Evolutionary Origin of Human PALB2 Germline Pathogenic Variants
Abstract
1. Introduction
2. Results
2.1. Cross Species Conservation Analysis
2.2. Ancient Human Genomic Analysis
3. Discussion
4. Methods and Materials
4.1. Source of Human PALB2 Variants
4.2. Cross-Species Genomic Analysis
4.3. Ancient Human Genomic Analysis
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xia, B.; Sheng, Q.; Nakanishi, K.; Ohashi, A.; Wu, J.; Christ, N.; Liu, X.; Jasin, M.; Couch, F.J.; Livingston, D.M. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell 2006, 22, 719–729. [Google Scholar] [CrossRef]
- Buisson, R.; Dion-Côté, A.M.; Coulombe, Y.; Launay, H.; Cai, H.; Stasiak, A.Z.; Stasiak, A.; Xia, B.; Masson, J.Y. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat. Struct. Mol. Biol. 2010, 17, 1247–1254. [Google Scholar] [CrossRef]
- Rahman, N.; Seal, S.; Thompson, D.; Kelly, P.; Renwick, A.; Elliott, A.; Reid, S.; Spanova, K.; Barfoot, R.; Chagtai, T.; et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat. Genet. 2007, 39, 165–167. [Google Scholar] [CrossRef]
- Ohmoto, A.; Yachida, S.; Morizane, C. Genomic Features and Clinical Management of Patients with Hereditary Pancreatic Cancer Syndromes and Familial Pancreatic Cancer. Int. J. Mol. Sci. 2019, 20, 561. [Google Scholar] [CrossRef]
- Yang, X.; Leslie, G.; Doroszuk, A.; Schneider, S.; Allen, J.; Decker, B.; Dunning, A.M.; Redman, J.; Scarth, J.; Plaskocinska, I.; et al. Cancer Risks Associated with Germline PALB2 Pathogenic Variants: An International Study of 524 Families. J. Clin. Oncol. 2020, 38, 674–685. [Google Scholar] [CrossRef]
- Xia, B.; Dorsman, J.C.; Ameziane, N.; de Vries, Y.; Rooimans, M.A.; Sheng, Q.; Pals, G.; Errami, A.; Gluckman, E.; Llera, J.; et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat. Genet. 2007, 39, 159–161. [Google Scholar] [CrossRef]
- Guo, M.; Wang, S.M. The BRCAness Landscape of Cancer. Cells 2022, 11, 3877. [Google Scholar] [CrossRef] [PubMed]
- Byrum, A.K.; Vindigni, A.; Mosammaparast, N. Defining and Modulating ‘BRCAness’. Trends Cell Biol. 2019, 29, 740–751. [Google Scholar] [CrossRef] [PubMed]
- Cortesi, L.; Rugo, H.S.; Jackisch, C. An Overview of PARP Inhibitors for the Treatment of Breast Cancer. Target Oncol. 2021, 16, 255–282. [Google Scholar] [CrossRef] [PubMed]
- Tutt, A.; Tovey, H.; Cheang, M.C.U.; Kernaghan, S.; Kilburn, L.; Gazinska, P.; Owen, J.; Abraham, J.; Barrett, S.; Barrett-Lee, P.; et al. Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: The TNT Trial. Nat. Med. 2018, 24, 628–637. [Google Scholar] [CrossRef] [PubMed]
- Bleuyard, J.Y.; Buisson, R.; Masson, J.Y.; Esashi, F. ChAM, a novel motif that mediates PALB2 intrinsic chromatin binding and facilitates DNA repair. EMBO Rep. 2012, 13, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Li, J.; Wang, S.M. Pathogenic variants in human DNA damage repair genes mostly arose in recent human history. Nat. Commun. 2023. submitted. [Google Scholar]
- Li, J.; Zhao, B.; Huang, T.; Qin, Z.; Wang, S.M. Human BRCA pathogenic variants were originated during recent human history. Life Sci. Alliance 2022, 5, e202101263. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Suleski, M.; Craig, J.M.; Kasprowicz, A.E.; Sanderford, M.; Li, M.; Stecher, G.; Hedges, S.B. TimeTree 5: An Expanded Resource for Species Divergence Times. Mol. Biol. Evol. 2022, 39, msac174. [Google Scholar] [CrossRef]
