Single-Cell Transcriptome and Pigment Biochemistry Analysis Reveals the Potential for the High Nutritional and Medicinal Value of Purple Sea Cucumbers
Abstract
1. Introduction
2. Results
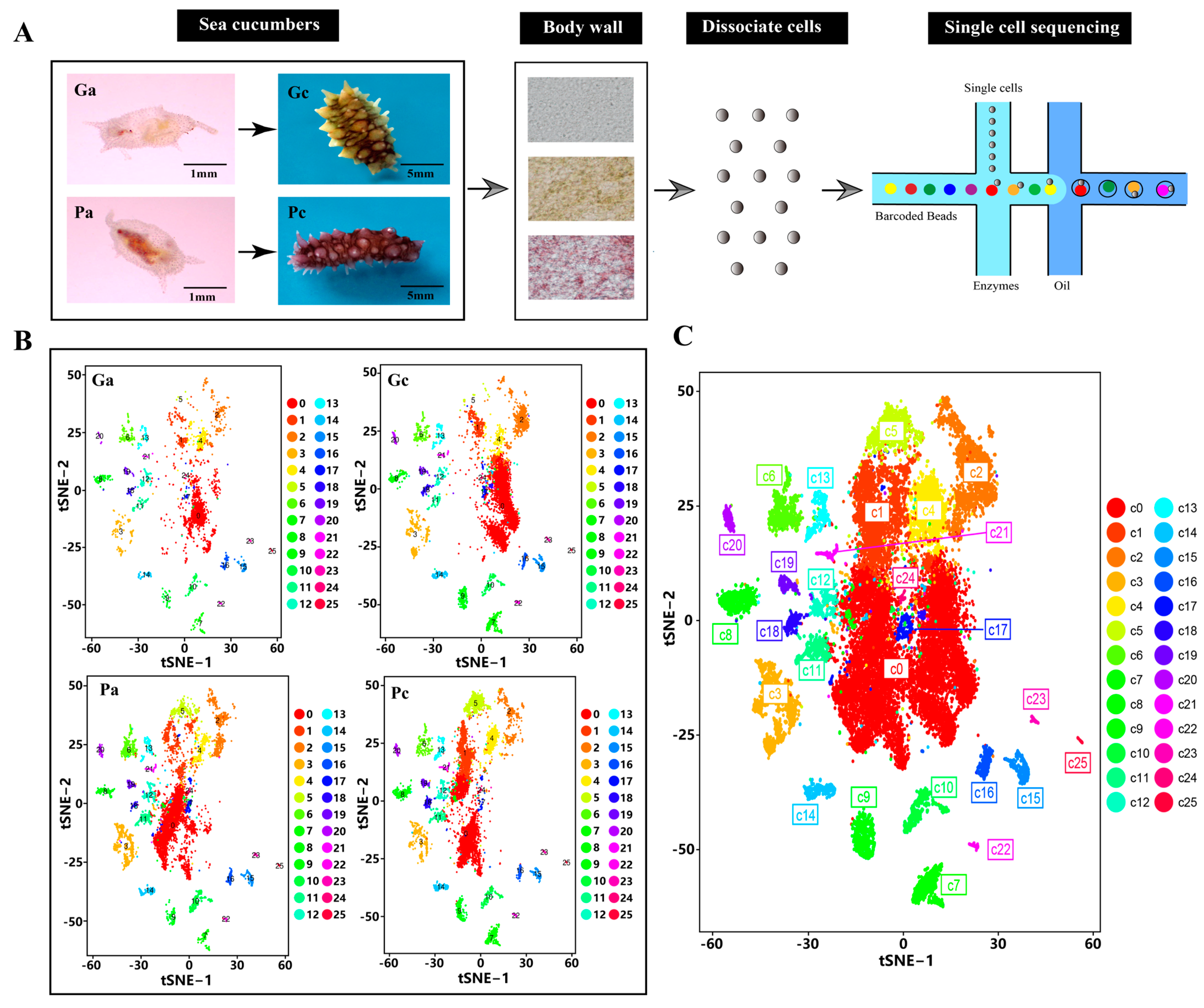
2.1. Transcriptional Clustering of Cell Populations in the Body Wall of the Sea Cucumber
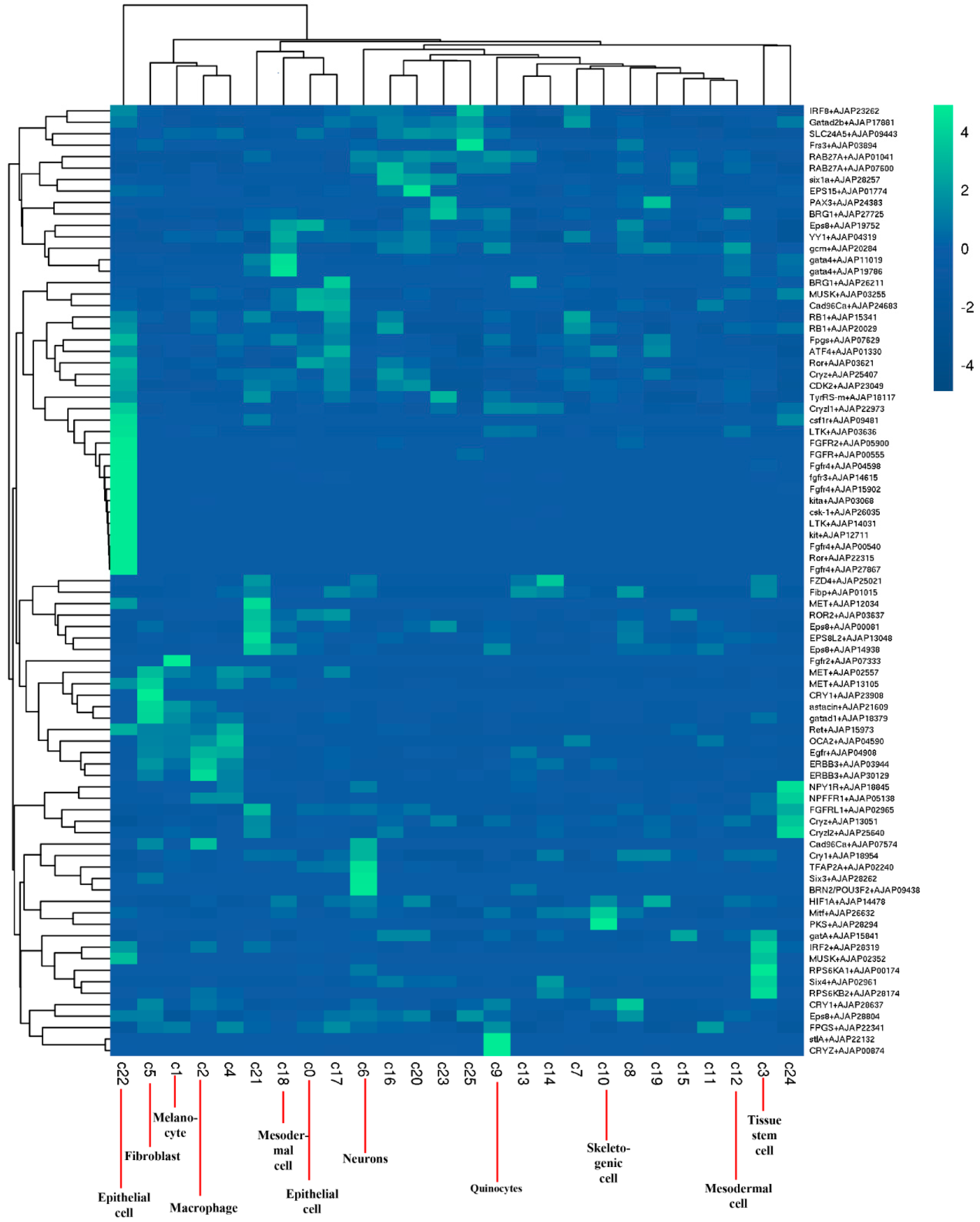
2.2. Identification of Cell Populations
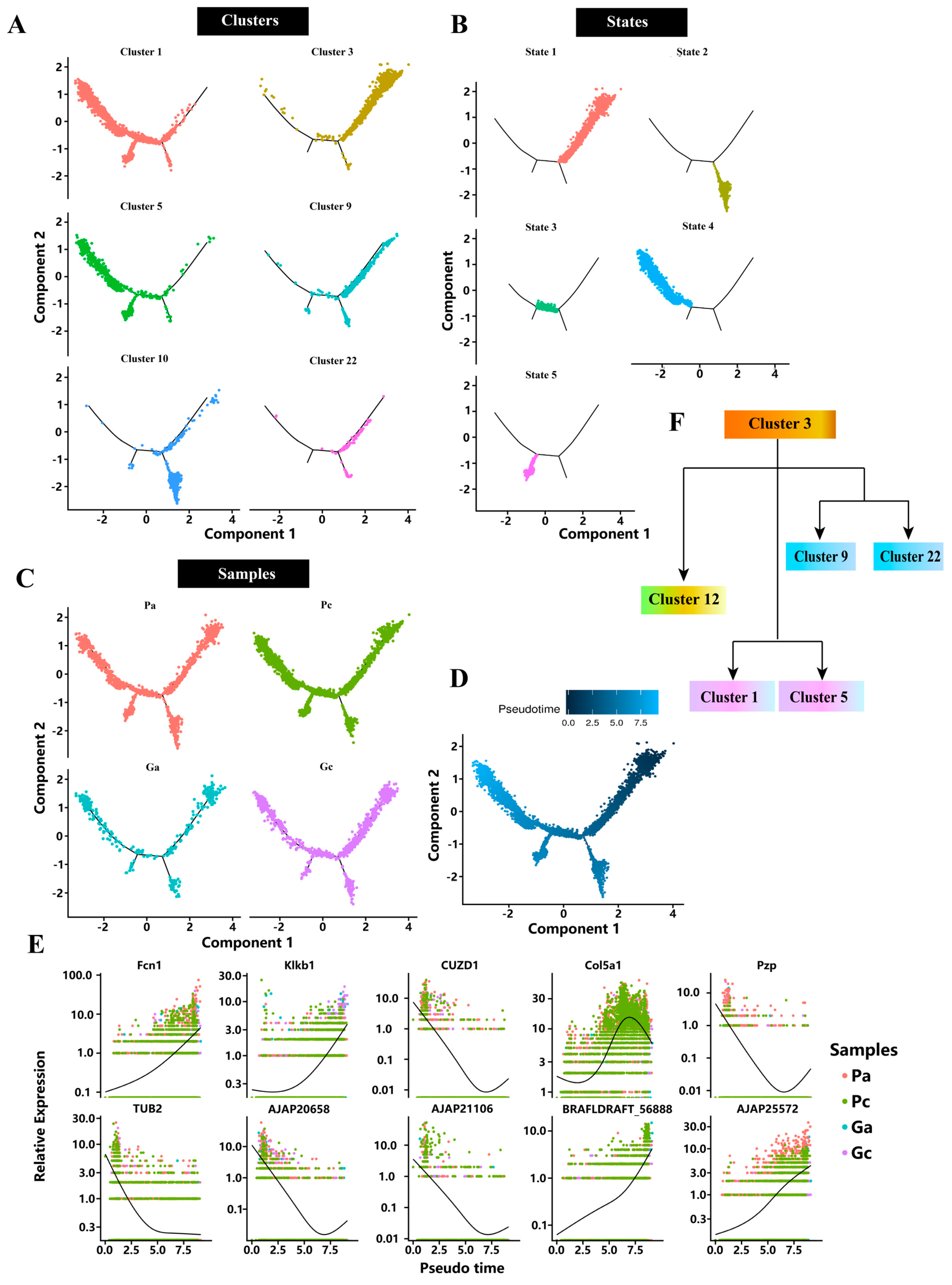
2.3. Pseudotime Analysis
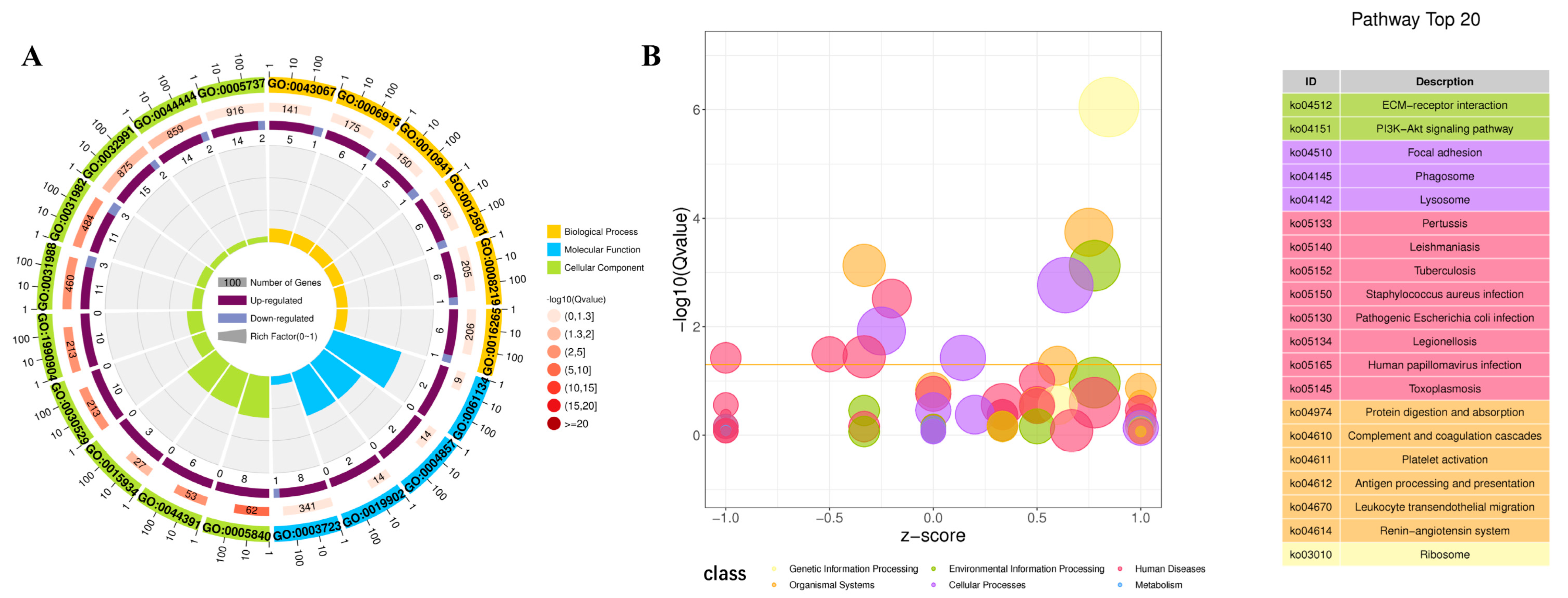
2.4. Analysis of DEGs at Different Development Stages
2.5. Analysis of DEGs in Different Color Morphs
2.6. Melanin and Quinone Pigments Content in Sea Cucumbers
3. Discussion
4. Materials and Methods
4.1. Animal Sampling
4.2. Body-Wall Dissociation and Single-Cell Suspension Preparation
4.3. Library Preparation and Sequencing
4.4. Quality Control and Data Processing
4.5. t-Distribution Stochastic Neighbor Embedding (t-SNE) Visualization and SingleR Analysis
4.6. Analysis of Differentially Expressed Genes (DEGs)
4.7. Pseudotime Analysis
4.8. Pigment Content Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Protas, M.E.; Patel, N.H. Evolution of Coloration Patterns. In Annual Review of Cell and Developmental Biology; FAnnual Reviews: San Mateo, CA, USA, 2008; Volume 24, pp. 425–446. [Google Scholar]
- Roulin, A. The evolution, maintenance and adaptive function of genetic colour polymorphism in birds. Biol. Rev. 2004, 79, 815–848. [Google Scholar] [CrossRef] [PubMed]
- Hoekstra, H.E. Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity 2006, 97, 222. [Google Scholar] [CrossRef] [PubMed]
- Okorie, O.E.; Ko, S.H.; Go, S.; Lee, S.; Bae, J.Y.; Han, K.M.; Bai, S.C. Preliminary Study of the Optimum Dietary Ascorbic Acid Level in Sea Cucumber, Apostichopus japonicus (Selenka). J. World Aquac. Soc. 2008, 39, 758–765. [Google Scholar] [CrossRef]
- Jiang, S.; Dong, S.; Gao, Q.; Wang, F.; Tian, X. Comparative study on nutrient composition and growth of green and red sea cucumber, Apostichopus japonicus (Selenka, 1867), under the same culture conditions. Aquac. Res. 2013, 44, 317–320. [Google Scholar] [CrossRef]
- Zhang, W.; Cao, Z.; Li, Y.; Zhao, H.; Huang, J.; Liang, Z.; Huang, L. Taxonomic status of the three color variants in sea cucumber (Apostichopus japonicus): Evidence from mitochondrial phylogenomic analyses. Mitochondrial DNA 2015, 27, 2330–2333. [Google Scholar] [CrossRef] [PubMed]
