Interactome of Arabidopsis ATG5 Suggests Functions beyond Autophagy
Abstract
1. Introduction
2. Results
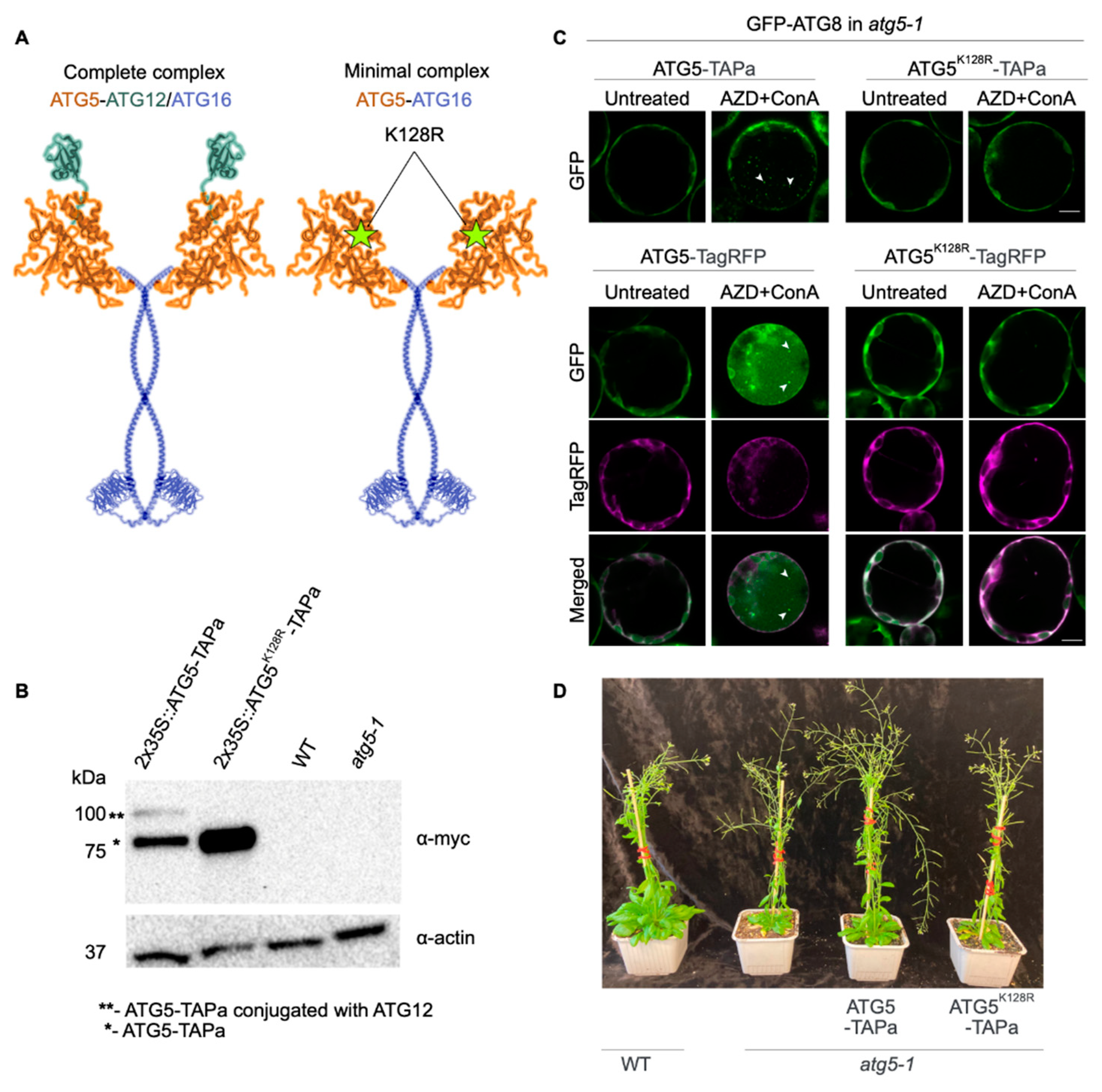
2.1. Generation and Characterization of Arabidopsis ATG5-TAP Lines
2.2. Interactomes of ATG5 and Its Complete and Minimal Complexes
2.2.1. Affinity Purification
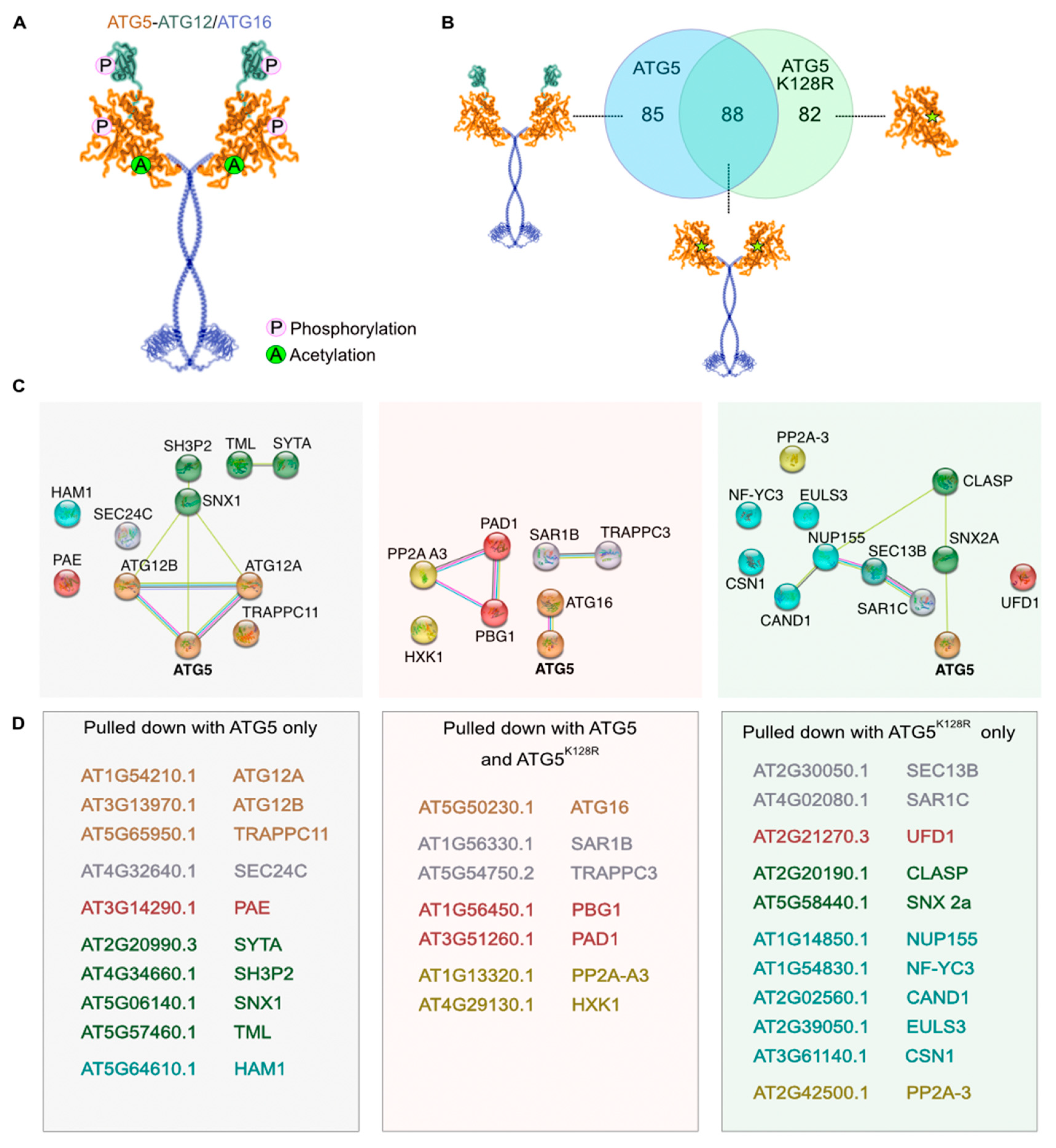
2.2.2. Detection of Post-Translational Modifications
2.2.3. Criteria for Identifying ATG5 Interactomes
2.2.4. Putative Interactors Acting as Stress Sensors
2.2.5. Putative Interactors belonging to Endomembrane Trafficking System
2.2.6. Putative Interactors belonging to Ubiquitin-Proteasome System
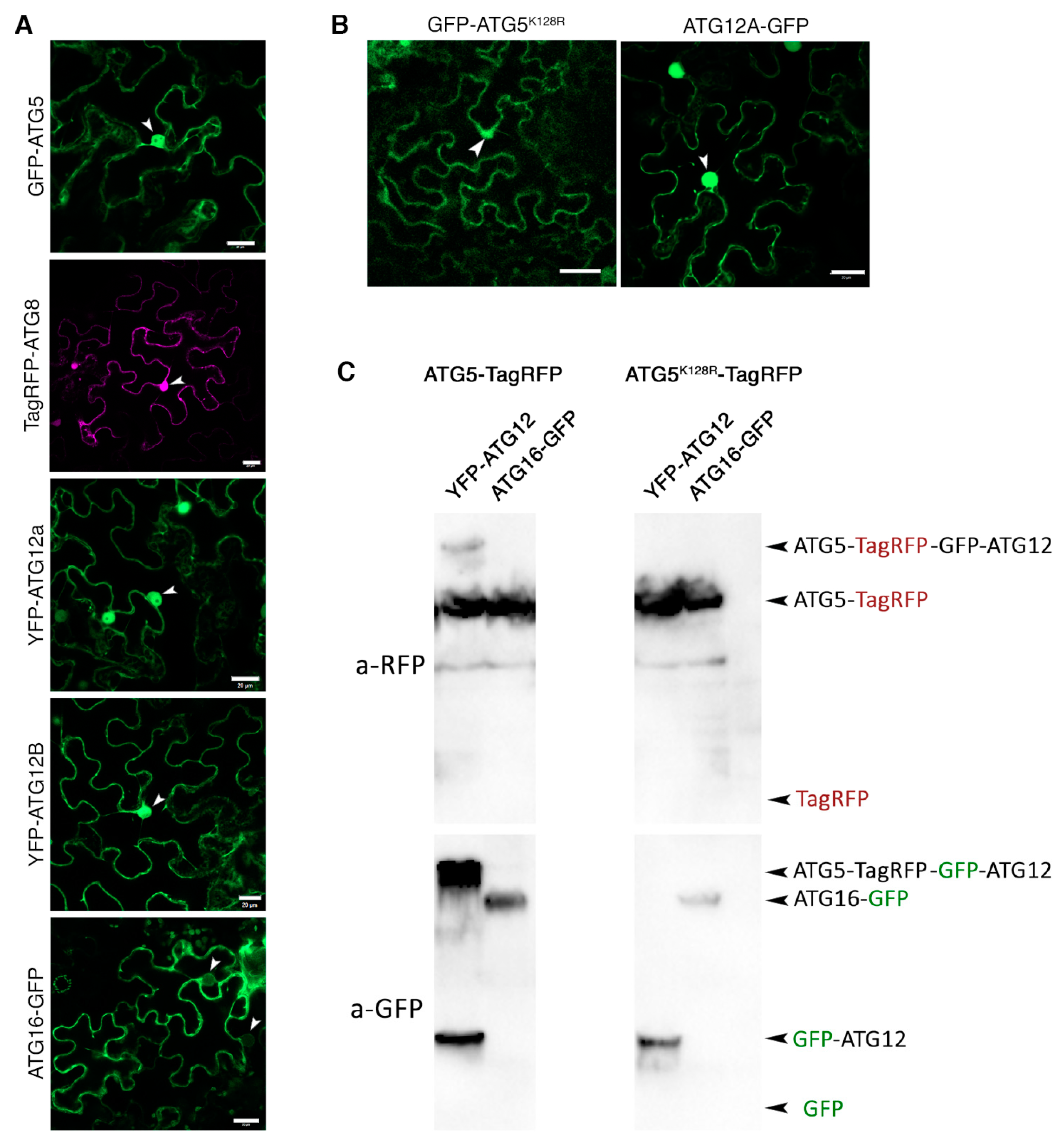
2.2.7. Putative Interactors of Nuclear-Localized ATG5 and ATG12
3. Discussion
3.1. Evolutionary Context of Autophagy
3.2. Closely Related Components of Endomembrane Trafficking Pathways Show Selectivity towards Different ATG5 Baits
3.3. Cross-Talk of ATG5 and Clathrin-Mediated Trafficking Is Conserved
4. Methods
4.1. Plasmids Construction
4.2. Protoplast and Plant Transformation
4.2.1. Transient Expression of Arabidopsis Protoplasts
4.2.2. Transient Expression in Nicotiana Benthamiana
4.2.3. Arabidopsis Thaliana Growth and Transformation
4.3. Immunoblotting
4.4. Tandem Affinity Purification
4.5. Mass Spectrometry
4.5.1. Liquid Chromatography and Mass Spectrometry Assay
4.5.2. LC-MS/MS Data Analysis
4.5.3. Statistical Analysis of Interactors—MSqRobSum
4.5.4. STRING Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parzych, K.R.; Klionsky, D.J. Vacuolar hydrolysis and efflux: Current knowledge and unanswered questions. Autophagy 2019, 15, 212–227. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Li, X.; Yang, M.; Shao, Q.; Zhao, Y.; Ma, C.; Wang, P. Autophagy: An Intracellular Degradation Pathway Regulating Plant Survival and Stress Response. Front. Plant Sci. 2020, 11, 164. [Google Scholar] [CrossRef] [PubMed]
- Ketelaar, T.; Voss, C.; Dimmock, S.A.; Thumm, M.; Hussey, P.J. Arabidopsis homologues of the autophagy protein Atg8 are a novel family of microtubule binding proteins. FEBS Lett. 2004, 567, 302–306. [Google Scholar] [CrossRef]