- Foulkes, W.D.; Ghadirian, P.; Akbari, M.R.; Hamel, N.; Giroux, S.; Sabbaghian, N.; Darnel, A.; Royer, R.; Poll, A.; Fafard, E.; et al. Identification of a novel truncating PALB2 mutation and analysis of its contribution to early-onset breast cancer in French-Canadian women. Breast Cancer Res. 2007, 9, R83. [Google Scholar] [CrossRef] [PubMed]
- Erkko, H.; Xia, B.; Nikkilä, J.; Schleutker, J.; Syrjäkoski, K.; Mannermaa, A.; Kallioniemi, A.; Pylkäs, K.; Karppinen, S.M.; Rapakko, K.; et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature 2007, 446, 316–319. [Google Scholar] [CrossRef]
- Patterson, N.; Isakov, M.; Booth, T.; Büster, L.; Fischer, C.E.; Olalde, I.; Ringbauer, H.; Akbari, A.; Cheronet, O.; Bleasdale, M.; et al. Large-scale migration into Britain during the Middle to Late Bronze Age. Nature 2022, 601, 588–594. [Google Scholar] [CrossRef]
- Mathieson, I.; Alpaslan-Roodenberg, S.; Posth, C.; Szécsényi-Nagy, A.; Rohland, N.; Mallick, S.; Olalde, I.; Broomandkhoshbacht, N.; Candilio, F.; Cheronet, O.; et al. The genomic history of southeastern Europe. Nature 2018, 555, 197–203. [Google Scholar] [CrossRef]
- Antonio, M.L.; Gao, Z.; Moots, H.M.; Lucci, M.; Candilio, F.; Sawyer, S.; Oberreiter, V.; Calderon, D.; Devitofranceschi, K.; Aikens, R.C.; et al. Ancient Rome: A genetic crossroads of Europe and the Mediterranean. Science 2019, 366, 708–714. [Google Scholar] [CrossRef]
- Agranat-Tamir, L.; Waldman, S.; Martin, M.A.S.; Gokhman, D.; Mishol, N.; Eshel, T.; Cheronet, O.; Rohland, N.; Mallick, S.; Adamski, N.; et al. The Genomic History of the Bronze Age Southern Levant. Cell 2020, 181, 1146–1157.e1111. [Google Scholar] [CrossRef]
- Wang, C.C.; Yeh, H.Y.; Popov, A.N.; Zhang, H.Q.; Matsumura, H.; Sirak, K.; Cheronet, O.; Kovalev, A.; Rohland, N.; Kim, A.M.; et al. Genomic insights into the formation of human populations in East Asia. Nature 2021, 591, 413–419. [Google Scholar] [CrossRef]
- Lazaridis, I.; Nadel, D.; Rollefson, G.; Merrett, D.C.; Rohland, N.; Mallick, S.; Fernandes, D.; Novak, M.; Gamarra, B.; Sirak, K.; et al. Genomic insights into the origin of farming in the ancient Near East. Nature 2016, 536, 419–424. [Google Scholar] [CrossRef]
- Damgaard, P.B.; Marchi, N.; Rasmussen, S.; Peyrot, M.; Renaud, G.; Korneliussen, T.; Moreno-Mayar, J.V.; Pedersen, M.W.; Goldberg, A.; Usmanova, E.; et al. 137 ancient human genomes from across the Eurasian steppes. Nature 2018, 557, 369–374. [Google Scholar] [CrossRef]
- de Barros Damgaard, P.; Martiniano, R.; Kamm, J.; Moreno-Mayar, J.V.; Kroonen, G.; Peyrot, M.; Barjamovic, G.; Rasmussen, S.; Zacho, C.; Baimukhanov, N.; et al. The first horse herders and the impact of early Bronze Age steppe expansions into Asia. Science 2018, 360, eaar7711. [Google Scholar] [CrossRef]
- McColl, H.; Racimo, F.; Vinner, L.; Demeter, F.; Gakuhari, T.; Moreno-Mayar, J.V.; van Driem, G.; Gram Wilken, U.; Seguin-Orlando, A.; de la Fuente Castro, C.; et al. The prehistoric peopling of Southeast Asia. Science 2018, 361, 88–92. [Google Scholar] [CrossRef] [PubMed]
- Sikora, M.; Pitulko, V.V.; Sousa, V.C.; Allentoft, M.E.; Vinner, L.; Rasmussen, S.; Margaryan, A.; de Barros Damgaard, P.; de la Fuente, C.; Renaud, G.; et al. The population history of northeastern Siberia since the Pleistocene. Nature 2019, 570, 182–188. [Google Scholar] [CrossRef]