- Klein, A.M.; Mazutis, L.; Akartuna, I.; Tallapragada, N.; Veres, A.; Li, V.; Peshkin, L.; Weitz, D.A.; Kirschner, M.W. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 2015, 161, 1187–1201. [Google Scholar] [CrossRef]
- Hwang, B.; Lee, J.H.; Bang, D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp. Mol. Med. 2018, 50, 1–14. [Google Scholar] [CrossRef]
- Perillo, M.; Oulhen, N.; Foster, S.; Spurrell, M.; Calestani, C.; Wessel, G. Regulation of dynamic pigment cell states at single-cell resolution. Elife 2020, 9, e60388. [Google Scholar] [CrossRef]
- Paganos, P.; Voronov, D.; Musser, J.M.; Arendt, D.; Arnone, M.I. Single-cell RNA sequencing of the Strongylocentrotus purpuratus larva reveals the blueprint of major cell types and nervous system of a non-chordate deuterostome. Elife 2021, 10, e70416. [Google Scholar] [CrossRef]
- Massri, A.J.; Greenstreet, L.; Afanassiev, A.; Berrio, A.; Wray, G.A.; Schiebinger, G.; McClay, D.R. Developmental single-cell transcriptomics in the Lytechinus variegatus sea urchin embryo. Development 2021, 148, 198614. [Google Scholar] [CrossRef]
- Xing, L.; Sun, L.; Liu, S.; Li, X.; Miao, T.; Zhang, L.; Yang, H. Comparison of pigment composition and melanin content among white, light-green, dark-green, and purple morphs of sea cucumber, Apostichopus japonicus. Acta Oceanol. Sin. 2017, 36, 45–51. [Google Scholar] [CrossRef]
- Cordero, R.J.B.; Casadevall, A. Melanin. Curr. Biol. 2020, 30, R142–R143. [Google Scholar] [CrossRef] [PubMed]
- Sroyraya, M.; Kaewphalug, W.; Anantachoke, N.; Poomtong, T.; Sobhon, P.; Srimongkol, A.; Suphamungmee, W. Saponins enriched in the epidermal layer of Holothuria leucospilota body wall. Microsc. Res. Tech. 2018, 81, 1182–1190. [Google Scholar] [CrossRef] [PubMed]
- Schock, E.N.; York, J.R.; LaBonne, C. The developmental and evolutionary origins of cellular pluripotency in the vertebrate neural crest. Semin. Cell Dev. Biol. 2023, 138, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Brandon, A.A.; Almeida, D.; Powder, K.E. Neural crest cells as a source of microevolutionary variation. Semin. Cell Dev. Biol. 2023, 145, 42–51. [Google Scholar] [CrossRef]