- Nakatogawa, H.; Ichimura, Y.; Ohsumi, Y. Atg8, a Ubiquitin-like Protein Required for Autophagosome Formation, Mediates Membrane Tethering and Hemifusion. Cell 2007, 130, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Minina, E.A.; Moschou, P.N.; Vetukuri, R.R.; Sanchez-Vera, V.; Cardoso, C.; Liu, Q.; Elander, P.H.; Dalman, K.; Beganovic, M.; Yilmaz, J.L.; et al. Transcriptional stimulation of rate-limiting components of the autophagic pathway improves plant fitness. J. Exp. Bot. 2018, 69, 1415–1432. [Google Scholar] [CrossRef]
- Bu, F.; Yang, M.; Guo, X.; Huang, W.; Chen, L. Multiple Functions of ATG8 Family Proteins in Plant Autophagy. Front. Cell Dev. Biol. 2020, 8, 466. [Google Scholar] [CrossRef]
- Jia, M.; Liu, X.; Xue, H.; Wu, Y.; Shi, L.; Wang, R.; Chen, Y.; Xu, N.; Zhao, J.; Shao, J.; et al. Noncanonical ATG8-ABS3 interaction controls senescence in plants. Nat. Plants 2019, 5, 212–224. [Google Scholar] [CrossRef]
- Codogno, P.; Meijer, A.J. Atg5: More than an autophagy factor. Nat. Cell Biol. 2006, 8, 1045–1047. [Google Scholar] [CrossRef]
- Yoshimoto, K.; Jikumaru, Y.; Kamiya, Y.; Kusano, M.; Consonni, C.; Panstruga, R.; Ohsumi, Y.; Shirasu, K. Autophagy Negatively Regulates Cell Death by Controlling NPR1-Dependent Salicylic Acid Signaling during Senescence and the Innate Immune Response in Arabidopsis. Plant Cell Online 2009, 21, 2914–2927. [Google Scholar] [CrossRef]
- Barros, J.A.; Cavalcanti, J.H.F.; Medeiros, D.B.; Nunes-Nesi, A.; Avin-Wittenberg, T.; Fernie, A.R.; Araújo, W.L. Autophagy deficiency compromises alternative pathways of respiration following energy deprivation in Arabidopsis thaliana. Plant Physiol. 2017, 175, 62–76. [Google Scholar] [CrossRef]
- Matsushita, M.; Suzuki, N.N.; Obara, K.; Fujioka, Y.; Ohsumi, Y.; Inagaki, F. Structure of Atg5.Atg16, a complex essential for autophagy. J. Biol. Chem. 2007, 282, 6763–6772. [Google Scholar] [CrossRef]
- Baines, K.; Yoshioka, K.; Takuwa, Y.; Lane, J.D. The ATG5 interactome links clathrin-mediated vesicular trafficking with the autophagosome assembly machinery. Autophagy Rep. 2022, 1, 88–118. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Romanov, J.; Walczak, M.; Ibiricu, I.; Schüchner, S.; Ogris, E.; Kraft, C.; Martens, S. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J. 2012, 31, 4304–4317. [Google Scholar] [CrossRef]
- Rubio, V.; Shen, Y.; Saijo, Y.; Liu, Y.; Gusmaroli, G.; Dinesh-Kumar, S.P.; Deng, X.W. An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 2005, 41, 767–778. [Google Scholar] [CrossRef]
