Adipocyte-Derived Small Extracellular Vesicles from Patients with Alzheimer Disease Carry miRNAs Predicted to Target the CREB Signaling Pathway in Neurons
Abstract
:1. Introduction
2. Results
2.1. Subject Demographics
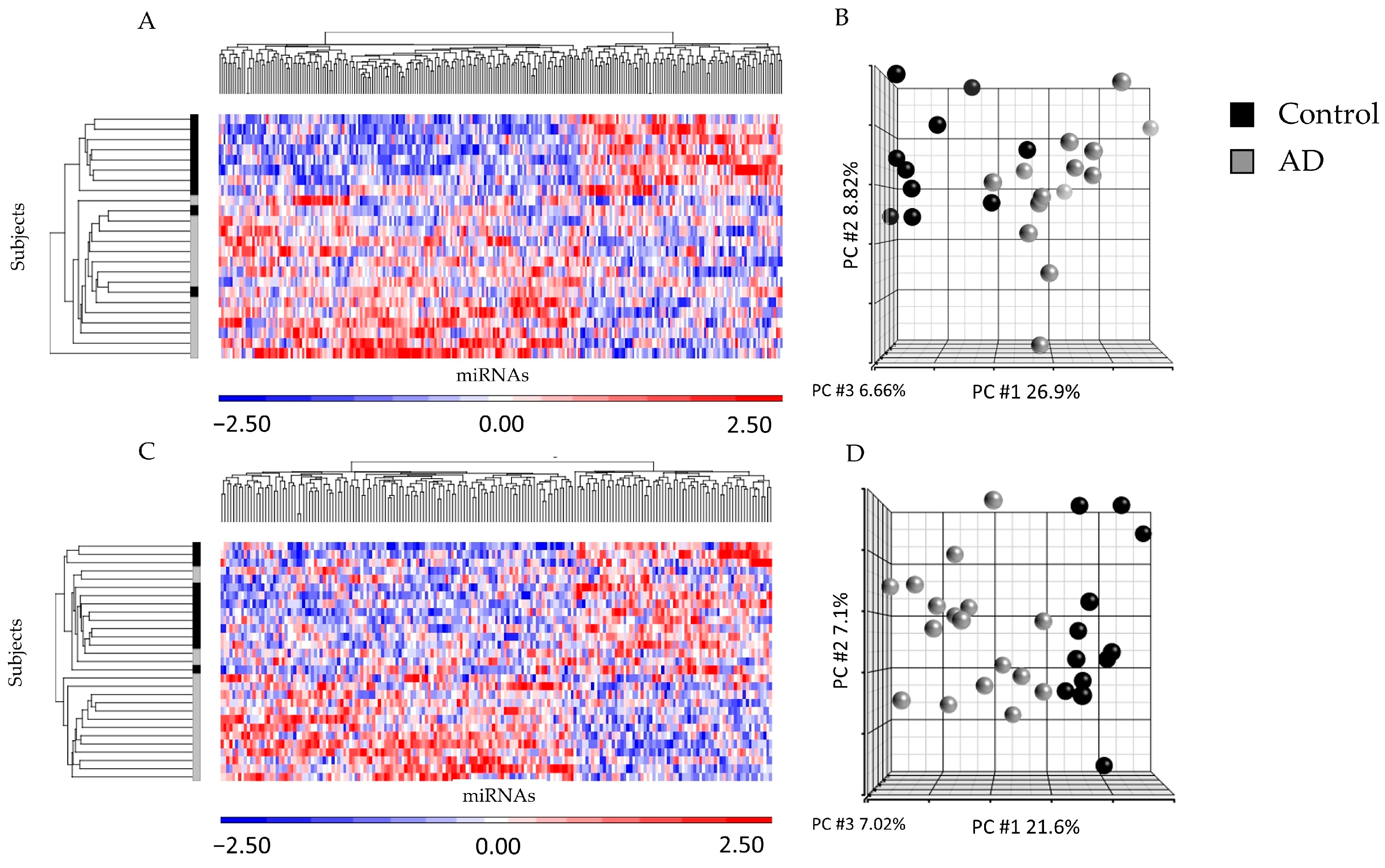
2.2. miRNA Profiling from ad-sEVs from Serum and CSF Identified Differences between Disease States
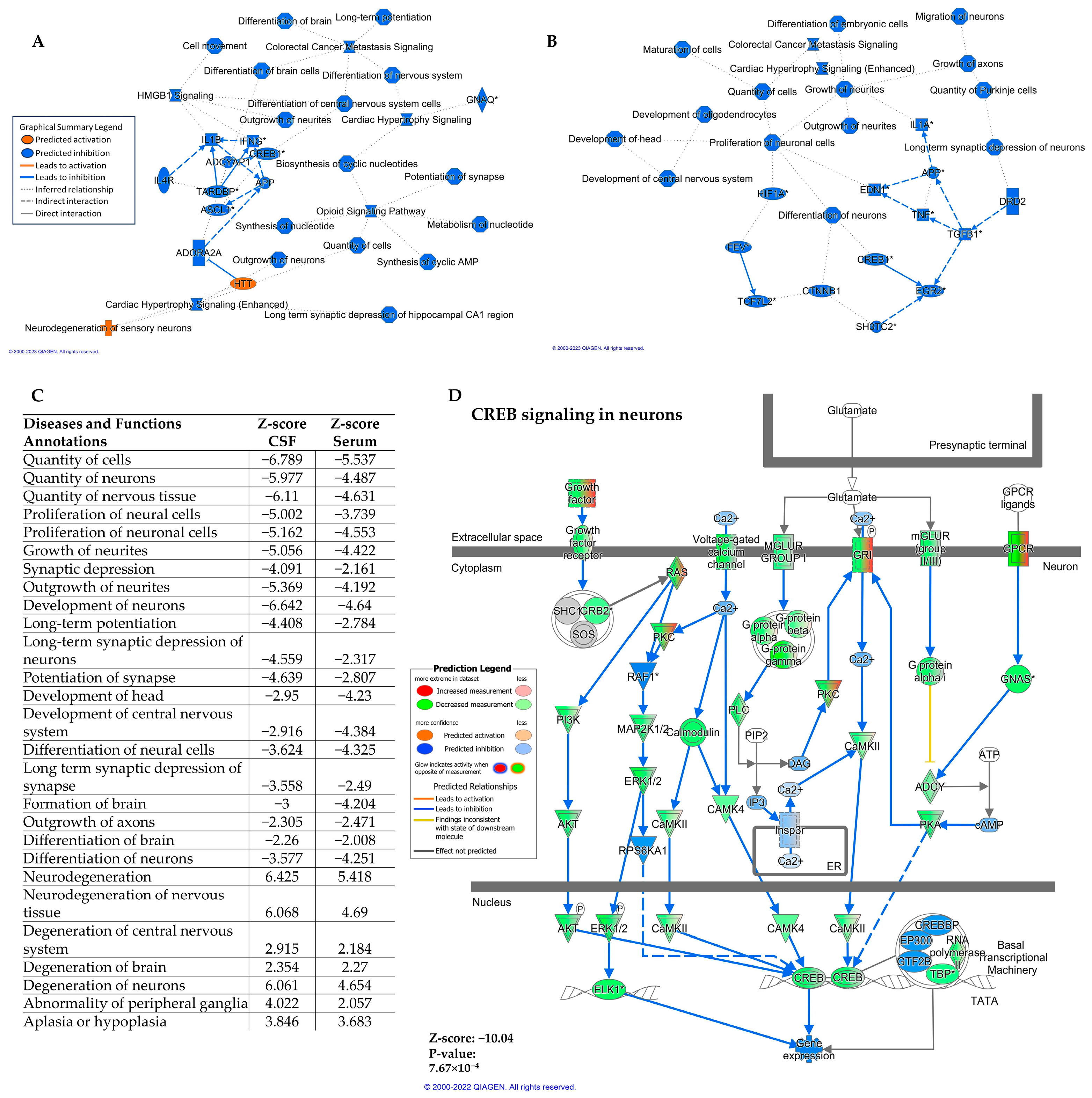
2.3. Differentially Expressed miRNA in Serum and CSF Are Predicted to Downregulate the CREB Signaling Pathway in Neurons
2.4. Identification and Functional Analysis of miRNA Clusters That Inversely Correlate with MMSE Scores
3. Discussion
4. Materials and Methods
4.1. Study Participants and Sample Collection
4.2. Adipocyte-Derived Small Extracellular Vesicle Isolation
4.3. RNA Extraction and Amplification
4.4. Adipocyte-Derived Small EV microRNA Profiles
4.5. Weighted Gene Co-Expression Network Analysis
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alzheimer’s Disease Facts and Figures. Alzheimers Dement. 2022, 2022, 700–789.