- Fu, Q.; Posth, C.; Hajdinjak, M.; Petr, M.; Mallick, S.; Fernandes, D.; Furtwängler, A.; Haak, W.; Meyer, M.; Mittnik, A.; et al. The genetic history of Ice Age Europe. Nature 2016, 534, 200–205. [Google Scholar] [CrossRef]
- Zhang, F.; Ning, C.; Scott, A.; Fu, Q.; Bjørn, R.; Li, W.; Wei, D.; Wang, W.; Fan, L.; Abuduresule, I.; et al. The genomic origins of the Bronze Age Tarim Basin mummies. Nature 2021, 599, 256–261. [Google Scholar] [CrossRef] [PubMed]
- Narasimhan, V.M.; Patterson, N.; Moorjani, P.; Rohland, N.; Bernardos, R.; Mallick, S.; Lazaridis, I.; Nakatsuka, N.; Olalde, I.; Lipson, M.; et al. The formation of human populations in South and Central Asia. Science 2019, 365, eaat7487. [Google Scholar] [CrossRef] [PubMed]
- Mathieson, I.; Lazaridis, I.; Rohland, N.; Mallick, S.; Patterson, N.; Roodenberg, S.A.; Harney, E.; Stewardson, K.; Fernandes, D.; Novak, M.; et al. Eight thousand years of natural selection in Europe. bioRxiv 2015. [Google Scholar] [CrossRef]
- Fowler, C.; Olalde, I.; Cummings, V.; Armit, I.; Büster, L.; Cuthbert, S.; Rohland, N.; Cheronet, O.; Pinhasi, R.; Reich, D. A high-resolution picture of kinship practices in an Early Neolithic tomb. Nature 2022, 601, 584–587. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Mayar, J.V.; Vinner, L.; de Barros Damgaard, P.; de la Fuente, C.; Chan, J.; Spence, J.P.; Allentoft, M.E.; Vimala, T.; Racimo, F.; Pinotti, T.; et al. Early human dispersals within the Americas. Science 2018, 362, eaav2621. [Google Scholar] [CrossRef] [PubMed]
- Flegontov, P.; Altınışık, N.E.; Changmai, P.; Rohland, N.; Mallick, S.; Adamski, N.; Bolnick, D.A.; Broomandkhoshbacht, N.; Candilio, F.; Culleton, B.J.; et al. Palaeo-Eskimo genetic ancestry and the peopling of Chukotka and North America. Nature 2019, 570, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Harney, É.; Cheronet, O.; Fernandes, D.M.; Sirak, K.; Mah, M.; Bernardos, R.; Adamski, N.; Broomandkhoshbacht, N.; Callan, K.; Lawson, A.M.; et al. A minimally destructive protocol for DNA extraction from ancient teeth. Genome Res. 2021, 31, 472–483. [Google Scholar] [CrossRef]
- Posth, C.; Nakatsuka, N.; Lazaridis, I.; Skoglund, P.; Mallick, S.; Lamnidis, T.C.; Rohland, N.; Nägele, K.; Adamski, N.; Bertolini, E.; et al. Reconstructing the Deep Population History of Central and South America. Cell 2018, 175, 1185–1197.e1122. [Google Scholar] [CrossRef]
- Gao, L.; Zhang, J. Why are some human disease-associated mutations fixed in mice? Trends Genet. 2003, 19, 678–681. [Google Scholar] [CrossRef]
- Reich, D.; Patterson, N.; Kircher, M.; Delfin, F.; Nandineni, M.R.; Pugach, I.; Ko, A.M.; Ko, Y.C.; Jinam, T.A.; Phipps, M.E.; et al. Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. Am. J. Hum. Genet. 2011, 89, 516–528. [Google Scholar] [CrossRef]
- Evans, D.G.; Burghel, G.J.; Smith, M.J. Differential rates of germline heterozygote and mosaic variants in NF2 may show varying propensity for meiotic or mitotic mutation. J. Med. Genet. 2023. [Google Scholar] [CrossRef] [PubMed]
- Hedges, S.B. The origin and evolution of model organisms. Nat. Rev. Genet. 2002, 3, 838–849. [Google Scholar] [CrossRef]