- Dauphinee, A.N.; Cardoso, C.; Dalman, K.; Ohlsson, J.A.; Fick, S.B.; Robert, S.; Hicks, G.R.; Bozhkov, P.V.; Minina, E.A. Chemical screening pipeline for identification of specific plant autophagy modulators. Plant Physiol. 2019, 181, 855–866. [Google Scholar] [CrossRef]
- Gutierrez-Beltran, E.; Elander, P.H.; Dalman, K.; Dayhoff, G.W.; Moschou, P.N.; Uversky, V.N.; Crespo, J.L.; Bozhkov, P.V. Tudor staphylococcal nuclease is a docking platform for stress granule components and is essential for SnRK1 activation in Arabidopsis. EMBO J. 2021, 40, e105043. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Schreiber, A.; Collins, B.C.; Davis, C.; Enchev, R.I.; Sedra, A.; D’antuono, R.; Aebersold, R.; Peter, M. Multilayered regulation of autophagy by the Atg1 kinase orchestrates spatial and temporal control of autophagosome formation. Mol. Cell 2021, 81, 5066–5081.e10. [Google Scholar] [CrossRef]
- Lee, I.H.; Finkel, T. Regulation of autophagy by the p300 acetyltransferase. J. Biol. Chem. 2009, 284, 6322–6328. [Google Scholar] [CrossRef]
- Lee, I.H.; Cao, L.; Mostoslavsky, R.; Lombard, D.B.; Liu, J.; Bruns, N.E.; Tsokos, M.; Alt, F.W.; Finkel, T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc. Natl. Acad. Sci. USA 2008, 105, 3374–3379. [Google Scholar] [CrossRef]
- Yeasmin, A.M.S.T.; Waliullah, T.M.; Kondo, A.; Kaneko, A.; Koike, N.; Ushimaru, T. Orchestrated action of PP2A antagonizes atg13 phosphorylation and promotes autophagy after the inactivation of TORC1. PLoS ONE 2016, 11, e0166636. [Google Scholar] [CrossRef]
- Bánréti, Á.; Lukácsovich, T.; Csikós, G.; Erdélyi, M.; Sass, M. PP2A regulates autophagy in two alternative ways in Drosophila. Autophagy 2012, 8, 623–636. [Google Scholar] [CrossRef]
- Khan, Z.H.; Agarwal, S.; Rai, A.; Memaya, M.B.; Mehrotra, S.; Mehrotra, R. Co-expression network analysis of protein phosphatase 2A (PP2A) genes with stress-responsive genes in Arabidopsis thaliana reveals 13 key regulators. Sci. Rep. 2020, 10, 21480. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Yu, L.-J.; Zhang, X.; Fan, B.; Wang, F.-Z.; Dai, Y.-S.; Qi, H.; Zhou, Y.; Xie, L.-J.; Xiao, S. Autophagy regulates glucose-mediated root meristem activity by modulating ROS production in Arabidopsis. Autophagy 2019, 15, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zeng, Y.; Jiang, L. COPII vesicles in plant autophagy and endomembrane trafficking. FEBS Lett. 2022, 596, 2314–2323. [Google Scholar] [CrossRef]