- Afsar, A.; Chacon Castro, M.D.C.; Soladogun, A.S.; Zhang, L. Recent Development in the Understanding of Molecular and Cellular Mechanisms Underlying the Etiopathogenesis of Alzheimer’s Disease. Int. J. Mol. Sci. 2023, 24, 7258. [Google Scholar] [CrossRef] [PubMed]
- de Wilde, M.C.; Overk, C.R.; Sijben, J.W.; Masliah, E. Meta-Analysis of Synaptic Pathology in Alzheimer’s Disease Reveals Selective Molecular Vesicular Machinery Vulnerability. Alzheimers Dement. 2016, 12, 633–644. [Google Scholar] [CrossRef]
- Gabrielli, M.; Prada, I.; Joshi, P.; Falcicchia, C.; D’Arrigo, G.; Rutigliano, G.; Battocchio, E.; Zenatelli, R.; Tozzi, F.; Radeghieri, A.; et al. Microglial Large Extracellular Vesicles Propagate Early Synaptic Dysfunction in Alzheimer’s Disease. Brain 2022, 145, 2849–2868. [Google Scholar] [CrossRef]
- Nilsson, J.; Cousins, K.A.Q.; Gobom, J.; Portelius, E.; Chen-Plotkin, A.; Shaw, L.M.; Grossman, M.; Irwin, D.J.; Trojanowski, J.Q.; Zetterberg, H.; et al. Cerebrospinal Fluid Biomarker Panel of Synaptic Dysfunction in Alzheimer’s Disease and Other Neurodegenerative Disorders. Alzheimers Dement. 2023, 19, 1775–1784. [Google Scholar] [CrossRef] [PubMed]
- Terry, R.D.; Masliah, E.; Salmon, D.P.; Butters, N.; DeTeresa, R.; Hill, R.; Hansen, L.A.; Katzman, R. Physical Basis of Cognitive Alterations in Alzheimer’s Disease: Synapse Loss Is the Major Correlate of Cognitive Impairment. Ann. Neurol. 1991, 30, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, M.B. Synaptic Signaling in Learning and Memory. Cold Spring Harb. Perspect. Biol. 2013, 8, a016824. [Google Scholar] [CrossRef]
- Teich, A.F.; Nicholls, R.E.; Puzzo, D.; Fiorito, J.; Purgatorio, R.; Fa Arancio, M. Synaptic Therapy in Alzheimer’s Disease: A CREB-Centric Approach. Neurotherapeutics 2015, 12, 29–41. [Google Scholar] [CrossRef]
- Biessels, G.J.; Staekenborg, S.; Brunner, E.; Brayne, C.; Scheltens, P. Risk of Dementia in Diabetes Mellitus: A Systematic Review. Lancet Neurol. 2006, 5, 64–74. [Google Scholar] [CrossRef]
- Reinke, C.; Buchmann, N.; Fink, A.; Tegeler, C.; Demuth, I.; Doblhammer, G. Diabetes Duration and the Risk of Dementia: A Cohort Study Based on German Health Claims Data. Age Ageing 2022, 51, afab231. [Google Scholar] [CrossRef]
- Erion, J.R.; Wosiski-Kuhn, M.; Dey, A.; Hao, S.; Davis, C.L.; Pollock, N.K.; Stranahan, A.M. Obesity Elicits Interleukin 1-Mediated Deficits in Hippocampal Synaptic Plasticity. J. Neurosci. 2014, 34, 2618–2631. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Ge, Q.; Wu, Y.; Zhang, J.; Gu, Q.; Han, J. Impairment of Long-Term Memory by a Short-Term High-Fat Diet via Hippo-Campal Oxidative Stress and Alterations in Synaptic Plasticity. Neuroscience 2020, 424, 24–33. [Google Scholar] [CrossRef]