- Crane, J.F.; Trainor, P.A. Neural crest stem and progenitor cells. Annu. Rev. Cell Dev. Biol. 2006, 22, 267–286. [Google Scholar] [CrossRef]
- Knecht, A.K.; Bronner-Fraser, M. Induction of the neural crest: A multigene process. Nat. Rev. Genet. 2002, 3, 453–461. [Google Scholar] [CrossRef]
- Le Douarin, N.M. The avian embryo as a model to study the development of the neural crest: A long and still ongoing story. Mech. Dev. 2004, 121, 1089–1102. [Google Scholar] [CrossRef]
- Mort, R.L.; Jackson, I.J.; Patton, E.E. The melanocyte lineage in development and disease. Development 2015, 142, 620–632. [Google Scholar] [CrossRef]
- Budi, E.H.; Patterson, L.B.; Parichy, D.M. Post-Embryonic Nerve-Associated Precursors to Adult Pigment Cells: Genetic Requirements and Dynamics of Morphogenesis and Differentiation. PLoS Genet. 2011, 7, e1002044. [Google Scholar] [CrossRef]
- Dooley, C.M.; Mongera, A.; Walderich, B.; Nüsslein-Volhard, C. On the embryonic origin of adult melanophores: The role of ErbB and Kit signalling in establishing melanophore stem cells in zebrafish. Development 2013, 140, 1003–1013. [Google Scholar] [CrossRef]
- Hurbain, I.; Geerts, W.J.; Boudier, T.; Marco, S.; Verkleij, A.J.; Marks, M.S.; Raposo, G. Electron tomography of early melanosomes: Implications for melanogenesis and the generation of fibrillar amyloid sheets. Proc. Natl. Acad. Sci. USA 2008, 105, 19726–19731. [Google Scholar] [CrossRef] [PubMed]
- Ghattavi, K.; Homaei, A.; Kamrani, E.; Kim, S.-K. Melanin pigment derived from marine organisms and its industrial applications. Dye. Pigment. 2022, 201, 110214. [Google Scholar] [CrossRef]
- Różanowska, M.; Sarna, T.; Land, E.J.; Truscott, T.G. Free radical scavenging properties of melanin: Interaction of eu-and pheo-melanin models with reducing and oxidising radicals. Free. Radic. Biol. Med. 1999, 26, 518–525. [Google Scholar] [CrossRef] [PubMed]
- Manning, J.T.; Bundred, P.E.; Henzi, P. Melanin and HIV in sub-Saharan Africa. J. Theor. Biol. 2003, 223, 131–133. [Google Scholar] [CrossRef]
- Sava, V.M.; Galkin, B.N.; Hong, M.Y.; Yang, P.C.; Huang, G.S. A novel melanin-like pigment derived from black tea leaves with immune-stimulating activity. Food Res. Int. 2001, 34, 337–343. [Google Scholar] [CrossRef]
