit from Poly (ADP Ribose) Polymerase Inhibitors in Ovarian Cancer. J. Clin. Oncol. 2010, 28, 3570–3576. [Google Scholar] [CrossRef]
- Giannini, A.; Di Dio, C.; Di Donato, V.; D’oria, O.; Salerno, M.G.; Capalbo, G.; Cuccu, I.; Perniola, G.; Muzii, L.; Bogani, G. PARP Inhibitors in Newly Diagnosed and Recurrent Ovarian Cancer. Am. J. Clin. Oncol. 2023, 46, 414–419. [Google Scholar] [CrossRef]
- Perrin-Vidoz, L.; Sinilnikova, O.M.; Stoppa-Lyonnet, D.; Lenoir, G.M.; Mazoyer, S. The Nonsense-Mediated MRNA Decay Pathway Triggers Degradation of Most BRCA1 MRNAs Bearing Premature Termination Codons. Hum. Mol. Genet. 2002, 11, 2805–2814. [Google Scholar] [CrossRef]
- Wang, Y.; Krais, J.J.; Bernhardy, A.J.; Nicolas, E.; Cai, K.Q.; Harrell, M.I.; Kim, H.H.; George, E.; Swisher, E.M.; Simpkins, F.; et al. RING Domain-Deficient BRCA1 Promotes PARP Inhibitor and Platinum Resistance. J. Clin. Investig. 2016, 126, 3145–3157. [Google Scholar] [CrossRef]
- Hollis, R.L.; Churchman, M.; Gourley, C. Distinct Implications of Different BRCA Mutations: Efficacy of Cytotoxic Chemotherapy, PARP Inhibition and Clinical Outcome in Ovarian Cancer. OncoTargets Ther. 2017, 10, 2539–2551. [Google Scholar] [CrossRef] [PubMed]
- Mylavarapu, S.; Das, A.; Roy, M. Role of BRCA Mutations in the Modulation of Response to Platinum Therapy. Front. Oncol. 2018, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Hyman, D.M.; Zhou, Q.; Iasonos, A.; Grisham, R.N.; Arnold, A.G.; Phillips, M.F.; Bhatia, J.; Levine, D.A.; Aghajanian, C.; Offit, K.; et al. Improved Survival for BRCA2-Associated Serous Ovarian Cancer Compared with Both BRCA-Negative and BRCA1-Associated Serous Ovarian Cancer. Cancer 2012, 118, 3703–3709. [Google Scholar] [CrossRef]
- Yang, D.; Khan, S.; Sun, Y.; Hess, K.; Shmulevich, I.; Sood, A.K.; Zhang, W. Association of BRCA1 and BRCA2 Mutations with Survival, Chemotherapy Sensitivity, and Gene Mutator Phenotype in Patients with Ovarian Cancer. JAMA 2011, 306, 1557–1565. [Google Scholar] [CrossRef]
- Kim, S.I.; Lee, M.; Kim, H.S.; Chung, H.H.; Kim, J.-W.; Park, N.H.; Song, Y.-S. Effect of BRCA Mutational Status on Survival Outcome in Advanced-Stage High-Grade Serous Ovarian Cancer. J. Ovarian Res. 2019, 12, 40. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Brosnan, M.; Sokol, E.S.; Xie, M.; Dry, J.R.; Harrington, E.A.; Barrett, J.C.; Hodgson, D. Landscape of Homologous Recombination Deficiencies in Solid Tumours: Analyses of Two Independent Genomic Datasets. BMC Cancer 2022, 22, 13. [Google Scholar] [CrossRef]
- Lim, Y.W.; Chen-Harris, H.; Mayba, O.; Lianoglou, S.; Wuster, A.; Bhangale, T.; Khan, Z.; Mariathasan, S.; Daemen, A.; Reeder, J.; et al. Germline genetic polymorphisms influence tumor gene expression and immune cell infiltration. Proc. Natl. Acad. Sci. USA 2018, 115, E11701–E11710. [Google Scholar] [CrossRef]

| Patients (n = 104) | ||
|---|---|---|
| Age at diagnosis | ||
| Median (range) | 63.4 (30.4–87.0) | |