- Itakura, E.; Kishi-Itakura, C.; Mizushima, N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 2012, 151, 1256–1269. [Google Scholar] [CrossRef]
- Aniento, F.; Hernández, V.S.d.M.; Dagdas, Y.; Rojas-Pierce, M.; Russinova, E. Molecular mechanisms of endomembrane trafficking in plants. Plant Cell 2022, 34, 146–173. [Google Scholar] [CrossRef] [PubMed]
- Minina, E.A.; Moschou, P.N.; Bozhkov, P.V. Limited and digestive proteolysis: Crosstalk between evolutionary conserved pathways. New Phytol. 2017, 215, 958–964. [Google Scholar] [CrossRef]
- Li, Y.; Li, S.; Wu, H. Ubiquitination-proteasome system (UPS) and autophagy two main protein degradation machineries in response to cell stress. Cells 2022, 11, 851. [Google Scholar] [CrossRef]
- Kocaturk, N.M.; Peker, N.; Eberhart, K.; Akkoc, Y.; Deveci, G.; Dengjel, J.; Gozuacik, D. Novel protein complexes containing autophagy and UPS components regulate proteasome-dependent PARK2 recruitment onto mitochondria and PARK2-PARK6 activity during mitophagy. Cell Death Dis. 2022, 13, 947. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yuan, J.; Li, Y.; Sun, H.; Ma, T.; Huai, J.; Yang, W.; Zhang, W.; Lin, R. The CDC48 complex mediates ubiquitin-dependent degradation of intra-chloroplast proteins in plants. Cell Rep. 2022, 39, 110664. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Fukao, Y.; Iwamoto, M.; Haraguchi, T.; Hara-Nishimura, I. Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell 2010, 22, 4084–4097. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Zhou, X.-J.; Zhang, H. Exploring the role of autophagy-related gene 5 (ATG5) yields important insights into autophagy in autoimmune/autoinflammatory diseases. Front. Immunol. 2018, 9, 2334. [Google Scholar] [CrossRef]
- Huang, R.; Xu, Y.; Wan, W.; Shou, X.; Qian, J.; You, Z.; Liu, B.; Chang, C.; Zhou, T.; Lippincott-Schwartz, J.; et al. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol. Cell 2015, 57, 456–467. [Google Scholar] [CrossRef]
- Li, F.; Zhang, M.; Zhang, C.; Zhou, X. Nuclear autophagy degrades a geminivirus nuclear protein to restrict viral infection in solanaceous plants. New Phytol. 2020, 225, 1746–1761. [Google Scholar] [CrossRef]
- Minina, E.A.; Sanchez-Vera, V.; Moschou, P.N.; Suarez, M.F.; Sundberg, E.; Weih, M.; Bozhkov, P.V. Autophagy mediates caloric restriction-induced lifespan extension in Arabidopsis. Aging Cell 2013, 12, 327–329. [Google Scholar] [CrossRef]