- Whitmer, R.A.; Gustafson, D.R.; Barrett-Connor, E.; Haan, M.N.; Gunderson, E.P.; Yaffe, K. Central Obesity and Increased Risk of Dementia More than Three Decades Later. Neurology 2008, 71, 1057–1064. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Joshi, P.; Ang, T.F.A.; Liu, C.; Auerbach, S.; Devine, S.; Au, R. Mid- to Late-Life Body Mass Index and Dementia Risk: 38 Years of Follow-up of the Framingham Study. Am. J. Epidemiol. 2021, 190, 2503–2510. [Google Scholar] [CrossRef]
- Yu, J.-T.; Xu, W.; Tan, C.-C.; Andrieu, S.; Suckling, J.; Evangelou, E.; Pan, A.; Zhang, C.; Jia, J.; Feng, L. Evidence-Based Preven-Tion of Alzheimer’s Disease: Systematic Review and Meta-Analysis of 243 Observational Prospective Studies and 153 Random-Ised Controlled Trials. J. Neurol. Neurosurg. Psychiatry 2020, 91, 1201–1209. [Google Scholar] [CrossRef]
- Norton, S.; Matthews, F.E.; Barnes, D.E.; Yaffe, K.; Brayne, C. Potential for Primary Prevention of Alzheimer’s Disease: An Analysis of Population-Based Data. Lancet Neurol. 2014, 13, 788–794. [Google Scholar] [CrossRef] [PubMed]
- Ou, M.-Y.; Zhang, H.; Tan, P.-C.; Zhou, S.-B.; Li, Q.-F. Adipose Tissue Aging: Mechanisms and Therapeutic Implications. Cell Death Dis. 2022, 13, 300. [Google Scholar] [CrossRef] [PubMed]
- Kershaw, E.E.; Flier, J.S. Adipose Tissue as an Endocrine Organ. J. Clin. Endocrinol. Metab. 2004, 89, 2548–2556. [Google Scholar] [CrossRef]
- Ferrante, S.C.; Nadler, E.P.; Pillai, D.K.; Hubal, M.J.; Wang, Z.; Wang, J.M.; Gordish-Dressman, H.; Koeck, E.; Sevilla, S.; Wiles, A.A.; et al. Adipocyte-Derived Exosomal MiRNAs: A Novel Mechanism for Obesity-Related Disease. Pediatr. Res. 2015, 77, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Hubal, M.J.; Nadler, E.P.; Ferrante, S.C.; Barberio, M.D.; Suh, J.-H.; Wang, J.; Dohm, G.L.; Pories, W.J.; Mietus-Snyder, M.; Freishtat, R.J. Circulating Adipocyte-Derived Exosomal MicroRNAs Associated with Decreased Insulin Resistance after Gastric Bypass. Obesity 2017, 25, 102–110. [Google Scholar] [CrossRef]
- Barberio, M.D.; Kasselman, L.J.; Playford, M.P.; Epstein, S.B.; Renna, H.A.; Goldberg, M.; DeLeon, J.; Voloshyna, I.; Barlev, A.; Salama, M.; et al. Cholesterol Efflux Alterations in Adolescent Obesity: Role of Adipose-Derived Extracellular Vesical MicroRNAs. J. Transl. Med. 2019, 17, 232. [Google Scholar] [CrossRef] [PubMed]
- Morris, K.V.; Witwer, K.W. The Evolving Paradigm of Extracellular Vesicles in Intercellular Signaling and Delivery of Therapeutic RNAs. Mol. Ther. 2022, 30, 2393–2394. [Google Scholar] [CrossRef] [PubMed]
- Thomou, T.; Mori, M.A.; Dreyfuss, J.M.; Konishi, M.; Sakaguchi, M.; Wolfrum, C.; Rao, T.N.; Winnay, J.N.; Garcia-Martin, R.; Grinspoon, S.K.; et al. Adipose-Derived Circulating MiRNAs Regulate Gene Expression in Other Tissues. Yearb. Pediatr. Endocrinol. 2018, 542, 450–455. [Google Scholar] [CrossRef] [PubMed]