- Geng, J.; Yu, S.-B.; Wan, X.; Wang, X.-J.; Shen, P.; Zhou, P.; Chen, X.-D. Protective action of bacterial melanin against spectrums by a sensitive plasmid-based DNA damage in full UV noncellular system. J. Biochem. Biophys. Methods 2008, 70, 1151–1155. [Google Scholar] [CrossRef]
- Liu, J.-H.; Tian, Y.-G.; Wang, Y.; Nie, S.-P.; Xie, M.-Y.; Zhu, S.; Wang, C.-Y.; Zhang, P. Characterization and in vitro antioxidation of papain hydrolysate from black-bone silky fowl (Gallus gallus domesticus Brisson) muscle and its fractions. Food Res. Int. 2011, 44, 133–138. [Google Scholar] [CrossRef]
- Sava, V.M.; Hung, Y.C.; Golkin, B.N.; Hong, M.Y.; Huang, G.S. Protective activity of melanin-like pigment derived from tea on Drosophila melasnogaster against the toxic effects of benzidine. Food Res. Int. 2002, 35, 619–626. [Google Scholar] [CrossRef]
- Wu, Y.; Shan, L.; Yang, S.; Ma, A. Identification and antioxidant activity of melanin isolated from Hypoxylon archeri, a companion fungus of Tremella fuciformis. J. Basic Microbiol. 2008, 48, 217–221. [Google Scholar] [CrossRef]
- Rosas, A.L.; Casadevall, A. Melanization decreases the susceptibility of Cryptococcus neoformans to enzymatic degradation. Mycopathologia 2001, 151, 53–56. [Google Scholar] [CrossRef] [PubMed]
- Lamason, R.L.; Mohideen, M.-A.P.K.; Mest, J.R.; Wong, A.C.; Norton, H.L.; Aros, M.C.; Jurynec, M.J.; Mao, X.; Humphreville, V.R.; Humbert, J.E.; et al. SLC24A5, a Putative Cation Exchanger, Affects Pigmentation in Zebrafish and Humans. Science 2005, 310, 1782–1786. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Cui, F.; Yang, Y.; Liu, Y.; Qi, S.; Wang, C. Mechanisms of developmental toxicity in zebrafish embryos (Danio rerio) induced by boscalid. Sci. Total Environ. 2018, 634, 478–487. [Google Scholar] [CrossRef]
- Jablonski, N.G. The evolution of human skin pigmentation involved the interactions of genetic, environmental, and cultural variables. Pigment. Cell Melanoma Res. 2021, 34, 707–729. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.-J.; Huang, C.-H.; Liu, P.-C.; Lin, I.C.; Huang, Y.-L.; Chen, A.-Y.; Chiu, H.-P.; Shih, S.-R.; Lin, L.-H.; Lien, S.-P.; et al. Zinc finger protein ZFP36L1 inhibits influenza A virus through translational repression by targeting HA, M and NS RNA transcripts. Nucleic Acids Res. 2020, 48, 7371–7384. [Google Scholar] [CrossRef]
