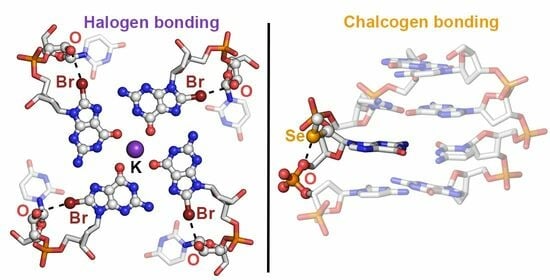
On the Importance of Halogen and Chalcogen Bonds in the Solid State of Nucleic Acids: A Combined Crystallographic and Theoretical Perspective
Abstract
:1. Introduction
2. Results and Discussion
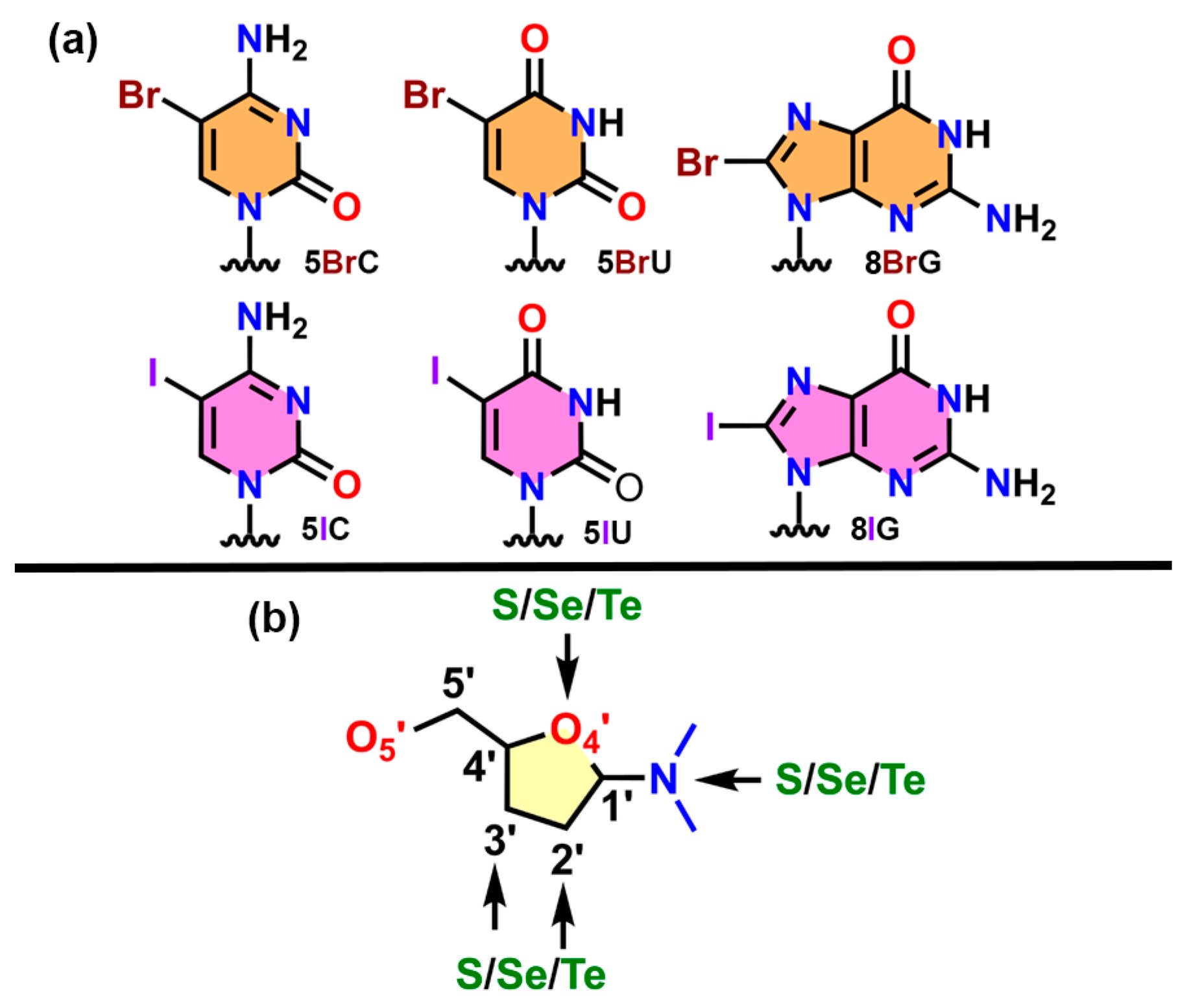
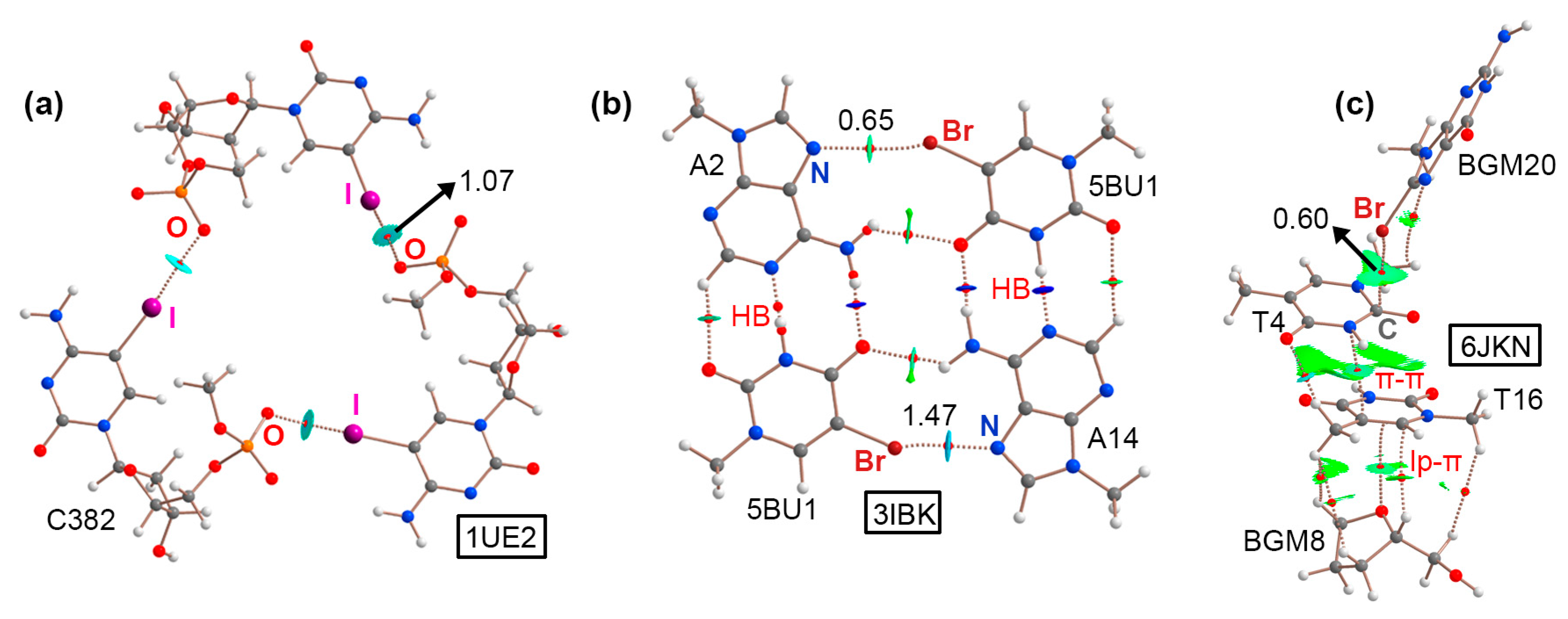
2.1. Results from the PDB Survey: Br and I Halogen Bonds
2.2. Results from the PDB Survey: S, Se, and Te Chalcogen Bonds
2.3. QTAIM and NCIplot Analyses
2.4. NBO Analysis
3. Materials and Methods
3.1. Protein Data Bank Search Criteria
- -
- Only isolated nucleic acid X-ray structures were considered;
- -
- Structures with disorder were not considered;
- -
- In the case of halogens, only the incorporation of Br/I in the nucleobase (U, C, and G) was taken into account, while for chalcogens, the incorporation of S/Se/Te was considered in both the nucleobase and the sugar moiety (see Figure 1 above);
- -
- Any type of nucleic acid structure (both canonical and noncanonical) was considered.
3.2. QM Calculations on Selected Structures
- -
- In 3IBK, the amino group from the guanine ring was replaced by a −H atom;
- -
- In the 3LTU, 3HG8, and 3FA1 structures, the Ch–X (Ch = S, Se, and Te; X = Me and H) moiety was replaced by a –H atom.