- Rome, S.; Blandin, A.; Le Lay, S. Adipocyte-Derived Extracellular Vesicles: State of the Art. Int. J. Mol. Sci. 2021, 22, 1788. [Google Scholar] [CrossRef] [PubMed]
- Koeck, E.S.; Iordanskaia, T.; Sevilla, S.; Ferrante, S.C.; Hubal, M.J.; Freishtat, R.J.; Nadler, E.P. Adipocyte Exosomes Induce Transforming Growth Factor Beta Pathway Dysregulation in Hepatocytes: A Novel Paradigm for Obesity-Related Liver Disease. J. Surg. Res. 2014, 192, 268–275. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal Analysis Approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Koffie, R.M.; Hyman, B.T.; Spires-Jones, T.L. Alzheimer’s Disease: Synapses Gone Cold. Mol. Neurodegener. 2011, 6, 63. [Google Scholar] [CrossRef]
- Fiandaca, M.S.; Kapogiannis, D.; Mapstone, M.; Boxer, A.; Eitan, E.; Schwartz, J.B.; Abner, E.L.; Petersen, R.C.; Federoff, H.J.; Miller, B.L. Identification of Preclinical Alzheimer’s Disease by a Profile of Pathogenic Proteins in Neurally Derived Blood Exo-Somes: A Case-Control Study. Alzheimers Dement. 2015, 11, 600–607. [Google Scholar] [CrossRef]
- Saman, S.; Kim, W.; Raya, M.; Visnick, Y.; Miro, S.; Saman, S.; Jackson, B.; McKee, A.C.; Alvarez, V.E.; Lee, N.C.Y.; et al. Exosome-Associated Tau Is Secreted in Tauopathy Models and Is Selectively Phosphorylated in Cerebrospinal Fluid in Early Alzheimer Disease. J. Biol. Chem. 2012, 287, 3842–3849. [Google Scholar] [CrossRef]
- Sinha, S.; Ansell-Schultz, M.; Civitelli, A.; Hildesjö, L.; Larsson, C.; Lannfelt, M.; Ingelsson, L.; Hallbeck, M. Alzheimer’s Disease Pathology Propagation by Exosomes Containing Toxic Amyloid-Beta Oligomers. Acta Neuropathol. 2018, 136, 41–56. [Google Scholar] [CrossRef]
- Deng, Z.-B.; Poliakov, A.; Hardy, R.W.; Clements, R.; Liu, C.; Liu, Y.; Wang, J.; Xiang, X.; Zhang, S.; Zhuang, X.; et al. Adipose Tissue Exosome-like Vesicles Mediate Activation of Macrophage-Induced Insulin Resistance. Diabetes 2009, 58, 2498–2505. [Google Scholar] [CrossRef] [PubMed]
- Wadey, R.M.; Connolly, K.D.; Mathew, D.; Walters, G.; Rees, D.A.; James, P.E. Inflammatory Adipocyte-Derived Extracellular Vesicles Promote Leukocyte Attachment to Vascular Endothelial Cells. Atherosclerosis 2019, 283, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Tian, X.; Li, J.; Liu, D.; Ye, D.; Xie, Z.; Han, Y.; Zou, M.-H. A High-Fat Diet Attenuates AMPK A1 in Adipocytes to Induce Exosome Shedding and Nonalcoholic Fatty Liver Development in Vivo. Diabetes 2021, 70, 577–588. [Google Scholar] [CrossRef] [PubMed]
- Amidfar, M.; de Oliveira, J.; Kucharska, E.; Budni, J.; Kim, Y.-K. The Role of CREB and BDNF in Neurobiology and Treatment of Alzheimer’s Disease. Life Sci. 2020, 257, 118020. [Google Scholar] [CrossRef] [PubMed]
- Yiu, A.P.; Rashid, A.J.; Josselyn, S.A. Increasing CREB Function in the CA1 Region of Dorsal Hippocampus Rescues the Spatial Memory Deficits in a Mouse Model of Alzheimer’s Disease. Neuropsychopharmacology 2011, 36, 2169–2186. [Google Scholar] [CrossRef] [PubMed]