- Henn, B.M.; Cavalli-Sforza, L.L.; Feldman, M.W. The great human expansion. Proc. Natl. Acad. Sci. USA 2012, 109, 17758–17764. [Google Scholar] [CrossRef]
- Gignoux, C.R.; Henn, B.M.; Mountain, J.L. Rapid, global demographic expansions after the origins of agriculture. Proc. Natl. Acad. Sci. USA 2011, 108, 6044–6049. [Google Scholar] [CrossRef] [PubMed]
- Lou, D.I.; McBee, R.M.; Le, U.Q.; Stone, A.C.; Wilkerson, G.K.; Demogines, A.M.; Sawyer, S.L. Rapid evolution of BRCA1 and BRCA2 in humans and other primates. BMC Evol. Biol. 2014, 14, 155. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.M.; Fan, S.; Ma, Y. BRCA1 regulation of transcription. Cancer Lett. 2006, 236, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Pao, G.M.; Zhu, Q.; Perez-Garcia, C.G.; Chou, S.J.; Suh, H.; Gage, F.H.; O’Leary, D.D.; Verma, I.M. Role of BRCA1 in brain development. Proc. Natl. Acad. Sci. USA 2014, 111, E1240–E1248. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.R.; Hanson, H.A.; Hollingshaus, M.S. BRCA1 and BRCA2 mutations and female fertility. Curr. Opin. Obs. Gynecol. 2013, 25, 207–213. [Google Scholar] [CrossRef]
- Maxwell, K.N.; Domchek, S.M.; Nathanson, K.L.; Robson, M.E. Population Frequency of Germline BRCA1/2 Mutations. J. Clin. Oncol. 2016, 34, 4183–4185. [Google Scholar] [CrossRef]
- Dong, H.; Chandratre, K.; Qin, Y.; Zhang, J.; Tian, X.; Rong, C.; Wang, N.; Guo, M.; Zhao, G.; Wang, S.M. Prevalence of BRCA1/BRCA2 pathogenic variation in Chinese Han population. J. Med. Genet. 2021, 58, 565–569. [Google Scholar] [CrossRef]
- Qin, Z.; Kuok, C.N.; Dong, H.; Jiang, L.; Zhang, L.; Guo, M.; Leong, H.K.; Wang, L.; Meng, G.; Wang, S.M. Can population BRCA screening be applied in non-Ashkenazi Jewish populations? Experience in Macau population. J. Med. Genet. 2021, 58, 587–591. [Google Scholar] [CrossRef]
- Chian, J.; Sinha, S.; Qin, Z.; Wang, S.M. BRCA1 and BRCA2 Variation in Taiwanese General Population and the Cancer Cohort. Front. Mol. Biosci. 2021, 8, 685174. [Google Scholar] [CrossRef]
- Dorling, L.; Carvalho, S.; Allen, J.; González-Neira, A.; Luccarini, C.; Wahlström, C.; Pooley, K.A.; Parsons, M.T.; Fortuno, C.; Wang, Q.; et al. Breast Cancer Risk Genes—Association Analysis in More than 113,000 Women. N. Engl. J. Med. 2021, 384, 428–439. [Google Scholar] [CrossRef]
- Fu, W.; O’Connor, T.D.; Jun, G.; Kang, H.M.; Abecasis, G.; Leal, S.M.; Gabriel, S.; Rieder, M.J.; Altshuler, D.; Shendure, J.; et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature 2013, 493, 216–220. [Google Scholar] [CrossRef] [PubMed]
- Blanchette, M.; Kent, W.J.; Riemer, C.; Elnitski, L.; Smit, A.F.; Roskin, K.M.; Baertsch, R.; Rosenbloom, K.; Clawson, H.; Green, E.D.; et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004, 14, 708–715. [Google Scholar] [CrossRef] [PubMed]
- Hubisz, M.J.; Pollard, K.S.; Siepel, A. PHAST and RPHAST: Phylogenetic analysis with space/time models. Brief. Bioinform. 2011, 12, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Ramani, R.; Krumholz, K.; Huang, Y.F.; Siepel, A. PhastWeb: A web interface for evolutionary conservation scoring of multiple sequence alignments using phastCons and phyloP. Bioinformatics 2019, 35, 2320–2322. [Google Scholar] [CrossRef]
- GetBase. Available online: https://github.com/Skylette14/GetBase (accessed on 4 February 2023).