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schneider, H.J. Supramolecular Systems in Biomedical Fields, 1st ed.; RSC Publishing: Cambridge, UK, 2013. [Google Scholar]
- Lehn, J.M. Supramolecular Chemistry: Concepts and Perspectives, 1st ed.; Wiley VCH: Weinheim, Germany, 1995. [Google Scholar]
- Cragg, P.J. Supramolecular Chemistry: From Biological Inspiration to Biomedical Applications, 1st ed.; Springer: Dordrecht, The Netherlands, 2010. [Google Scholar]
- Steed, A.W.; Atwood, J.L. Supramolecular Chemistry, 1st ed.; John Wily & Sons, Ltd.: Chichester, UK, 2009. [Google Scholar]
- Williams, D.H.; Stephens, E.; O’Brien, D.P.; Zhou, M. Understanding Noncovalent Interactions: Ligand Binding Energy and Catalytic Efficiency from Ligand-Induced Reductions in Motion within Receptors and Enzymes. Angew. Chem. Int. Ed. 2004, 43, 6596–6616. [Google Scholar] [CrossRef] [PubMed]
- Vargas Jentzsch, A.; Emery, D.; Mareda, J.; Nayak, S.K.; Metrangolo, P.; Resnati, G.; Sakai, N.; Matile, S. Transmembrane anion transport mediated by halogen-bond donors. Nat. Commun. 2012, 3, 905. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, G.J.; Choudhary, A.; Raines, R.T.; Woolfson, D.N. n→π* interactions in proteins. Nat. Chem. Biol. 2010, 6, 615–620. [Google Scholar] [CrossRef] [PubMed]
- Gilli, G.; Gilli, P. The Nature of the Hydrogen Bond; Oxford University Press: Oxford, UK, 2009. [Google Scholar]
- Anderson, L.N.; Aquino, F.W.; Raeber, A.E.; Chen, X.; Wong, B.M. Halogen Bonding Interactions: Revised Benchmarks and a New Assessment of Exchange vs Dispersion. J. Chem. Theory Comput. 2018, 14, 180–190. [Google Scholar] [CrossRef]
- Wolters, L.P.; Schyman, P.; Pavan, M.J.; Jorgensen, W.L.; Bickelhaupt, F.M.; Kozuch, S. The many faces of halogen bonding: A review of theoretical models and methods. WIREs Comput. Mol. Sci. 2014, 4, 523–540. [Google Scholar] [CrossRef]
- Cavallo, G.; Metrangolo, P.; Milani, R.; Pilati, T.; Priimagi, A.; Resnati, G.; Terraneo, G. The Halogen Bond. Chem. Rev. 2016, 116, 2478–2601. [Google Scholar] [CrossRef]
- Szell, P.M.J.; Gabriel, S.A.; Caron-Poulin, E.; Jeannin, O.; Fourmigué, M.; Bryce, D.L. Cosublimation: A Rapid Route Toward Otherwise Inaccessible Halogen-Bonded Architectures. Cryst. Growth Des. 2018, 18, 6227–6238. [Google Scholar] [CrossRef]
- Erakovic, M.; Cincic, D.; Molcanov, K.; Stilinovic, V. A Crystallographic Charge Density Study of the Partial Covalent Nature of Strong N⋅⋅⋅Br Halogen Bonds. Angew. Chem. Int. Ed. 2019, 58, 15702–15706. [Google Scholar] [CrossRef]
- Puttreddy, R.; Rautiainen, J.M.; Maekelae, T.; Rissanen, K. Strong N-X⋅⋅⋅O-N Halogen Bonds: A Comprehensive Study on N-Halosaccharin Pyridine N-Oxide Complexes. Angew. Chem. Int. Ed. 2019, 58, 18610–18618. [Google Scholar] [CrossRef]
- Vioglio, P.C.; Chierotti, M.R.; Gobetto, R. Solid-state nuclear magnetic resonance as a tool for investigating the halogen bond. CrystEngComm 2016, 18, 9173–9184. [Google Scholar] [CrossRef]
