Epithelial-to-Mesenchymal Transition Gene Signature in Circulating Melanoma Cells: Biological and Clinical Relevance
Abstract
1. Introduction
2. Results
2.1. Optimization and Validation of qRT-PCR Assay on Control Cell Lines and Healthy Donors
2.2. Detection of In Vivo Isolated CMCs
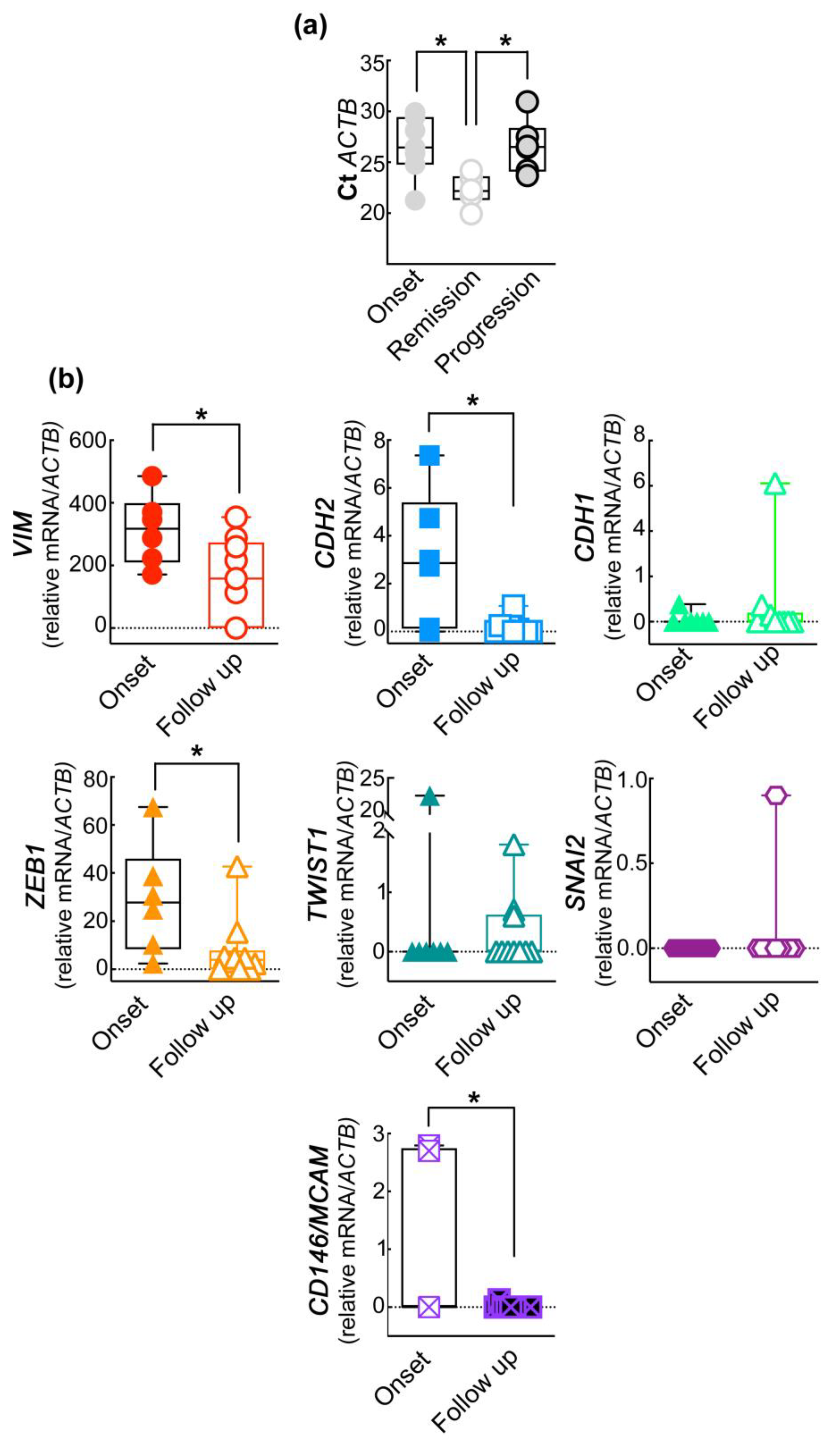
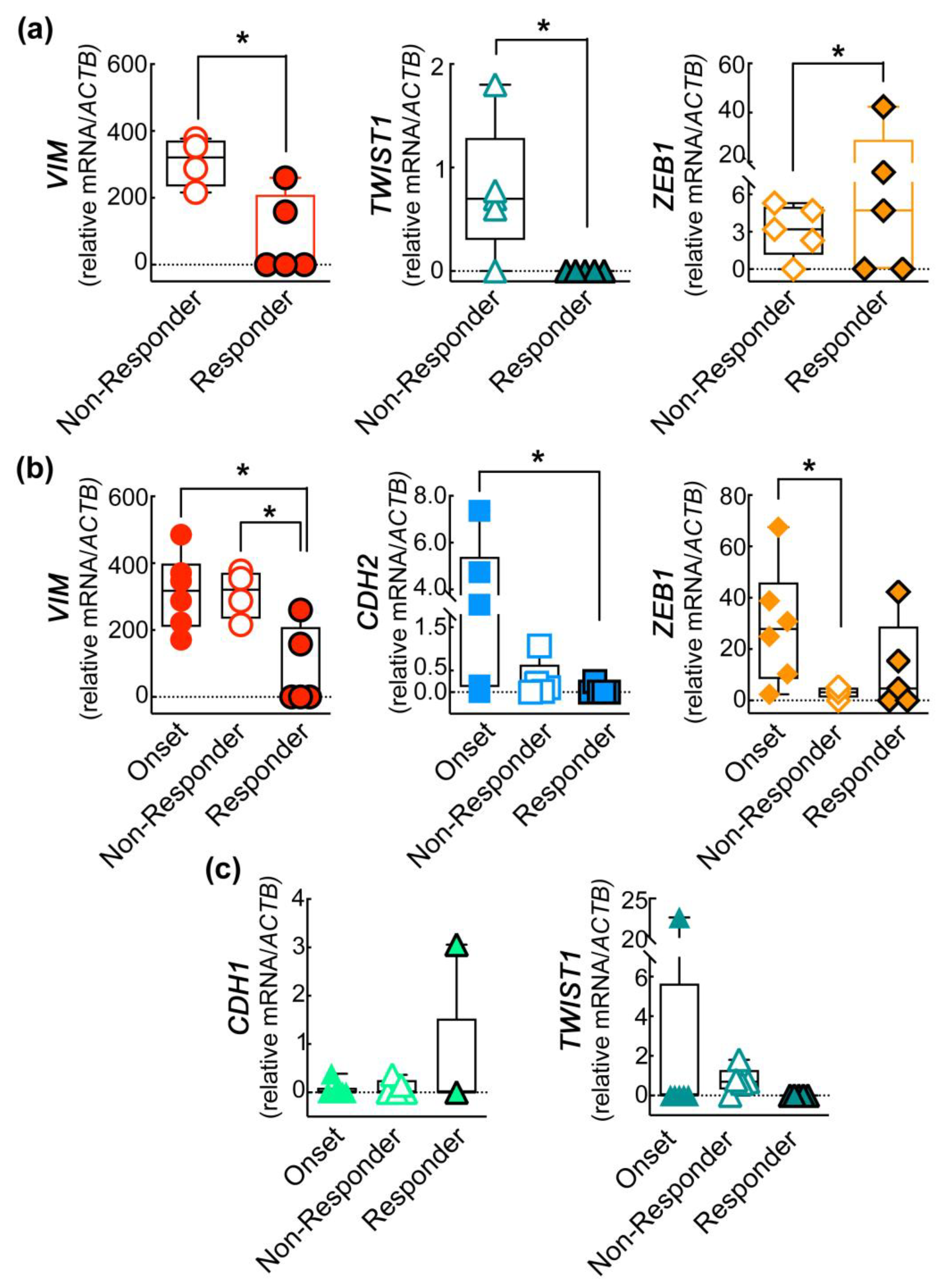
2.3. The Expression of EMT Genes of CD45−CD146+ABCB5+ CMCs Subpopulation Differs in Melanoma Patients Undergoing Treatment
2.4. The EMT Gene Signature of CD45−CD146+ABCB+ CMCs Subpopulation Correlates with the Clinical Progression of the Disease
3. Discussion
4. Materials and Methods
4.1. Patients and Healthy Donors
4.2. Selection of Reference Genes Panel
4.3. Cell Lines
4.4. In Vivo CMCs Enrichment
4.5. Optimization and Validation of the qRT-PCR Assay for the EMT Profile of Enriched CMCs
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Laga, A.C.; Murphy, G.F. Cellular Heterogeneity in Vertical Growth Phase Melanoma. Arch. Pathol. Lab. Med. 2010, 134, 1750–1757. [Google Scholar] [CrossRef] [PubMed]
- Chin, L. The genetics of malignant melanoma: Lessons from mouse and man. Nat. Rev. Cancer 2003, 3, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Luzzi, K.J.; MacDonald, I.C.; Schmidt, E.E.; Kerkvliet, N.; Morris, V.L.; Chambers, A.F.; Groom, A.C. Multistep nature of metastatic inefficiency: Dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. Am. J. Pathol. 1998, 153, 865–873. [Google Scholar] [CrossRef]
- Ghossein, R.A.; Bhattacharya, S.; Rosai, J. Molecular detection of micrometastases and circulating tumor cells in solid tumors. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 1999, 5, 1950–1960. [Google Scholar]
- Haass, N.K.; Smalley, K.S.M.; Li, L.; Herlyn, M. Adhesion, migration and communication in melanocytes and melanoma. Pigment. Cell Res. 2005, 18, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Mocellin, S.; Keilholz, U.; Rossi, C.R.; Nitti, D. Circulating tumor cells: The “leukemic phase” of solid cancers. Trends Mol. Med. 2006, 12, 130–139. [Google Scholar] [CrossRef]
- Paterlini-Brechot, P.; Benali, N.L. Circulating tumor cells (CTC) detection: Clinical impact and future directions. Cancer Lett. 2007, 253, 180–204. [Google Scholar] [CrossRef]