- Bartolotti, N.; Bennett, D.A.; Lazarov, O. Reduced PCREB in Alzheimer’s Disease Prefrontal Cortex Is Reflected in Peripheral Blood Mononuclear Cells. Mol. Psychiatry 2016, 21, 1158–1166. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto-Sasaki, M.; Ozawa, H.; Saito, T.; Rösler, M.; Riederer, P. Impaired Phosphorylation of Cyclic AMP Response Element Binding Protein in the Hippocampus of Dementia of the Alzheimer Type. Brain Res. 1999, 824, 300–303. [Google Scholar] [CrossRef]
- Pugazhenthi, S.; Wang, M.; Pham, S.; Sze, C.-I.; Eckman, C.B. Downregulation of CREB Expression in Alzheimer’s Brain and in Aβ-Treated Rat Hippocampal Neurons. Mol. Neurodegener. 2011, 6, 60. [Google Scholar] [CrossRef]
- Vitolo, O.V.; Sant’Angelo, A.; Costanzo, V.; Battaglia, F.; Arancio, O.; Shelanski, M. Amyloid Beta -Peptide Inhibition of the PKA/CREB Pathway and Long-Term Potentiation: Reversibility by Drugs That Enhance CAMP Signaling. Proc. Natl. Acad. Sci. USA 2002, 99, 13217–13221. [Google Scholar] [CrossRef]
- Chen, Z.; Sui, G.; Wang, L.; Yang, C.; Wang, F. High-Fat Diet Induced Hippocampal CREB Dysfunction, Cognitive Impairment and Depression-like Behaviors via Downregulation of Interleukin-2 in the Mice. Metab. Brain Dis. 2022, 37, 1163–1174. [Google Scholar] [CrossRef]
- Wang, J.; Li, L.; Zhang, Z.; Zhang, X.; Zhu, Y.; Zhang, C.; Bi, Y. Extracellular Vesicles Mediate the Communication of Adipose Tissue with Brain and Promote Cognitive Impairment Associated with Insulin Resistance. Cell Metab. 2022, 34, 1264–1279.e8. [Google Scholar] [CrossRef] [PubMed]
- Garzon, D.; Yu, G.; Fahnestock, M. A New Brain-Derived Neurotrophic Factor Transcript and Decrease in Brain-Derived Neu-Rotrophic Factor Transcripts 1, 2 and 3 in Alzheimer’s Disease Parietal Cortex. J. Neurochem. 2002, 82, 1058–1064. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Wuu, J.; Mufson, E.J.; Fahnestock, M. Precursor Form of Brain-Derived Neurotrophic Factor and Mature Brain-Derived Neurotrophic Factor Are Decreased in the Pre-Clinical Stages of Alzheimer’s Disease. J. Neurochem. 2005, 93, 1412–1421. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Liu, J.; Duan, F.; Cong, L.; Qi, X. The Role of the MicroRNA Regulatory Network in Alzheimer’s Disease: A Bioinformatics Analysis. Arch. Med. Sci. 2022, 18, 206–222. [Google Scholar] [CrossRef] [PubMed]
- Gomaa, A.A.; Farghaly, H.S.M.; Ahmed, A.M.; Hemida, F.K. Intermittent Treatment with Apremilast, a Phosphodiesterase-4 In-Hibitor, Ameliorates Alzheimer’s-like Pathology and Symptoms through Multiple Targeting Actions in Aged T2D Rats. Int. Immunopharmacol. 2023, 117, 109927. [Google Scholar] [CrossRef] [PubMed]