- Prenzler, F.; Fragasso, A.; Schmitt, A.; Munz, B. Functional analysis of ZFP36 proteins in keratinocytes. Eur. J. Cell Biol. 2016, 95, 277–284. [Google Scholar] [CrossRef]
- Michelini, S.; Amato, B.; Ricci, M.; Serrani, R.; Veselenyiova, D.; Kenanoglu, S.; Kurti, D.; Dautaj, A.; Baglivo, M.; Compagna, R.; et al. SVEP1 is important for morphogenesis of lymphatic system: Possible implications in lymphedema. Lymphology 2021, 54, 12–22. [Google Scholar] [CrossRef]
- Samuelov, L.; Li, Q.; Bochner, R.; Najor, N.A.; Albrecht, L.; Malchin, N.; Goldsmith, T.; Grafi-Cohen, M.; Vodo, D.; Fainberg, G.; et al. SVEP1 plays a crucial role in epidermal differentiation. Exp. Dermatol. 2017, 26, 423–430. [Google Scholar] [CrossRef]
- Lv, C.; Zhao, G.; Loennerdal, B. Bioavailability of iron from plant and animal ferritins. J. Nutr. Biochem. 2015, 26, 532–540. [Google Scholar] [CrossRef]
- Tribl, F.; Asan, E.; Arzberger, T.; Meyer, H.; Gerlach, M.; Riederer, P.; Marcus, K. Ferritin is located within neuromelanin granules, an iron storing organelle in dopaminergic neurons of the human brain. Mol. Cell. Proteom. 2006, 5, S159. [Google Scholar]
- Kominami, T.; Takata, H.; Takaichi, M. Behavior of pigment cells in gastrula-stage embryos of Hemicentrotus pulcherrimus and Scaphechinus mirabilis. Dev. Growth Differ. 2001, 43, 699–707. [Google Scholar] [CrossRef] [PubMed]
- Calestani, C.; Rast, J.P.; Davidson, E.H. Isolation of pigment cell specific genes in the sea urchin embryo by differential macroarray screening. Development 2003, 130, 4587–4596. [Google Scholar] [CrossRef] [PubMed]
- Oulhen, N.; Wessel, G.M. Albinism as a Visual, In Vivo Guide for CRISPR/Cas9 Functionality in the Sea Urchin Embryo. Mol. Reprod. Dev. 2016, 83, 1046–1047. [Google Scholar] [CrossRef]
- Wessel, G.M.; Kiyomoto, M.; Shen, T.-L.; Yajima, M. Genetic manipulation of the pigment pathway in a sea urchin reveals distinct lineage commitment prior to metamorphosis in the bilateral to radial body plan transition. Sci. Rep. 2020, 10, 1973. [Google Scholar] [CrossRef]
- Materna, S.C.; Davidson, E.H. A comprehensive analysis of Delta signaling in pre-gastrular sea urchin embryos. Dev. Biol. 2012, 364, 77–87. [Google Scholar] [CrossRef] [PubMed]