- Braeuer, R.R.; Watson, I.R.; Wu, C.-J.; Mobley, A.K.; Kamiya, T.; Shoshan, E.; Bar-Eli, M. Why is melanoma so metastatic? Pigment. Cell Melanoma Res. 2014, 27, 19–36. [Google Scholar] [CrossRef]
- Thomas, Q.D.; Colard-Thomas, J.; Delansay, D.; Leenhardt, F.; Solassol, J.; Vendrell, J.A.; Quantin, X. Case report: Liquid biopsy, the sooner the better? Front. Oncol. 2022, 12, 1089108. [Google Scholar] [CrossRef]
- Di Sario, G.; Rossella, V.; Famulari, E.S.; Maurizio, A.; Lazarevic, D.; Giannese, F.; Felici, C. Enhancing clinical potential of liquid biopsy through a multi-omic approach: A systematic review. Front. Genet. 2023, 14, 1152470. [Google Scholar] [CrossRef]
- Ricciardi, E.; Giordani, E.; Ziccheddu, G.; Falcone, I.; Giacomini, P.; Fanciulli, M.; Russillo, M.; Cerro, M.; Ciliberto, G.; Morrone, A.; et al. Metastatic Melanoma: Liquid Biopsy as a New Precision Medicine Approach. Int. J. Mol. Sci. 2023, 24, 4014. [Google Scholar] [CrossRef]
- Mellado, B.; Gutierrez, L.; Castel, T.; Colomer, D.; Fontanillas, M.; Castro, J.; Estapé, J. Prognostic significance of the detection of circulating malignant cells by reverse transcriptase-polymerase chain reaction in long-term clinically disease-free melanoma patients. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 1999, 5, 1843–1848. [Google Scholar]
- Palmieri, G.; Ascierto, P.A.; Perrone, F.; Satriano, S.M.R.; Ottaiano, A.; Daponte, A.; Napolitano, M.; Caracò, C.; Mozzillo, N.; Melucci, M.T.; et al. Prognostic value of circulating melanoma cells detected by reverse transcriptase-polymerase chain reaction. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2003, 21, 767–773. [Google Scholar] [CrossRef]
- Klein, W.M.; Wu, B.P.; Zhao, S.; Wu, H.; Klein-Szanto, A.J.P.; Tahan, S.R. Increased expression of stem cell markers in malignant melanoma. Mod. Pathol. 2007, 20, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Schatton, T.; Murphy, G.F.; Frank, N.Y.; Yamaura, K.; Waaga-Gasser, A.M.; Gasser, M.; Zhan, Q.; Jordan, S.; Duncan, L.M.; Weishaupt, C.; et al. Identification of cells initiating human melanomas. Nature 2008, 451, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Ziman, M. Circulating Melanoma Cells. In Breakthroughs in Melanoma Research; Tanaka, Y., Ed.; IntechOpen: Rijeka, Croatia, 2011. [Google Scholar]