- McKhann, G.; Drachman, D.; Folstein, M.; Katzman, R.; Price, D.; Stadlan, E.M. Clinical Diagnosis of Alzheimer’s Disease: Report of the NINCDS-ADRDA Work Group under the Auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 1984, 34, 939–944. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Sokal, I.; Quinn, J.F.; Leverenz, J.B.; Brodey, M.; Schellenberg, G.D.; Kaye, J.A.; Raskind, M.A.; Zhang, J.; Peskind, E.R.; et al. CSF Tau/Abeta42 Ratio for Increased Risk of Mild Cognitive Impairment: A Follow-up Study. Neurology 2007, 69, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Millard, S.P.; Peskind, E.R.; Zhang, J.; Yu, C.-E.; Leverenz, J.B.; Mayer, C.; Shofer, J.S.; Raskind, M.A.; Quinn, J.F.; et al. Cross-Sectional and Longitudinal Relationships between Cerebrospinal Fluid Biomarkers and Cognitive Function in People without Cognitive Impairment from across the Adult Life Span. JAMA Neurol. 2014, 71, 742–751. [Google Scholar] [CrossRef]
- Siqueira, I.R.; de Souza Rodrigues, A.; Flores, M.S.; Vieira Cunha, E.L.; Goldberg, M.; Harmon, B.; Batabyal, R.; Freishtat, R.J.; Cechinel, L.R. Circulating Extracellular Vesicles and Particles Derived From Adipocytes: The Potential Role in Spreading MicroRNAs Associated With Cellular Senescence. Front. Aging 2022, 3, 867100. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Zhang, B.; Horvath, S. A General Framework for Weighted Gene Co-Expression Network Analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 17. [Google Scholar] [CrossRef]
- Bergeron, D.; Flynn, K.; Verret, L.; Poulin, S.; Bouchard, R.W.; Bocti, C.; Fülöp, T.; Lacombe, G.; Gauthier, S.; Nasreddine, Z.; et al. Multicenter Validation of an MMSE-MoCA Conversion Table. J. Am. Geriatr. Soc. 2017, 65, 1067–1072. [Google Scholar] [CrossRef]

| Alzheimer Disease (n = 18) | Controls (n = 14) | p-Value | |
|---|---|---|---|
| Age (years) | 73 ± 8 | 74 ± 9 | 0.83 |
| Male sex (n, %) | 9 (50%) | 7 (50%) | 0.88 |
| Race/Ethnicity | |||
| White (n, %) | 17 (94%) | 13 (93%) | n/a |
| Asian (n, %) | 1 (6%) | 1 (7%) | n/a |
| BMI, (kg/m2) | 24.8 ± 3.5 | 25.8 ± 4.7 | 0.52 |
| CDR® Score * | 1.25 ± 0.67 | 0.04 ± 0.13 | <0.001 |
| MMSE | 18.8 ± 5.4 | n/a | n/a |
| MoCA | n/a | 27.6 ± 2.5 | n/a |
| CSF Aβ1–42, (pg/mL) * | 134.5 ± 38.0 | 809.6 ± 241.8 | <0.001 |
| CSF Total Tau, (pg/mL) | 88.4 ± 62.2 | 75.7 ± 24.0 | 0.43 |
| Pathway Name | CSF z-Score | Serum z-Score |
|---|---|---|
| CREB Signaling in Neurons | −11.465 | −7.832 |
| Synaptogenesis Signaling Pathway | −7.717 | −3.578 |
| Gustation Pathway | −6.638 | −4.323 |
| GNRH Signaling | −6.38 | −4.523 |
| Ephrin Receptor Signaling | −6.14 | −2.846 |
| Oxytocin In Brain Signaling Pathway | −6.114 | −3.781 |
| SNARE Signaling Pathway | −5.921 | −2.795 |
| Synaptic Long-Term Depression | −5.897 | −3.111 |
| Cholecystokinin/Gastrin-mediated Signaling | −5.88 | −4.768 |