- Su, W.; Bao, Y.; Yu, X.; Xia, X.; Liu, C.; Yin, W. Autophagy and its regulators in response to stress in plants. Int. J. Mol. Sci. 2020, 21, 8889. [Google Scholar] [CrossRef] [PubMed]
- Bieluszewski, T.; Sura, W.; Dziegielewski, W.; Bieluszewska, A.; Lachance, C.; Kabza, M.; Szymanska-Lejman, M.; Abram, M.; Wlodzimierz, P.; De Winne, N.; et al. NuA4 and H2A.Z control environmental responses and autotrophic growth in Arabidopsis. Nat. Commun. 2022, 13, 277. [Google Scholar] [CrossRef] [PubMed]
- Dubiel, M.; Beeckman, T.; Smagghe, G.; Van Damme, E.J.M. Arabidopsis lectin EULS3 is involved in ABA signaling in roots. Front. Plant Sci. 2020, 11, 437. [Google Scholar] [CrossRef]
- Hwang, K.; Susila, H.; Nasim, Z.; Jung, J.-Y.; Ahn, J.H. Arabidopsis ABF3 and ABF4 Transcription Factors Act with the NF-YC Complex to Regulate SOC1 Expression and Mediate Drought-Accelerated Flowering. Mol. Plant 2019, 12, 489–505. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Hama, Y.; Mizushima, N. The evolution of autophagy proteins—Diversification in eukaryotes and potential ancestors in prokaryotes. J. Cell Sci. 2021, 134, jcs233742. [Google Scholar] [CrossRef] [PubMed]
- Nakasone, A.; Fujiwara, M.; Fukao, Y.; Biswas, K.K.; Rahman, A.; Kawai-Yamada, M.; Narumi, I.; Uchimiya, H.; Oono, Y. SMALL ACIDIC PROTEIN1 acts with RUB modification components, the COP9 signalosome, and AXR1 to regulate growth and development of Arabidopsis. Plant Physiol. 2012, 160, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.H.; Shen, Y.P.; Sullivan, J.A.; Rubio, V.; Xiong, Y.; Sun, T.-P.; Deng, X.W. Arabidopsis CAND1, an unmodified CUL1-interacting protein, is involved in multiple developmental pathways controlled by ubiquitin/proteasome-mediated protein degradation. Plant Cell 2004, 16, 1870–1882. [Google Scholar] [CrossRef]
- Huotari, J.; Helenius, A. Endosome maturation. EMBO J. 2011, 30, 3481–3500. [Google Scholar] [CrossRef]
- Di Sansebastiano, G.P.; Barozzi, F.; Piro, G.; Denecke, J.; de Marcos Lousa, C. Trafficking routes to the plant vacuole: Connecting alternative and classical pathways. J. Exp. Bot. 2018, 69, 79–90. [Google Scholar] [CrossRef]
- Hershko, A.; Heller, H.; Elias, S.; Ciechanover, A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J. Biol. Chem. 1983, 258, 8206–8214. [Google Scholar] [CrossRef]