- Politzer, P.; Murray, J.S.; Clark, T. Halogen Bonding and Other σ-Hole Interactions: A Perspective. Phys. Chem. Chem. Phys. 2013, 15, 11178–11189. [Google Scholar] [CrossRef] [PubMed]
- Clark, T. σ-Holes. WIREs Comput. Mol. Sci. 2013, 3, 13–20. [Google Scholar] [CrossRef]
- Politzer, P.; Murray, J.S.; Lane, P. σ-Hole Bonding and Hydrogen Bonding: Competitive Interactions. Int. J. Quantum Chem. 2007, 107, 3046–3052. [Google Scholar] [CrossRef]
- Shinada, N.K.; de Brevern, A.G.; Schmidtke, P. Halogens in Protein–Ligand Binding Mechanism: A Structural Perspective. J. Med. Chem. 2019, 62, 9341–9356. [Google Scholar] [CrossRef] [PubMed]
- Sirimulla, S.; Bailey, J.B.; Vegesna, R.; Narayan, M. Halogen Interactions in Protein–Ligand Complexes: Implications of Halogen Bonding for Rational Drug Design. J. Chem. Inf. Model. 2013, 53, 2781–2791. [Google Scholar] [CrossRef]
- Pähler, A.; Smith, J.L.; Hendrickson, W.A. A Probability Representation for Phase Information from Multiwavelength Anomalous Dispersion. Acta Crystallogr. A 1990, 46, 537–540. [Google Scholar] [CrossRef]
- Carter, M.; Voth, A.R.; Scholfield, M.R.; Rummel, B.; Sowers, L.C.; Ho, P.S. Enthalpy–Entropy Compensation in Biomolecular Halogen Bonds Measured in DNA Junctions. Biochemistry 2013, 52, 4891–4903. [Google Scholar] [CrossRef]
- Voth, A.R.; Hays, F.A.; Ho, P.S. Directing macromolecular conformation through halogen bonds. Proc. Nat. Acad. Sci. USA 2007, 104, 6188–6193. [Google Scholar] [CrossRef]
- Parker, A.J.; Stewart, J.; Donald, K.J.; Parish, C.A. Halogen Bonding in DNA Base Pairs. J. Am. Chem. Soc. 2012, 134, 5165–5172. [Google Scholar] [CrossRef]
- Xu, L.; Sang, P.; Zou, J.-W.; Xu, M.-B.; Li, X.-M.; Yu, Q.-S. Evaluation of nucleotide C–Br···O–P contacts from ONIOM calculations: Theoretical insight into halogen bonding in nucleic acids. ChemPhysLett 2011, 509, 175–180. [Google Scholar] [CrossRef]
- Ennifar, E.; Bernacchi, S.; Wolff, P.; Dumas, P. Influence of C-5 halogenation of uridines on hairpin versus duplex RNA folding. RNA 2007, 13, 1445–1452. [Google Scholar] [CrossRef] [PubMed]
- Kolář, M.H.; Tabarrini, O. Halogen Bonding in Nucleic Acid Complexes. J. Med. Chem. 2017, 60, 8681–8690. [Google Scholar] [CrossRef] [PubMed]
- Auffinger, P.; Hays, F.A.; Westhof, E.; Shing Ho, P. Halogen bonds in biological molecules. Proc. Natl. Acad. Sci. USA 2004, 101, 16789–16794. [Google Scholar] [CrossRef] [PubMed]
- Frontera, A.; Bauzá, A. Halogen Bonds in Protein Nucleic Acid Recognition. J. Chem. Theory Comput. 2020, 16, 4744–4752. [Google Scholar] [CrossRef] [PubMed]
- Piña, M.N.P.; Frontera, A.; Bauzá, A. Quantifying Intramolecular Halogen Bonds in Nucleic Acids: A Combined Protein Data Bank and Theoretical Study. ACS Chem. Biol. 2020, 15, 1942–1948. [Google Scholar] [CrossRef]
- Gomila, R.M.; Frontera, A.; Bauzá, A. A Comprehensive Ab Initio Study of Halogenated A···U and G···C Base Pair Geometries and Energies. Int. J. Mol. Sci. 2023, 24, 5530. [Google Scholar] [CrossRef]
- Tiekink, E.R.T. Therapeutic potential of selenium and tellurium compounds: Opportunities yet unrealized. Dalton Trans. 2012, 41, 6390–6395. [Google Scholar] [CrossRef]