- Luo, X.; Mitra, D.; Sullivan, R.J.; Wittner, B.S.; Kimura, A.M.; Pan, S.; Hoang, M.P.; Brannigan, B.W.; Lawrence, D.P.; Flaherty, K.T.; et al. Isolation and molecular characterization of circulating melanoma cells. Cell Rep. 2014, 7, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Karakousis, G.; Yang, R.; Xu, X. Circulating Melanoma Cells as a Predictive Biomarker. J. Investig. Dermatol. 2013, 133, 1460–1462. [Google Scholar] [CrossRef]
- Khoja, L.; Lorigan, P.; Zhou, C.; Lancashire, M.; Booth, J.; Cummings, J.; Clack, G.; Hughes, A.; Dive, C. Biomarker utility of circulating tumor cells in metastatic cutaneous melanoma. J. Investig. Dermatol. 2013, 133, 1582–1590. [Google Scholar] [CrossRef]
- Hall, C.S.; Ross, M.; Bowman Bauldry, J.B.; Upshaw, J.; Karhade, M.G.; Royal, R.; Patel, S.; Lucci, A. Circulating Tumor Cells in Stage IV Melanoma Patients. J. Am. Coll. Surg. 2018, 227, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Rao, C.; Bui, T.; Connelly, M.; Doyle, G.; Karydis, I.; Middleton, M.R.; Clack, G.; Malone, M.; Coumans, F.A.; Terstappen, L.W. Circulating melanoma cells and survival in metastatic melanoma. Int. J. Oncol. 2011, 38, 755–760. [Google Scholar]
- Klinac, D.; Gray, E.S.; Freeman, J.B.; Reid, A.; Bowyer, S.; Millward, M.; Ziman, M. Monitoring changes in circulating tumour cells as a prognostic indicator of overall survival and treatment response in patients with metastatic melanoma. BMC Cancer 2014, 14, 423. [Google Scholar] [CrossRef]
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef]
- Seo, J.; Ha, J.; Kang, E.; Cho, S. The role of epithelial–mesenchymal transition-regulating transcription factors in anti-cancer drug resistance. Arch. Pharm. Res. 2021, 44, 281–292. [Google Scholar] [CrossRef]
- Khattak, M.A.; Reid, A.; Freeman, J.; Pereira, M.; McEvoy, A.; Lo, J.; Frank, M.H.; Meniawy, T.; Didan, A.; Spencer, I.; et al. PD-L1 Expression on Circulating Tumor Cells May Be Predictive of Response to Pembrolizumab in Advanced Melanoma: Results from a Pilot Study. Oncologist 2019, 25, e520–e527. [Google Scholar] [CrossRef]
- Rapanotti, M.C.; Campione, E.; Spallone, G.; Orlandi, A.; Bernardini, S.; Bianchi, L. Minimal residual disease in melanoma: Circulating melanoma cells and predictive role of MCAM/MUC18/MelCAM/CD146. Cell Death Discov. 2017, 3, 17005. [Google Scholar] [CrossRef]