- Harris, R.S. Improved Pairwise Alignment of Genomic DNA. Ph.D. Thesis, The Pennsylvania State University, University Park, PA, USA, 2007. [Google Scholar]
- Mallick, S.; Reich, D. The Allen Ancient DNA Resource (AADR): A curated compendium of ancient human resources. Harv. Dataverse 2023, V8. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.11–11.10.33. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]

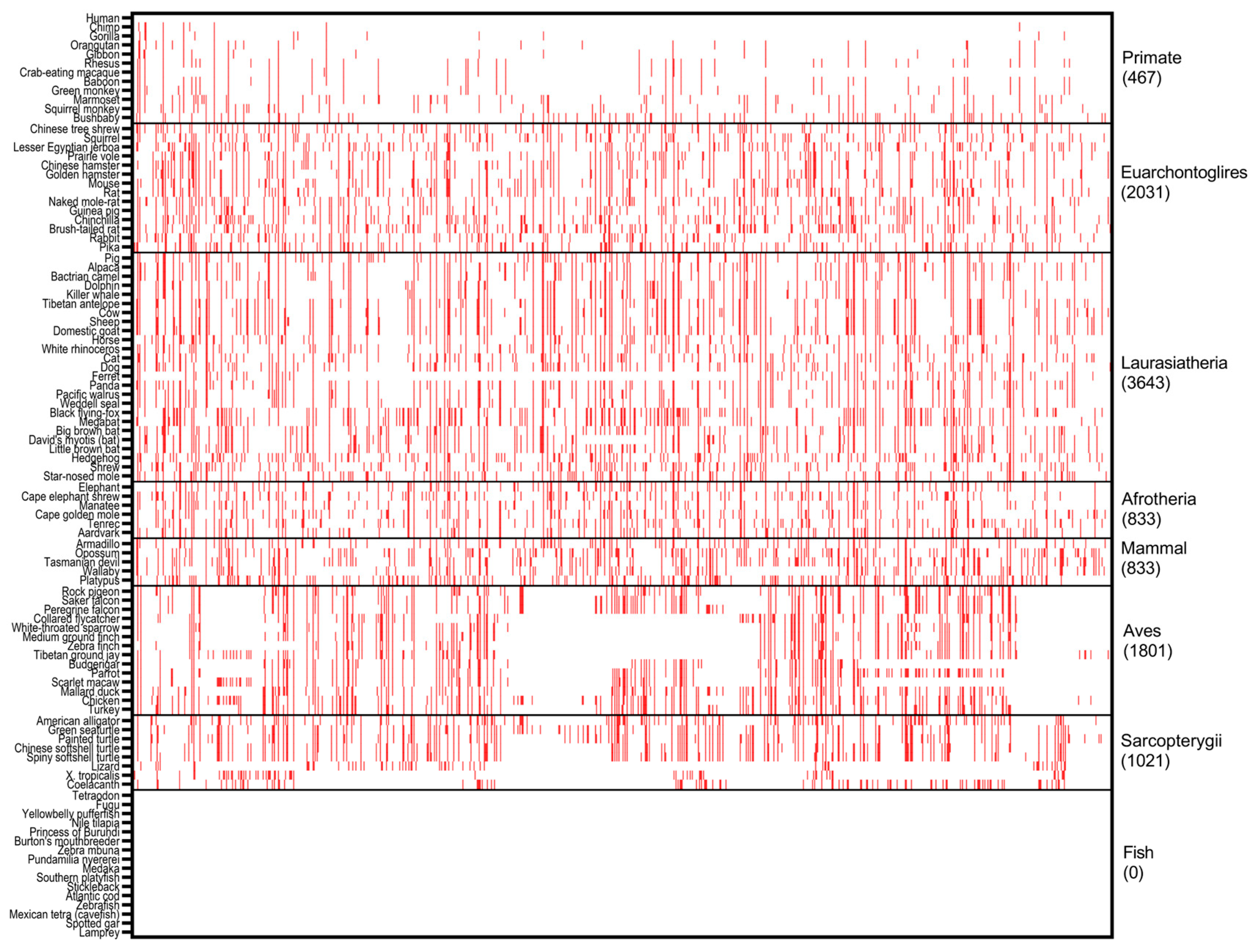

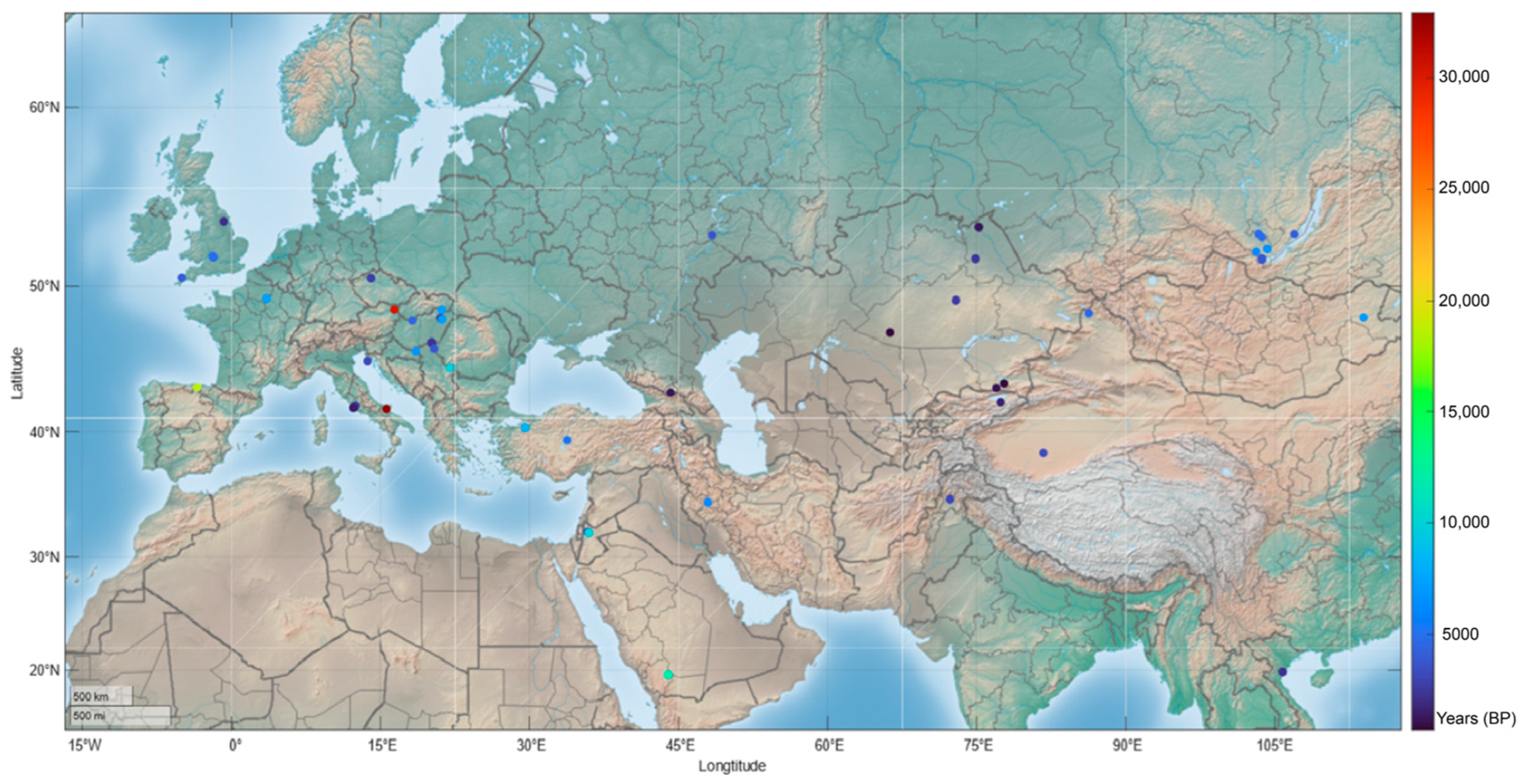
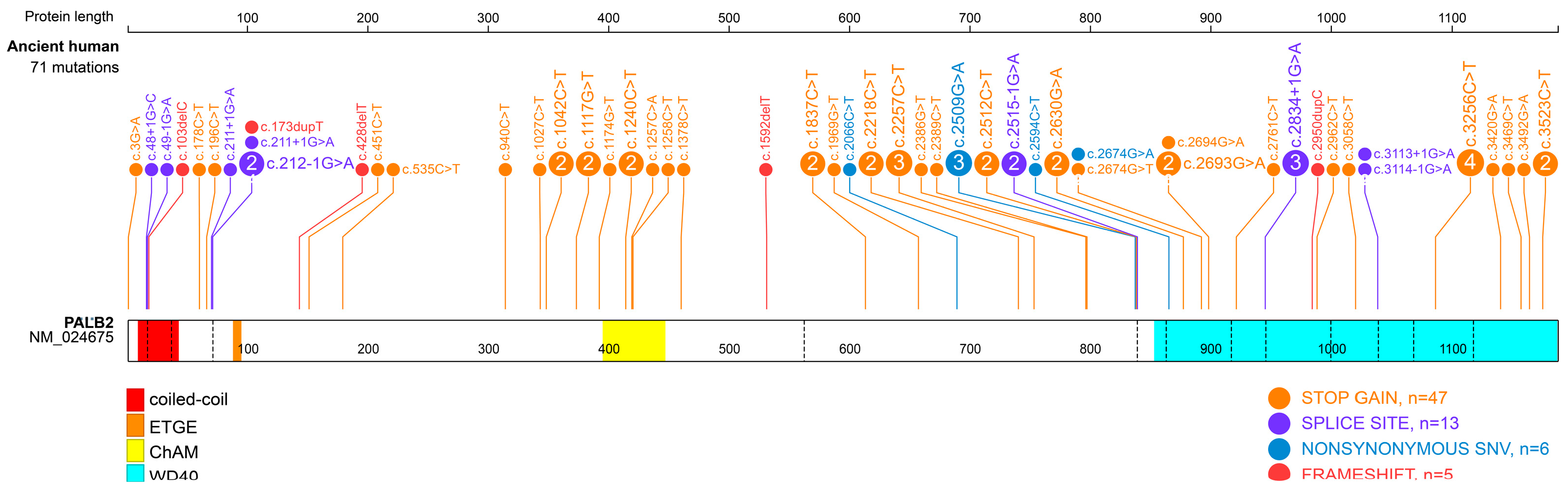
| Carrier Number | cDNA | Protein | Mutation Type | Domain | Arisen Time (BP) * | References |
|---|---|---|---|---|---|---|
| 4 | c.3256C>T | p.Arg1086Ter | stopgain | WD40 | 9860 | [17,18,19] |
| 3 | c.2257C>T | p.Arg753Ter | stopgain | - | 3325 | [17,19,20] |
| 3 | c.2509G>A | p.Glu837Lys | nonsynonymous SNV | - | 7050 | [19,21,22] |
| 3 | c.2834+1G>A | - | splice site | WD40 | 6319 | [23,24,25] |
| 2 | c.212-1G>A | - | splice site | ETGE | 2275 | [25,26] |
| 2 | c.211+1G>A | - | splice site | ETGE | 10,050 | [27] |
| 2 | c.1042C>T | p.Gln348Ter | stopgain | - | 6319 | [24,28] |
| 2 | c.1117G>T | p.Glu373Ter | stopgain | - | 3157 | [19,29] |