- McClay, D.R.; Peterson, R.E.; Range, R.C.; Winter-Vann, A.M.; Ferkowicz, M.J. A micromere induction signal is activated by beta-catenin and acts through Notch to initiate specification of secondary mesenchyme cells in the sea urchin embryo. Development 2000, 127, 5113–5122. [Google Scholar] [CrossRef]
- Sherwood, D.R.; McClay, D.R. LvNotch signaling mediates secondary mesenchyme specification in the sea urchin embryo. Development 1999, 126, 1703–1713. [Google Scholar] [CrossRef]
- Hausen, B.; Schiedermair, I. The sensitizing capacity of chimaphilin, a naturally-occurring quinone. Contact Dermat. 1988, 19, 180–183. [Google Scholar] [CrossRef]
- Daqian, W.; Chuandong, W.; Xinhua, Q.; Songtao, A.; Kerong, D. Chimaphilin inhibits proliferation and induces apoptosis in multidrug resistant osteosarcoma cell lines through insulin-like growth factor-I receptor (IGF-IR) signaling. Chem. Biol. Interact. 2015, 237, 25–30. [Google Scholar] [CrossRef]
- Balogun, B.A.; Aliyu, N.O.; Ajiboye, T.O. Menadione perturbs oxidative stress biomarkers and testicular function indices of rats. J. Biochem. Mol. Toxicol. 2019, 33, e22282. [Google Scholar] [CrossRef]
- Kim, E.-H.; Kim, M.-K.; Yun, H.-Y.; Baek, K.J.; Kwon, N.S.; Park, K.-C.; Kim, D.-S. Menadione (Vitamin K3) decreases melanin synthesis through ERK activation in Mel-Ab cells. Eur. J. Pharmacol. 2013, 718, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhang, H.; Xu, Y.; Zhang, J.; Zhu, W.; Zhang, Y.; Chen, L.; Hua, W.; Mao, Y. Juglone induces apoptosis of tumor stem-like cells through ROS-p38 pathway in glioblastoma. BMC Neurol. 2017, 17, 70. [Google Scholar] [CrossRef]
- Dos S Moreira, C.; Santos, T.B.; Freitas, R.H.; Pacheco, P.A.; da Rocha, D.R. Juglone: A Versatile Natural Platform for Obtaining New Bioactive Compounds. Curr. Top. Med. Chem. 2021, 21, 2018–2045. [Google Scholar] [CrossRef]
- Jha, B.K.; Jung, H.J.; Seo, I.; Suh, S.I.; Suh, M.H.; Baek, W.K. Juglone induces cell death of Acanthamoeba through increased production of reactive oxygen species. Exp. Parasitol. 2015, 159, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Arifeen, W.-U.; Rehman, F.-U.; Adeel, S.; Zuber, M.; Ahmad, M.N.; Ahmad, T. Environmental friendly extraction of walnut bark-based juglone natural colorant for dyeing studies of wool fabric. Environ. Sci. Pollut. Res. 2021, 28, 49958–49966. [Google Scholar] [CrossRef] [PubMed]