- Rapanotti, M.C.; Campione, E.; Suarez Viguria, T.M.; Spallone, G.; Costanza, G.; Rossi, P.; Orlandi, A.; Valenti, P.; Bernardini, S.; Bianchi, L. Stem-Mesenchymal Signature Cell Genes Detected in Heterogeneous Circulating Melanoma Cells Correlate with Disease Stage in Melanoma Patients. Front. Mol. Biosci. 2020, 7, 92. [Google Scholar] [CrossRef] [PubMed]
- Rapanotti, M.C.; Viguria, T.M.S.; Spallone, G.; Terrinoni, A.; Rossi, P.; Costanza, G.; Campione, E.; Lombardo, P.; Pathirannehalage, C.D.; Orlandi, A.; et al. Minimal Residual Disease in Melanoma:molecular characterization of in transit cutaneous metastases and Circulating Melanoma Cells recognizes an expression panel potentially related to disease progression. Cancer Treat. Res. Commun. 2020, 25, 100262. [Google Scholar] [CrossRef]
- Rapanotti, M.C.; Cugini, E.; Nuccetelli, M.; Terrinoni, A.; Di Raimondo, C.; Lombardo, P.; Costanza, G.; Cosio, T.; Rossi, P.; Orlandi, A.; et al. Molecular Sciences MCAM/MUC18/CD146 as a Multifaceted Warning Marker of Melanoma Progression in Liquid Biopsy. Int. J. Mol. Sci. 2021, 22, 12416. [Google Scholar] [CrossRef]
- Rapanotti, M.C.; Bianchi, L.; Ricozzi, I.; Campione, E.; Pierantozzi, A.; Orlandi, A.; Orlandi, A.; Chimenti, S.; Federici, G.; Bernardini, S. Melanoma-associated markers expression in blood: MUC-18 is associated with advanced stages in melanoma patients. Br. J. Dermatol. 2009, 160, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Rapanotti, M.C.; Ricozzi, I.; Campione, E.; Orlandi, A.; Bianchi, L. Blood MUC-18/MCAM expression in patients with melanoma: A suitable marker of poor outcome. Br. J. Dermatol. 2013, 169, 221–222. [Google Scholar] [CrossRef] [PubMed]
- Rapanotti, M.C.; Suarez Viguria, T.M.; Costanza, G.; Ricozzi, I.; Pierantozzi, A.; Di Stefani, A.; Campione, E.; Bernardini, S.; Chimenti, S.; Orlandi, A.; et al. Sequential molecular analysis of circulating MCAM/MUC18 expression: A promising disease biomarker related to clinical outcome in melanoma. Arch. Dermatol. Res. 2014, 306, 527–537. [Google Scholar] [CrossRef] [PubMed]
- De Luca, A.; Carpanese, D.; Rapanotti, M.C.; Suarez Viguria, T.M.; Forgione, M.A.; Rotili, D.; Fulci, C.; Iorio, E.; Quintieri, L.; Chimenti, S.; et al. The nitrobenzoxadiazole derivative MC3181 blocks melanoma invasion and metastasis. Oncotarget 2017, 8, 15520. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.J.; Saab, K.R.; Ma, J.; Schatton, T.; Pütz, P.; Zhan, Q.; Murphy, G.F.; Gasser, M.; Waaga-Gasser, A.M.; Frank, N.Y.; et al. ABCB5 Maintains Melanoma-Initiating Cells through a Proinflammatory Cytokine Signaling Circuit. Cancer Res. 2014, 74, 4196–4207. [Google Scholar] [CrossRef] [PubMed]