| 2 | c.1240C>T | p.Arg414Ter | stopgain | ChAM | 3740 | [23,30] |
| 2 | c.1837C>T | p.Gln613Ter | stopgain | - | 32,895 | [26,27] |
| 2 | c.2218C>T | p.Gln740Ter | stopgain | - | 8315 | [23,30] |
| 2 | c.2512C>T | p.Gln838Ter | stopgain | - | 3871 | [23,24] |
| 2 | c.2515-1G>A | - | splice site | - | 4106 | [17,24] |
| 2 | c.2630G>A | p.Trp877Ter | stopgain | WD40 | 5677 | [17,31] |
| 2 | c.2693G>A | p.Trp898Ter | stopgain | WD40 | >10,000 | [24,32] |
| 2 | c.3523C>T | p.Gln1175Ter | stopgain | WD40 | 6713 | [24] |
| 1 | c.3G>A | p.Met1Ile | startloss | - | 31,630 | [26] |
| 1 | c.48+1G>C | - | splice site | coiled-coil | 30,260 | [27] |
| 1 | c.49-1G>A | - | splice site | coiled-coil | 689 | [23] |
| 1 | c.103delC | p.Leu35PhefsTer18 | frameshift deletion | coiled-coil | 2758 | [23] |
| 1 | c.173dupT | p.Leu58PhefsTer16 | frameshift insertion | - | 2554 | [23] |
| 1 | c.178C>T | p.Gln60Ter | stopgain | - | 6750 | [22] |
| 1 | c.196C>T | p.Gln66Ter | stopgain | - | 4158 | [24] |
| 1 | c.428delT | p.Leu143ArgfsTer34 | frameshift deletion | - | 7178 | [30] |
| 1 | c.451C>T | p.Gln151Ter | stopgain | - | 8790 | [22] |
| 1 | c.535C>T | p.Gln179Ter | stopgain | - | 4885 | [32] |
| 1 | c.940C>T | p.Gln314Ter | stopgain | - | 1613 | [23] |