- Xavier, M.R.; Santos, M.M.S.; Queiroz, M.G.; de Lima Silva, M.S.; Goes, A.J.S.; De Morais, M.A., Jr. Lawsone, a 2-hydroxy-1,4-naphthoquinone from Lawsonia inermis (henna), produces mitochondrial dysfunctions and triggers mitophagy in Saccharomyces cerevisiae. Mol. Biol. Rep. 2020, 47, 1173–1185. [Google Scholar] [CrossRef]
- Meazza, G.; Dayan, F.E.; Wedge, D.E. Activity of quinones on Colletotrichum species. J. Agric. Food Chem. 2003, 51, 3824–3828. [Google Scholar] [CrossRef]
- Zwirchmayr, N.S.; Hosoya, T.; Henniges, U.; Gille, L.; Bacher, M.; Furtmüller, P.; Rosenau, T. Degradation of the Cellulosic Key Chromophore 5,8-Dihydroxy-[1,4]-naphthoquinone by Hydrogen Peroxide under Alkaline Conditions. J. Org. Chem. 2017, 82, 11558–11565. [Google Scholar] [CrossRef]
- Kalinin, V.I. Echinoderms Metabolites: Structure, Functions, and Biomedical Perspectives. Mar. Drugs 2021, 19, 125. [Google Scholar] [CrossRef]
- Rahman, M.M.; Islam, M.R.; Akash, S.; Shohag, S.; Ahmed, L.; Supti, F.A.; Rauf, A.; Aljohani, A.S.M.; Al Abdulmonem, W.; Khalil, A.A.; et al. Naphthoquinones and derivatives as potential anticancer agents: An updated review. Chem. Biol. Interact. 2022, 368, 110198. [Google Scholar] [CrossRef]
- Xu, D.; Chengdong, F.; Ying, T. The Growing seedling Technique of Apostichoupus japonicus. Hebei Fisheries 2009, 5, 33–35. (In Chinese) [Google Scholar]
- Chung, N.C.; Storey, J.D. Statistical significance of variables driving systematic variation in high-dimensional data. Bioinformatics 2015, 31, 545–554. [Google Scholar] [CrossRef]
- McDavid, A.; Finak, G.; Chattopadyay, P.K.; Dominguez, M.; Lamoreaux, L.; Ma, S.S.; Roederer, M.; Gottardo, R. Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics 2013, 29, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Aran, D.; Looney, A.P.; Liu, L.; Wu, E.; Fong, V.; Hsu, A.; Chak, S.; Naikawadi, R.P.; Wolters, P.J.; Abate, A.R.; et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019, 20, 163–172. [Google Scholar] [CrossRef]
- Butler, A.; Hoffman, P.; Smibert, P.; Papalexi, E.; Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018, 36, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., III; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902. [Google Scholar] [CrossRef] [PubMed]