- Aakeroy, C.B.; Bryce, D.L.; Desiraju, G.R.; Frontera, A.; Legon, A.C.; Nicotra, F.; Rissanen, K.; Scheiner, S.; Terraneo, G.; Metrangolo, P.; et al. Definition of the chalcogen bond (IUPAC Recommendations 2019). Pure Appl. Chem. 2019, 91, 1889–1892. [Google Scholar] [CrossRef]
- Pascoe, D.J.; Ling, K.B.; Cockroft, S.L. The Origin of Chalcogen-Bonding Interactions. J. Am. Chem. Soc. 2017, 139, 15160–15167. [Google Scholar] [CrossRef]
- Carugo, O.; Resnati, G.; Metrangolo, P. Chalcogen Bonds Involving Selenium in Protein Structures. ACS Chem. Biol. 2021, 16, 1622–1627. [Google Scholar] [CrossRef]
- Carugo, O. Interplay between hydrogen and chalcogen bonds in cysteine. Proteins 2023, 91, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Carugo, O. Chalcogen bonds formed by protein sulfur atoms in proteins. A survey of high-resolution structures deposited in the protein data bank. J. Biomol. Struct. 2022; ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Wotjkowiak, K.; Michalczyk, M.; Zierkiewicz, W.; Jezierska, A.; Panek, J.J. Chalcogen Bond as a Factor Stabilizing Ligand Conformation in the Binding Pocket of Carbonic Anhydrase IX Receptor Mimic. Int. J. Mol. Sci. 2022, 23, 13701. [Google Scholar] [CrossRef]
- Piña, M.N.P.; Frontera, A.; Bauzá, A. Charge Assisted S/Se Chalcogen Bonds in SAM Riboswitches: A Combined PDB and ab Initio Study. ACS Chem. Biol. 2021, 16, 1701–1708. [Google Scholar] [CrossRef] [PubMed]
- Fernández Riveras, J.A.; Frontera, A.; Bauzá, A. Selenium chalcogen bonds are involved in protein–carbohydrate recognition: A combined PDB and theoretical study. Phys. Chem. Chem. Phys. 2021, 23, 17656–17662. [Google Scholar] [CrossRef] [PubMed]
- Wilds, C.J.; Pattanayek, R.; Pan, C.; Wawrzak, Z.; Egli, M. Selenium-Assisted Nucleic Acid Crystallography: Use of Phosphoroselenoates for MAD Phasing of a DNA Structure. J. Am. Chem. Soc. 2002, 124, 14910–14916. [Google Scholar] [CrossRef]
- Sheng, J.; Huang, Z. Selenium Derivatization of Nucleic Acids for Phase and Structure Determination in Nucleic Acid X-ray Crystallography. Int. J. Mol. Sci. 2008, 9, 258–271. [Google Scholar] [CrossRef]
- Teplova, M.; Wilds, C.J.; Wawrzak, Z.; Tereshko, V.; Du, Q.; Carrasco, N.; Huang, Z.; Egli, M. Covalent incorporation of selenium into oligonucleotides for X-ray crystal structure determination via MAD: Proof of principle. Biochimie 2002, 84, 849–858. [Google Scholar] [CrossRef]
- Sharma, K.D.; Kathuria, P.; Wetmore, S.D.; Sharma, P. Can modified DNA base pairs with chalcogen bonding expand the genetic alphabet? A combined quantum chemical and molecular dynamics simulation study. Phys. Chem. Chem. Phys. 2020, 22, 23754–23765. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Sunami, T.; Kondo, J.; Hirao, I.; Watanabe, K.; Miura, K.; Takénaka, A. Structure of d(GCGAAAGC) (hexagonal form): A base-intercalated duplex as a stable structure. Acta Cryst. 2004, 60, 90–96. [Google Scholar]