- Nunoura, T.; Takaki, Y.; Kakuta, J.; Nishi, S.; Sugahara, J.; Kazama, H.; Chee, G.-J.; Hattori, M.; Kanai, A.; Atomi, H.; et al. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res. 2011, 39, 3204–3223. [Google Scholar] [CrossRef]
- Ji, C.H.; Kwon, Y.T. Crosstalk and interplay between the ubiquitin-proteasome system and autophagy. Mol. Cells 2017, 40, 441. [Google Scholar]
- Yamasaki, A.; Noda, N.N. Structural Biology of the Cvt Pathway. J. Mol. Biol. 2017, 429, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Nakano, A. The Golgi apparatus and its next-door neighbors. Front. Cell Dev. Biol. 2022, 10, 884360. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Eskelinen, E.-L. The vacuole vs. the lysosome. Autophagy 2014, 10, 185–187. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.G. The Golgi Apparatus and the Plant Secretory Pathway; Blackwell: Oxford, UK, 2003; Volume 9. [Google Scholar]
- Contento, A.L.; Bassham, D.C. Structure and function of endosomes in plant cells. J. Cell Sci. 2012, 125, 3511–3518. [Google Scholar] [CrossRef]
- Yorimitsu, T.; Sato, K.; Takeuchi, M. Molecular mechanisms of Sar/Arf GTPases in vesicular trafficking in yeast and plants. Front. Plant Sci. 2014, 5, 411. [Google Scholar] [CrossRef]
- Chung, K.P.; Zeng, Y.; Jiang, L. COPII paralogs in plants: Functional redundancy or diversity? Trends Plant Sci. 2016, 21, 758–769. [Google Scholar] [CrossRef]
- Hanton, S.L.; Chatre, L.; Matheson, L.A.; Rossi, M.; Held, M.A.; Brandizzi, F. Plant Sar1 isoforms with near-identical protein sequences exhibit different localisations and effects on secretion. Plant Mol. Biol. 2008, 67, 283–294. [Google Scholar] [CrossRef]
- Liang, X.; Li, S.-W.; Gong, L.-M.; Li, S.; Zhang, Y. COPII components Sar1b and Sar1c play distinct yet interchangeable roles in pollen development. Plant Physiol. 2020, 183, 974–985. [Google Scholar] [CrossRef]
- Kim, J.H.; Lee, H.N.; Huang, X.; Jung, H.; Otegui, M.S.; Li, F.; Chung, T. FYVE2, a phosphatidylinositol 3-phosphate effector, interacts with the COPII machinery to control autophagosome formation in Arabidopsis. Plant Cell 2022, 34, 351–373. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zeng, Y.; Cao, W.; Zhang, W.; Cheng, L.; Yin, H.; Wu, Q.; Wang, X.; Huang, Y.; Lau, W.C.Y.; et al. A distinct giant coat protein complex II vesicle population in Arabidopsis thaliana. Nat. Plants 2021, 7, 1335–1346. [Google Scholar] [CrossRef] [PubMed]