- Finak, G.; McDavid, A.; Yajima, M.; Deng, J.; Gersuk, V.; Shalek, A.K.; Slichter, C.K.; Miller, H.W.; McElrath, M.J.; Prlic, M.; et al. MAST: A flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015, 16, 278. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Trapnell, C.; Cacchiarelli, D.; Grimsby, J.; Pokharel, P.; Li, S.; Morse, M.; Lennon, N.J.; Livak, K.J.; Mikkelsen, T.S.; Rinn, J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014, 32, 381–386. [Google Scholar] [CrossRef]
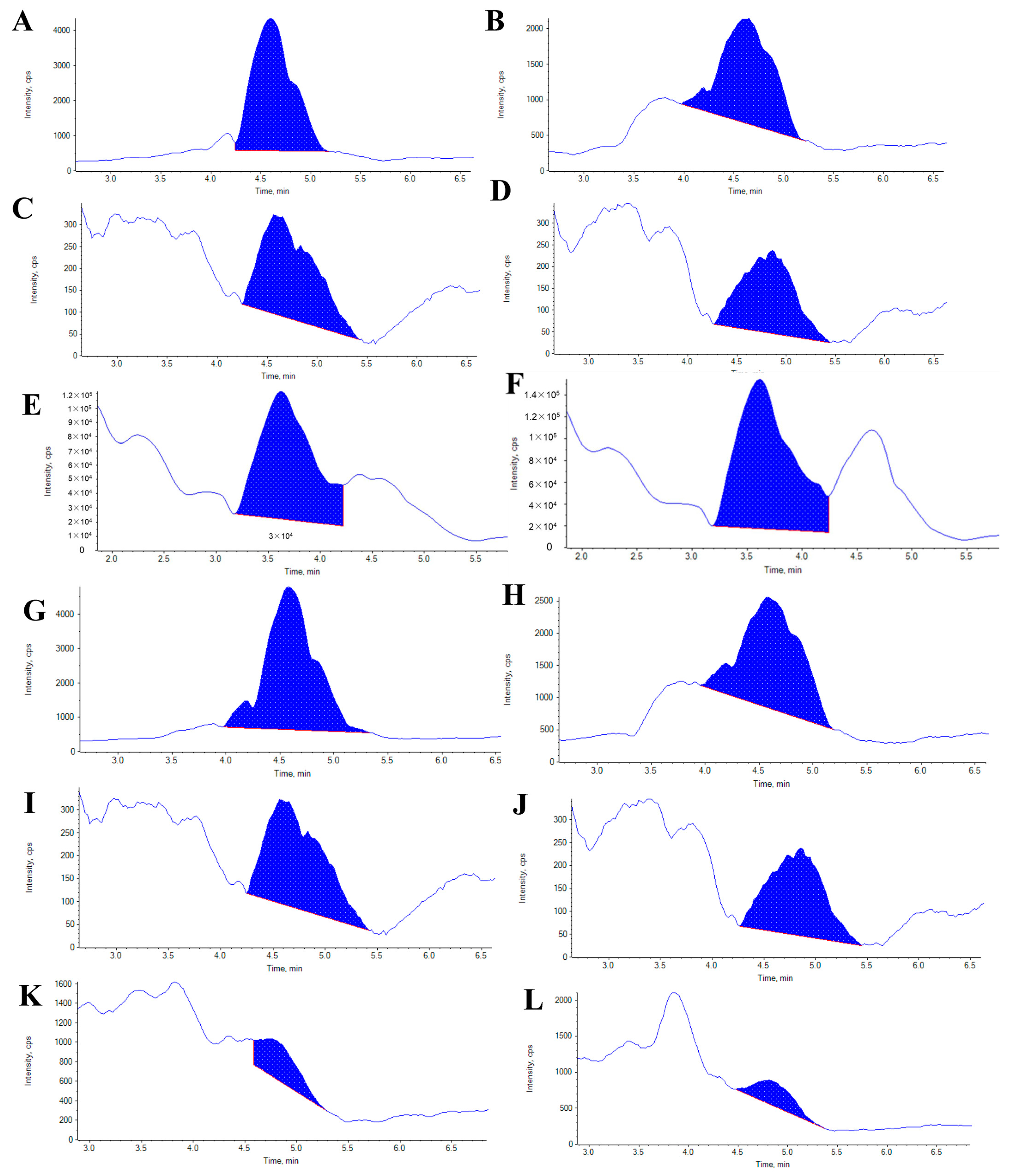
| Pigment | Purple A. japonicus | Green A. japonicus |
|---|---|---|
| Melanin | 41.21± 1.22 | 19.12 ± 1.11 |
| Chimaphilin (2,7-dimethyl-1,4-naphthoquinone) | 57.88 ± 4.98 | 31.26 ± 1.53 |
| Menadione (2-methyl-1,4-naphthoquinone) | 40.33 ± 0.39 | 53.15 ± 1.95 |
| Juglone (5-hydroxy-1,4-naphthoquinone) | 10.15 ± 1.12 | 1.32 ± 0.17 |
| Lawsone (2-hydroxy-1,4-naphthoquinone) | 27.69 ± 1.43 | 8.23 ± 0.57 |
| 5,8-dihydroxy-1,4-naphthoquinone | 23.68 ± 2.63 | 16.20 ± 1.31 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xing, L.; Wang, L.; Liu, S.; Sun, L.; Wessel, G.M.; Yang, H. Single-Cell Transcriptome and Pigment Biochemistry Analysis Reveals the Potential for the High Nutritional and Medicinal Value of Purple Sea Cucumbers. Int. J. Mol. Sci. 2023, 24, 12213. https://doi.org/10.3390/ijms241512213
Xing L, Wang L, Liu S, Sun L, Wessel GM, Yang H. Single-Cell Transcriptome and Pigment Biochemistry Analysis Reveals the Potential for the High Nutritional and Medicinal Value of Purple Sea Cucumbers. International Journal of Molecular Sciences. 2023; 24(15):12213. https://doi.org/10.3390/ijms241512213
Chicago/Turabian StyleXing, Lili, Lingyu Wang, Shilin Liu, Lina Sun, Gary M. Wessel, and Hongsheng Yang. 2023. "Single-Cell Transcriptome and Pigment Biochemistry Analysis Reveals the Potential for the High Nutritional and Medicinal Value of Purple Sea Cucumbers" International Journal of Molecular Sciences 24, no. 15: 12213. https://doi.org/10.3390/ijms241512213
APA StyleXing, L., Wang, L., Liu, S., Sun, L., Wessel, G. M., & Yang, H. (2023). Single-Cell Transcriptome and Pigment Biochemistry Analysis Reveals the Potential for the High Nutritional and Medicinal Value of Purple Sea Cucumbers. International Journal of Molecular Sciences, 24(15), 12213. https://doi.org/10.3390/ijms241512213