| <60 | 41 | |
| ≥60 | 63 | |
| FIGO Stage | ||
| I | 6 | |
| II | 3 | |
| III | 36 | |
| IV | 19 | |
| Unknown | 40 | |
| Type of specimen | ||
| Biopsy | 28 | |
| Surgical specimen | 70 | |
| Unknown | 6 | |
| Tumor cell content | ||
| <50% | 16 | |
| ≥50% | 85 | |
| Unknown | 3 | |
| Clinical follow-up | ||
| Yes | 91 | |
| No | 13 | |
| PARP inhibitor administration | ||
| Yes | 58 | |
| No | 33 | |
| Unknown | 13 | |
| Germline status | ||
| Known | 80 | |
| Unknown | 24 | |
| Mutation present | ||
| At the germline level | 47 | |
| At the somatic level only | 33 | |
| Genetic result not available | 24 | |
| Gene mutated | ||
| BRCA1 | 69 | |
| BRCA2 | 35 | |
| Number of Samples | Univariate Analysis | ||||
|---|---|---|---|---|---|
| HR | 95% CI | p a | |||
| PFS to first-line platinum-based chemotherapy | |||||
| Gene affected (BRCA1; BRCA2) | 66 | 1.30 | 0.74–2.27 | 0.38 | |
| BRCA genetic status (germline; somatic) | 66 | 1.09 | 0.63–1.91 | 0.77 | |
| PFS to second-line PARP inhibitor treatment | |||||
| Gene affected (BRCA1; BRCA2) | 49 | 1.17 | 0.32–4.24 | 0.80 | |
| BRCA genetic status (germline; somatic) | 49 | 3.42 | 1.01–11.63 | 0.049 | |
| OS | |||||
| Gene affected (BRCA1; BRCA2) | 66 | 1.51 | 0.43–5.39 | 0.52 | |
| BRCA genetic status (germline; somatic) | 66 | 2.40 | 0.68–8.48 | 0.17 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vendrell, J.A.; Ban, I.O.; Solassol, I.; Audran, P.; Cabello-Aguilar, S.; Topart, D.; Lindet-Bourgeois, C.; Colombo, P.-E.; Legouffe, E.; D’Hondt, V.; et al. Differential Sensitivity of Germline and Somatic BRCA Variants to PARP Inhibitor in High-Grade Serous Ovarian Cancer. Int. J. Mol. Sci. 2023, 24, 14181. https://doi.org/10.3390/ijms241814181
Vendrell JA, Ban IO, Solassol I, Audran P, Cabello-Aguilar S, Topart D, Lindet-Bourgeois C, Colombo P-E, Legouffe E, D’Hondt V, et al. Differential Sensitivity of Germline and Somatic BRCA Variants to PARP Inhibitor in High-Grade Serous Ovarian Cancer. International Journal of Molecular Sciences. 2023; 24(18):14181. https://doi.org/10.3390/ijms241814181
Chicago/Turabian StyleVendrell, Julie A., Iulian O. Ban, Isabelle Solassol, Patricia Audran, Simon Cabello-Aguilar, Delphine Topart, Clothilde Lindet-Bourgeois, Pierre-Emmanuel Colombo, Eric Legouffe, Véronique D’Hondt, and et al. 2023. "Differential Sensitivity of Germline and Somatic BRCA Variants to PARP Inhibitor in High-Grade Serous Ovarian Cancer" International Journal of Molecular Sciences 24, no. 18: 14181. https://doi.org/10.3390/ijms241814181
APA StyleVendrell, J. A., Ban, I. O., Solassol, I., Audran, P., Cabello-Aguilar, S., Topart, D., Lindet-Bourgeois, C., Colombo, P.-E., Legouffe, E., D’Hondt, V., Fabbro, M., & Solassol, J. (2023). Differential Sensitivity of Germline and Somatic BRCA Variants to PARP Inhibitor in High-Grade Serous Ovarian Cancer. International Journal of Molecular Sciences, 24(18), 14181. https://doi.org/10.3390/ijms241814181