- Zavodszky, E.; Seaman, M.N.; Moreau, K.; Jimenez-Sanchez, M.; Breusegem, S.Y.; Harbour, M.E.; Rubinsztein, D.C. Mutation in VPS35 associated with Parkinson’s disease impairs WASH complex association and inhibits autophagy. Nat. Commun. 2014, 5, 3828. [Google Scholar] [CrossRef]
- Rodriguez-Furlan, C.; Domozych, D.; Qian, W.; Enquist, P.-A.; Li, X.; Zhang, C.; Schenk, R.; Winbigler, H.S.; Jackson, W.; Raikhel, N.V.; et al. Interaction between VPS35 and RABG3f is necessary as a checkpoint to control fusion of late compartments with the vacuole. Proc. Natl. Acad. Sci. USA 2019, 116, 21291–21301. [Google Scholar] [CrossRef] [PubMed]
- Pourcher, M.; Santambrogio, M.; Thazar, N.; Thierry, A.-M.; Fobis-Loisy, I.; Miège, C.; Jaillais, Y.; Gaude, T. Analyses of sorting nexins reveal distinct retromer-subcomplex functions in development and protein sorting in Arabidopsis thaliana. Plant Cell 2010, 22, 3980–3991. [Google Scholar] [CrossRef] [PubMed]
- Heucken, N.; Ivanov, R. The retromer, sorting nexins and the plant endomembrane protein trafficking. J. Cell Sci. 2018, 131, jcs203695. [Google Scholar] [CrossRef]
- Ambrose, C.; Ruan, Y.; Gardiner, J.; Tamblyn, L.M.; Catching, A.; Kirik, V.; Marc, J.; Overall, R.; Wasteneys, G.O. CLASP interacts with sorting nexin 1 to link microtubules and auxin transport via PIN2 recycling in Arabidopsis thaliana. Dev. Cell 2013, 24, 649–659. [Google Scholar] [CrossRef]
- Cui, Y.; Parashar, S.; Zahoor, M.; Needham, P.G.; Mari, M.; Zhu, M.; Chen, S.; Ho, H.-C.; Reggiori, F.; Farhan, H.; et al. A COPII subunit acts with an autophagy receptor to target endoplasmic reticulum for degradation. Science (1979) 2019, 365, 53–60. [Google Scholar] [CrossRef]
- Wang, P.; Pleskot, R.; Zang, J.; Winkler, J.; Wang, J.; Yperman, K.; Zhang, T.; Wang, K.; Gong, J.; Guan, Y.; et al. Plant AtEH/Pan1 proteins drive autophagosome formation at ER-PM contact sites with actin and endocytic machinery. Nat. Commun. 2019, 10, 5132. [Google Scholar] [CrossRef]
- Zhuang, X.; Wang, H.; Lam, S.K.; Gao, C.; Wang, X.; Cai, Y.; Jiang, L. A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell 2013, 25, 4596–4615. [Google Scholar] [CrossRef]