- Shepard, W.; Cruse, W.B.T.; Fourme, R.; de la Fortelle, E.; Prangé, T. A zipper-like duplex in DNA: The crystal structure of d(GCGAAAGCT) at 2.1 å resolution. Structure 1998, 6, 849–861. [Google Scholar] [CrossRef] [PubMed]
- Hirao, I.; Naraoka, T.; Kanamori, S.; Nakamura, M.; Miura, K. Synthetic oligodeoxyribonucleotides showing abnormal mobilities on polyacrylamide gel electrophoresis. Biochem. Int. 1988, 16, 157–162. [Google Scholar]
- Hirao, I.; Nishimura, Y.; Naraoka, T.; Watanabe, K.; Arata, Y.; Miura, K. Extraordinary stable structure of short single-stranded DNA fragments containing a specific base sequence: D(GCGAAAGC). Nucleic Acids Res. 1989, 17, 2223–2231. [Google Scholar] [CrossRef]
- Hirao, I.; Nishimura, Y.; Tagawa, Y.-I.; Watanabe, K.; Miura, K.-I. Extraordinarily stable mini-hairpins: Electrophoretical and thermal properties of the various sequence variants of d(GCFAAAGC)and their effect on DNA sequencing. Nucleic Acids Res. 1992, 20, 3891–3896. [Google Scholar] [CrossRef]
- Cruse, W.B.T.; Saludjian, P.; Leroux, Y.; Léger, G.; El Manouni, D.; Prangé, T. A continuous transition from A to B DNA in the 1:1 complex between nogalamycin and the hexamer dCCCGGG. J. Biol. Chem. 1996, 271, 15558–15566. [Google Scholar] [CrossRef]
- Cruse, W.B.T.; Saludjian, P.; Biala, E.; Strazewski, P.; Prangé, T.; Kennard, O. Structure of a mispaired RNA double helix at 1.6 Å resolution and implications for the prediction of RNA secondary structure. Proc. Natl Acad. Sci. USA 1994, 91, 4160–4164. [Google Scholar] [CrossRef]
- Wang, Y.-H.; Lu, Y.-X.; Zou, J.-W.; Yu, Q.-S. Theoretical investigation on charge-assisted halogen bonding interactions in the complexes of bromocarbons with some anions. Int. J. Quantum Chem. 2007, 108, 90–99. [Google Scholar] [CrossRef]
- Collie, G.W.; Haider, S.M.; Neidle, S.; Parkinson, G.N. A crystallographic and modelling study of a human telomeric RNA (TERRA) quadruplex. Nucleic Acids Res. 2010, 38, 5569–5580. [Google Scholar] [CrossRef]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef]
- Neidle, S.; Balasubramanian, S. Quadruplex Nucleic Acids, 1st ed.; Royal Society of Chemistry: Cambridge UK, 2006. [Google Scholar]
- Geng, Y.; Liu, C.; Zhou, B.; Cai, Q.; Miao, H.; Shi, X.; Xu, N.; You, Y.; Fung, C.P.; Din, R.U.; et al. The crystal structure of an antiparallel chair-type G-quadruplex formed by Bromo-substituted human telomeric DNA. Nucleic Acids Res. 2019, 47, 5395–5404. [Google Scholar] [CrossRef] [PubMed]
- Prasanna, M.D.; Guru Row, T.N. C–halogen···π interactions and their influence on molecular conformation and crystal packing: A database study. Cryst. Eng. 2000, 3, 135–154. [Google Scholar] [CrossRef]
- Moroder, H.; Kreutz, C.; Lang, K.; Serganov, A.; Micura, R. Synthesis, Oxidation Behavior, Crystallization and Structure of 2′-Methylseleno Guanosine Containing RNAs. J. Am. Chem. Soc. 2006, 128, 9909–9918. [Google Scholar] [CrossRef] [PubMed]
- Ota, M.; Takahashi, H.; Nogi, Y.; Kagotani, Y.; Saito-Tarashima, N.; Kondo, J.; Minakawa, N. Synthesis and properties of fully-modified 4′-selenoRNA, an endonuclease-resistant RNA analog. Bioorg. Med. Chem. 2022, 76, 117093. [Google Scholar] [CrossRef]