| Regulation of Actin-based Motility by Rho | −5.692 | −2.711 |
| NGF Signaling | −5.516 | −3.528 |
| Opioid Signaling Pathway | −5.07 | −2.828 |
| ERBB Signaling | −5.013 | −3 |
| Neurovascular Coupling Signaling Pathway | −4.93 | −3.75 |
| GPCR-Mediated Nutrient Sensing in Enteroendocrine Cells | −4.907 | −2.496 |
| GDNF Family Ligand-Receptor Interactions | −4.747 | −3.578 |
| Neurotrophin/TRK Signaling | −4.382 | −4.025 |
| Agrin Interactions at Neuromuscular Junction | −4.315 | −2.183 |
| Endocannabinoid Developing Neuron Pathway | −4.258 | −2.785 |
| Neuropathic Pain Signaling In Dorsal Horn Neurons | −4.117 | −2.357 |
| CNTF Signaling | −3.674 | −3.317 |
| Ephrin B Signaling | −3.651 | −2.357 |
| ERB2-ERBB3 Signaling | −3.402 | −2.183 |
| ERBB4 Signaling | −3.272 | −2.673 |
| Oxytocin In Spinal Neurons Signaling Pathway | −3.207 | −2.53 |
| Neuroinflammation Signaling Pathway | −3.161 | −3.597 |
| CDK5 Signaling | −3.015 | −4.017 |
| Dopamine Receptor Signaling | −2.714 | −2.236 |
| Dopamine-DARPP32 Feedback in cAMP Signaling | −2.534 | −3.333 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Batabyal, R.A.; Bansal, A.; Cechinel, L.R.; Authelet, K.; Goldberg, M.; Nadler, E.; Keene, C.D.; Jayadev, S.; Domoto-Reilly, K.; Li, G.; et al. Adipocyte-Derived Small Extracellular Vesicles from Patients with Alzheimer Disease Carry miRNAs Predicted to Target the CREB Signaling Pathway in Neurons. Int. J. Mol. Sci. 2023, 24, 14024. https://doi.org/10.3390/ijms241814024
Batabyal RA, Bansal A, Cechinel LR, Authelet K, Goldberg M, Nadler E, Keene CD, Jayadev S, Domoto-Reilly K, Li G, et al. Adipocyte-Derived Small Extracellular Vesicles from Patients with Alzheimer Disease Carry miRNAs Predicted to Target the CREB Signaling Pathway in Neurons. International Journal of Molecular Sciences. 2023; 24(18):14024. https://doi.org/10.3390/ijms241814024
Chicago/Turabian StyleBatabyal, Rachael A., Ankush Bansal, Laura Reck Cechinel, Kayla Authelet, Madeleine Goldberg, Evan Nadler, C. Dirk Keene, Suman Jayadev, Kimiko Domoto-Reilly, Gail Li, and et al. 2023. "Adipocyte-Derived Small Extracellular Vesicles from Patients with Alzheimer Disease Carry miRNAs Predicted to Target the CREB Signaling Pathway in Neurons" International Journal of Molecular Sciences 24, no. 18: 14024. https://doi.org/10.3390/ijms241814024
APA StyleBatabyal, R. A., Bansal, A., Cechinel, L. R., Authelet, K., Goldberg, M., Nadler, E., Keene, C. D., Jayadev, S., Domoto-Reilly, K., Li, G., Peskind, E., Hashimoto-Torii, K., Buchwald, D., & Freishtat, R. J. (2023). Adipocyte-Derived Small Extracellular Vesicles from Patients with Alzheimer Disease Carry miRNAs Predicted to Target the CREB Signaling Pathway in Neurons. International Journal of Molecular Sciences, 24(18), 14024. https://doi.org/10.3390/ijms241814024