| 1 | c.1027C>T | p.Gln343Ter | stopgain | - | 2255 | [25] |
| 1 | c.1174G>T | p.Glu392Ter | stopgain | - | 4161 | [23] |
| 1 | c.1257C>A | p.Cys419Ter | stopgain | ChAM | 3900 | [17] |
| 1 | c.1258C>T | p.Gln420Ter | stopgain | ChAM | 725 | [33] |
| 1 | c.1378C>T | p.Gln460Ter | stopgain | - | 3440 | [28] |
| 1 | c.1592delT | p.Leu531CysfsTer30 | frameshift deletion | - | 700 | [23] |
| 1 | c.1969G>T | p.Glu657Ter | stopgain | - | 2500 | [34] |
| 1 | c.2066C>T | p.Ser689Leu | nonsynonymous SNV | - | 6850 | [18] |
| 1 | c.2386G>T | p.Gly796Ter | stopgain | - | 1506 | [19] |
| 1 | c.2389C>T | p.Gln797Ter | stopgain | - | 4862 | [26] |
| 1 | c.2594C>T | p.Ser865Leu | nonsynonymous SNV | WD40 | 4940 | [31] |
| 1 | c.2674G>T | p.Glu892Ter | stopgain | WD40 | 8570 | [35] |
| 1 | c.2674G>A | p.Glu892Lys | nonsynonymous SNV | WD40 | 3740 | [30] |
| 1 | c.2694G>A | p.Trp898Ter | stopgain | WD40 | >10,000 | [32] |
| 1 | c.2761C>T | p.Gln921Ter | stopgain | WD40 | 2478 | [23] |
| 1 | c.2950dupC | p.Leu984ProfsTer3 | frameshift insertion | WD40 | 7221 | [30] |
| 1 | c.2962C>T | p.Gln988Ter | stopgain | WD40 | 1300 | [23] |
| 1 | c.3058C>T | p.Gln1020Ter | stopgain | WD40 | 1246 | [23] |