- Sheng, J.; Zhang, W.; Hassan, A.E.A.; Gan, J.; Soares, A.S.; Geng, S.; Ren, Y.; Huang, Z. Hydrogen bond formation between the naturally modified nucleobase and phosphate backbone. Nucleic Acids Res. 2012, 40, 8111–8118. [Google Scholar] [CrossRef]
- Jiang, S.; Gan, J.; Sheng, J.; Sun, H.; Huang, Z. Structure of Dickerson-Drew Dodecamer with 2′-MeSe-ara-G Modification. Available online: https://www.wwpdb.org/pdb?id=pdb_00004kw0 (accessed on 20 June 2023). [CrossRef]
- Olieric, V.; Rieder, U.; Lang, K.; Serganov, A.; Schulze-Briese, C.; Micura, R.; Dumas, P.; Ennifar, E. A fast selenium derivatization strategy for crystallization and phasing of RNA structures. RNA 2009, 15, 707–715. [Google Scholar] [CrossRef]
- Sheng, J.; Hassan, A.E.A.; Zhang, W.; Huang, Z. Crystal Structure of 5-SMe Derivatized DNA. Available online: https://www.wwpdb.org/pdb?id=pdb_00003hg8 (accessed on 20 June 2023). [CrossRef]
- Sheng, J.; Hassan, A.E.A.; Zhang, W.; Huang, Z. Crystal Structure of Tellurium Derivatized DNA. Available online: https://www.wwpdb.org/pdb?id=pdb_00003fa1 (accessed on 20 June 2023). [CrossRef]
- Bader, R.F.W. Atoms in Molecules. Acc. Chem. Res. 1985, 18, 9–15. [Google Scholar] [CrossRef]
- Weinhold, F.; Landis, C.R.; Glendening, E.D. What Is NBO Analysis and How Is It Useful? Int. Rev. Phys. Chem. 2016, 35, 399–440. [Google Scholar] [CrossRef]
- Álvarez, S. A cartography of the van der Waals territories. Dalton Trans. 2013, 42, 8617–8636. [Google Scholar] [CrossRef]
- Weigend, F.; Häser, M. RI-MP2: First derivatives and global consistency. Theor. Chem. Acc. 1997, 97, 331–340. [Google Scholar] [CrossRef]
- Weigend, F.; Ahlrichs, R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy. Phys. Chem. Chem. Phys. 2005, 7, 3297–3305. [Google Scholar] [CrossRef]
- Bauzá, A.; Alkorta, I.; Frontera, A.; Elguero, J. On the Reliability of Pure and Hybrid DFT Methods for the Evaluation of Halogen, Chalcogen, and Pnicogen Bonds Involving Anionic and Neutral Electron Donors. J. Chem. Theory Comput. 2013, 9, 5201–5210. [Google Scholar] [CrossRef] [PubMed]
- Ahlrichs, R.; Bar, M.; Haser, M.; Horn, H.; Kolmel, C. Electronic Structure Calculations on Workstation Computers—The Program System turbomole. Chem. Phys. Lett. 1989, 162, 165–169. [Google Scholar] [CrossRef]
- Peterson, K.A.; Figgen, D.; Goll, E.; Stoll, H.; Dolg, M. Systematically convergent basis sets with relativistic pseudopotentials. II. Small-core pseudopotentials and correlation consistent basis sets for the post-d group 16–18 elements. J. Chem. Phys. 2003, 119, 11113. [Google Scholar] [CrossRef]
- Becke, A.D. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys. Rev. A 1988, 38, 3098–3100. [Google Scholar] [CrossRef] [PubMed]
- Grimme, S.; Antony, J.; Ehrlich, S.; Krieg, H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010, 132, 154104. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16, revision, B.01; Gaussian, Inc.: Wallingford, CT, USA, 2016.
- Dennington, R.; Keith, T.A.; Millam, J.M. GaussView, version 6; Semichem Inc.: Shawnee Mission, KS, USA, 2016.