- Nascimbeni, A.C.; Giordano, F.; Dupont, N.; Grasso, D.; I Vaccaro, M.; Codogno, P.; Morel, E. ER–plasma membrane contact sites contribute to autophagosome biogenesis by regulation of local PI 3P synthesis. EMBO J. 2017, 36, 2018–2033. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.-H.; Shen, S.-C.; Lee, L.-Y.; Lee, S.-H.; Chan, M.-T.; Lin, C.-S. Tape-Arabidopsis Sandwich-a simpler Arabidopsis protoplast isolation method. Plant Methods 2009, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.-D.; Cho, Y.-H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.R.; Doelling, J.H.; Suttangkakul, A.; Vierstra, R.D. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 2005, 138, 2097–2110. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Krishnakumar, V.; Chan, A.P.; Thibaud-Nissen, F.; Schobel, S.; Town, C.D. Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017, 89, 789–804. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Cook, K.D. Oxidation artifacts in the electrospray mass spectrometry of Aβ peptide. Anal. Chem. 2007, 79, 2031–2036. [Google Scholar] [CrossRef] [PubMed]
- Sticker, A.; Goeminne, L.; Martens, L.; Clement, L. Robust summarization and inference in proteome-wide label-free quantification. Mol. Cell. Proteom. 2020, 19, 1209–1219. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elander, P.H.; Holla, S.; Sabljić, I.; Gutierrez-Beltran, E.; Willems, P.; Bozhkov, P.V.; Minina, E.A. Interactome of Arabidopsis ATG5 Suggests Functions beyond Autophagy. Int. J. Mol. Sci. 2023, 24, 12300. https://doi.org/10.3390/ijms241512300
Elander PH, Holla S, Sabljić I, Gutierrez-Beltran E, Willems P, Bozhkov PV, Minina EA. Interactome of Arabidopsis ATG5 Suggests Functions beyond Autophagy. International Journal of Molecular Sciences. 2023; 24(15):12300. https://doi.org/10.3390/ijms241512300
Chicago/Turabian StyleElander, Pernilla H., Sanjana Holla, Igor Sabljić, Emilio Gutierrez-Beltran, Patrick Willems, Peter V. Bozhkov, and Elena A. Minina. 2023. "Interactome of Arabidopsis ATG5 Suggests Functions beyond Autophagy" International Journal of Molecular Sciences 24, no. 15: 12300. https://doi.org/10.3390/ijms241512300
APA StyleElander, P. H., Holla, S., Sabljić, I., Gutierrez-Beltran, E., Willems, P., Bozhkov, P. V., & Minina, E. A. (2023). Interactome of Arabidopsis ATG5 Suggests Functions beyond Autophagy. International Journal of Molecular Sciences, 24(15), 12300. https://doi.org/10.3390/ijms241512300