| 1 | c.3114-1G>A | - | splice site | WD40 | 2279 | [23] |
| 1 | c.3113+1G>A | - | splice site | WD40 | 1646 | [26] |
| 1 | c.3420G>A | p.Trp1140Ter | stopgain | WD40 | 1246 | [23] |
| 1 | c.3469C>T | p.Gln1157Ter | stopgain | WD40 | 8315 | [30] |
| 1 | c.3492G>A | p.Trp1164Ter | stopgain | WD40 | 18,720 | [27] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chian, J.S.; Li, J.; Wang, S.M. Evolutionary Origin of Human PALB2 Germline Pathogenic Variants. Int. J. Mol. Sci. 2023, 24, 11343. https://doi.org/10.3390/ijms241411343
Chian JS, Li J, Wang SM. Evolutionary Origin of Human PALB2 Germline Pathogenic Variants. International Journal of Molecular Sciences. 2023; 24(14):11343. https://doi.org/10.3390/ijms241411343
Chicago/Turabian StyleChian, Jia Sheng, Jiaheng Li, and San Ming Wang. 2023. "Evolutionary Origin of Human PALB2 Germline Pathogenic Variants" International Journal of Molecular Sciences 24, no. 14: 11343. https://doi.org/10.3390/ijms241411343
APA StyleChian, J. S., Li, J., & Wang, S. M. (2023). Evolutionary Origin of Human PALB2 Germline Pathogenic Variants. International Journal of Molecular Sciences, 24(14), 11343. https://doi.org/10.3390/ijms241411343