- Todd, A.; Keith, T.K. AIMAll, version 13.05.06; Gristmill Software: Overland Park, KS, USA, 2013.
- Contreras-García, J.; Johnson, E.R.; Keinan, S.; Chaudret, R.; Piquemal, J.-P.; Beratan, D.N.; Yang, W. NCIPLOT: A Program for Plotting Noncovalent Interaction Regions. J. Chem. Theory Comput. 2011, 7, 625–632. [Google Scholar] [CrossRef]
- Johnson, E.R.; Keinan, S.; Mori-Sanchez, P.; Contreras-Garcia, J.; Cohen, A.J.; Yang, W. Revealing noncovalent interactions. J. Am. Chem. Soc. 2010, 132, 6498–6506. [Google Scholar] [CrossRef] [PubMed]

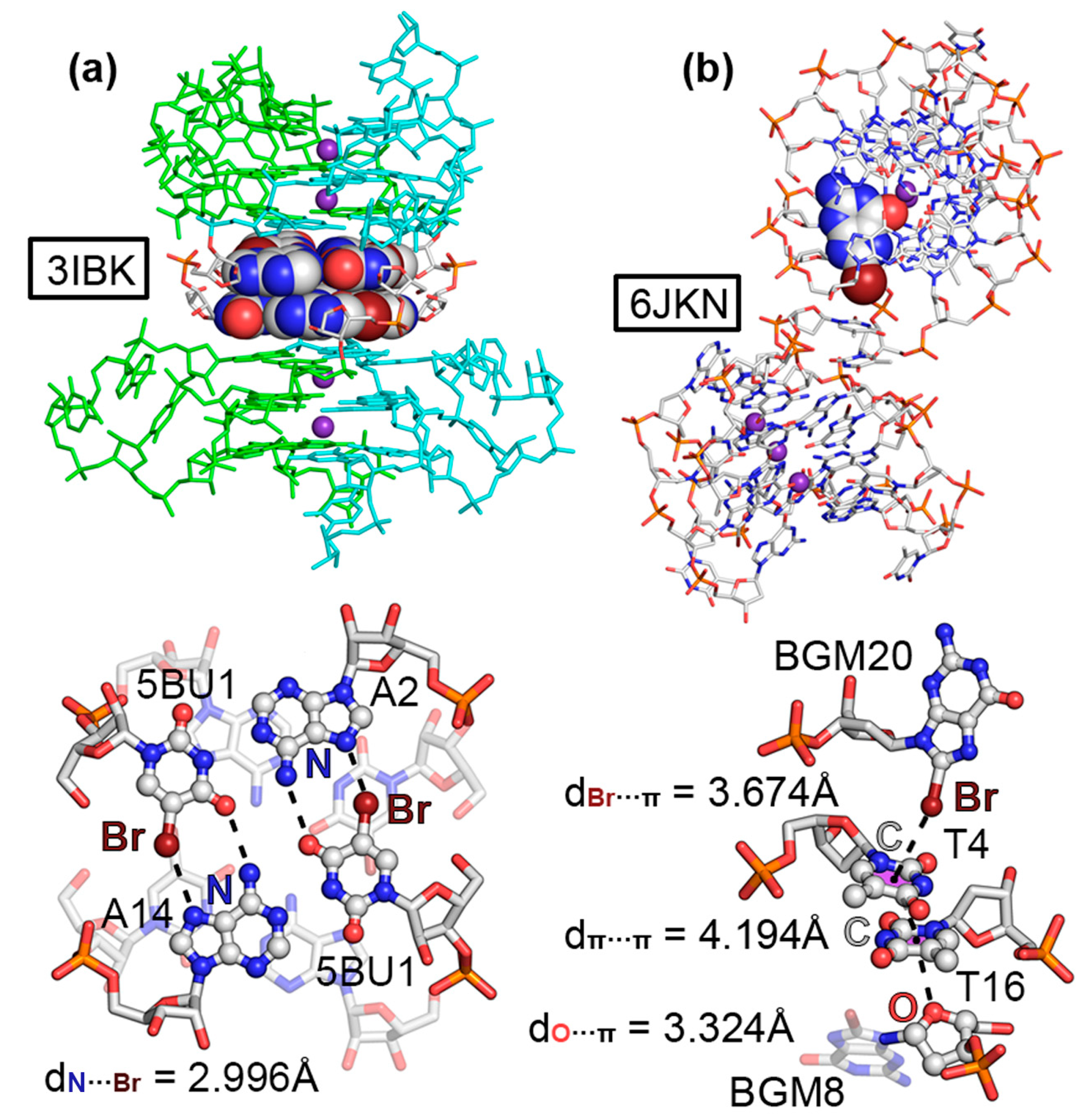



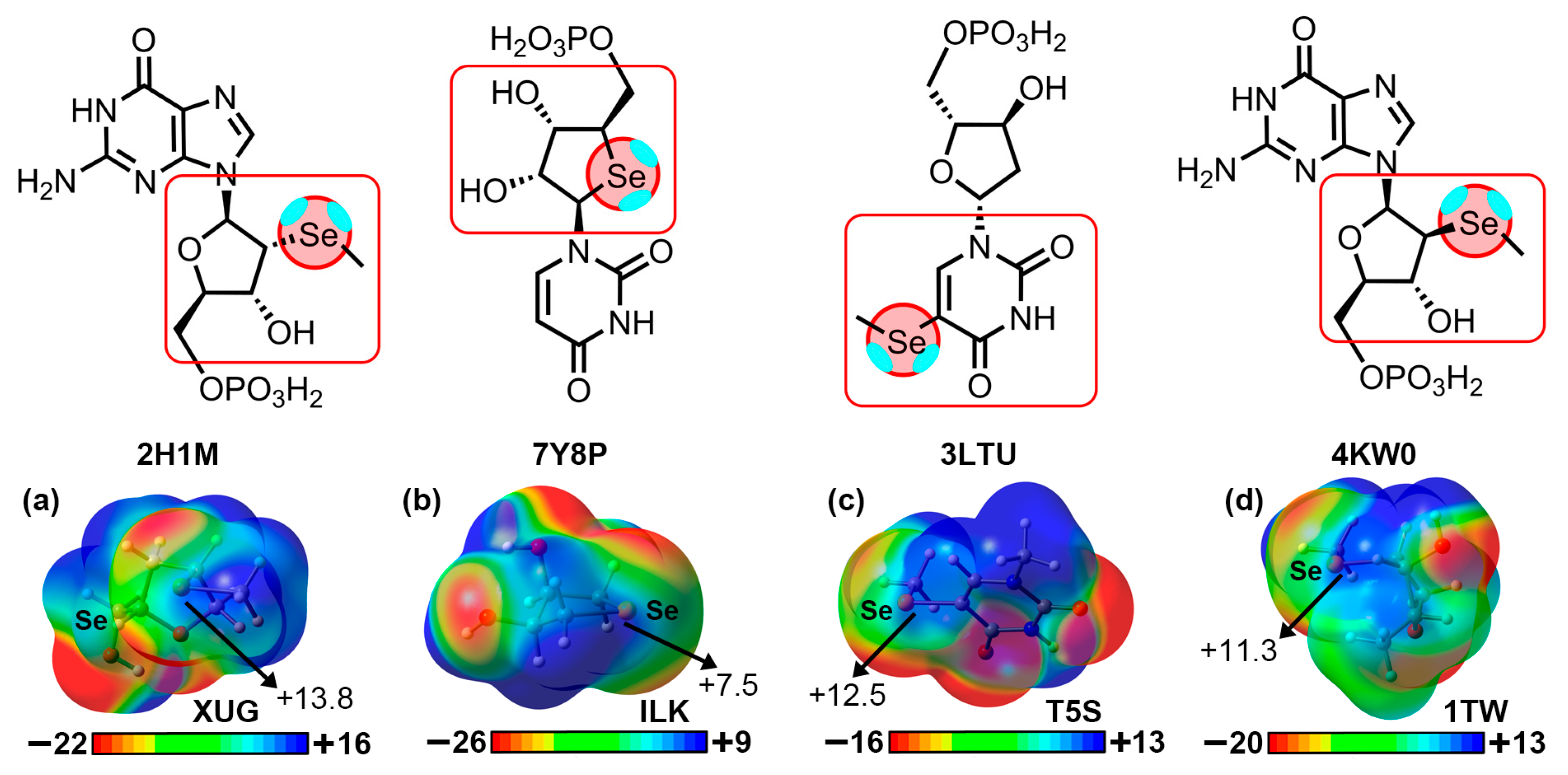

| PDBID | ΔEBSSE | d | α 1 |
|---|---|---|---|
| 376D (O···Br) | −12.8 | 3.093 | 162.2 |
| 3IBK (N···Br) | −6.0 (−2.2) | 2.996 | 157.2 |
| 6JKN (π···Br) | −2.2 | 3.674 | 174.4 |
| 1UE2 (O···I) | −16.6 | 3.144 | 168.3 |
| PDBID | ΔEBSSE | d | α 1 |
|---|---|---|---|
| 2H1M (Se···Se) | −3.7 | 3.958 | 170.4 |
| 7Y8P (O···Se) | −3.1 | 3.407 | 148.4 |
| 3LTU (π···Se) | −7.8 (−2.0) | 3.467 | 168.4 |
| 4KW0 (O···Se) | −4.3 | 3.089 | 172.6 |
| 3DW6 (N···Se) | −3.0 | 3.736 | 166.8 |
| 3HG8 (π···S) | −7.6 (−1.5) | 3.467 | 172.1 |
| 3FA1 (O···Te) | −15.8 | 3.528 | 170.8 |
| 3FA1 (π···Te) | −9.0 (−3.4) | 3.675 | 167.6 |
| PDBID | Donor | Acceptor | E(2) |
|---|---|---|---|
| 376D (O···Br) | LP O | BD* Br–C | 1.66 |
| 3IBK (N···Br) | LP N | BD* Br–C | 3.29 |
| 6JKN (π···Br) | BD C–O | BD* Br–C | 0.96 |
| 1UE2 (O···I) | LP O | BD* I–C | 2.81 |
| 2H1M (Se···Se) | LP Se | BD* Se–C | 0.85 |
| 7Y8P (O···Se) | LP O | BD* Se–C | 0.48 |
| 3LTU (π···Se) | BD C–C BD C–N | BD* Se–C BD* Se–C | 0.31 0.39 |
| 4KW0 (O···Se) | LP O | BD* Se–C | 1.97 |
| 3DW6 (N···Se) | LP N | BD* Se–C | 0.14 |
| 3HG8 (π···S) | BD C–N | BD* S–C | 0.40 |
| 3FA1 (O···Te) 3FA1 (π···Te) | LP O BD C–N | BD* Te–H BD* Te–H | 1.31 0.85 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piña, M.d.l.N.; Bauzá, A. On the Importance of Halogen and Chalcogen Bonds in the Solid State of Nucleic Acids: A Combined Crystallographic and Theoretical Perspective. Int. J. Mol. Sci. 2023, 24, 13035. https://doi.org/10.3390/ijms241713035
Piña MdlN, Bauzá A. On the Importance of Halogen and Chalcogen Bonds in the Solid State of Nucleic Acids: A Combined Crystallographic and Theoretical Perspective. International Journal of Molecular Sciences. 2023; 24(17):13035. https://doi.org/10.3390/ijms241713035
Chicago/Turabian StylePiña, María de las Nieves, and Antonio Bauzá. 2023. "On the Importance of Halogen and Chalcogen Bonds in the Solid State of Nucleic Acids: A Combined Crystallographic and Theoretical Perspective" International Journal of Molecular Sciences 24, no. 17: 13035. https://doi.org/10.3390/ijms241713035
APA StylePiña, M. d. l. N., & Bauzá, A. (2023). On the Importance of Halogen and Chalcogen Bonds in the Solid State of Nucleic Acids: A Combined Crystallographic and Theoretical Perspective. International Journal of Molecular Sciences, 24(17), 13035. https://doi.org/10.3390/ijms241713035