- Banyard, J.; Bielenberg, D.R. The role of EMT and MET in cancer dissemination. Connect. Tissue Res. 2015, 56, 403–413. [Google Scholar] [CrossRef]
- Yeung, K.T.; Yang, J. Epithelial–mesenchymal transition in tumor metastasis. Mol. Oncol. 2017, 11, 28–39. [Google Scholar] [CrossRef]
- Thiery, J.P. Epithelial–mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2002, 2, 442–454. [Google Scholar] [CrossRef]
- Scheel, C.; Weinberg, R.A. Cancer stem cells and epithelial–mesenchymal transition: Concepts and molecular links. Semin. Cancer Biol. 2012, 22, 396–403. [Google Scholar] [CrossRef]
- Tsai, J.H.; Donaher, J.L.; Murphy, D.A.; Chau, S.; Yang, J. Spatiotemporal Regulation of Epithelial-Mesenchymal Transition Is Essential for Squamous Cell Carcinoma Metastasis. Cancer Cell 2012, 22, 725–736. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Weinberg, R.A. Epithelial-to-mesenchymal transition in cancer: Complexity and opportunities. Front. Med. 2018, 12, 361–373. [Google Scholar] [CrossRef]
- Stemmler, M.P.; Eccles, R.L.; Brabletz, S.; Brabletz, T. Non-redundant functions of EMT transcription factors. Nat. Cell Biol. 2019, 21, 102–112. [Google Scholar] [CrossRef]
- Yang, J.; Weinberg, R.A. Epithelial-Mesenchymal Transition: At the Crossroads of Development and Tumor Metastasis. Dev. Cell 2008, 14, 818–829. [Google Scholar] [CrossRef] [PubMed]
- Giordano, A.; Gao, H.; Anfossi, S.; Cohen, E.; Mego, M.; Lee, B.-N.; Lee, B.N.; Tin, S.; De Laurentiis, M.; Parker, C.A.; et al. Epithelial–Mesenchymal Transition and Stem Cell Markers in Patients with HER2-Positive Metastatic Breast Cancer. Mol. Cancer Ther. 2012, 11, 2526–2534. [Google Scholar] [CrossRef] [PubMed]
- Kasimir-Bauer, S.; Hoffmann, O.; Wallwiener, D.; Kimmig, R.; Fehm, T. Expression of stem cell and epithelial-mesenchymal transition markers in primary breast cancer patients with circulating tumor cells. Breast Cancer Res. 2012, 14, R15. [Google Scholar] [CrossRef]
- Mego, M.; Mani, S.A.; Lee, B.-N.; Li, C.; Evans, K.W.; Cohen, E.N.; Gao, H.; Jackson, S.A.; Giordano, A.; Hortobagyi, G.N.; et al. Expression of epithelial–mesenchymal transition-inducing transcription factors in primary breast cancer: The effect of neoadjuvant therapy. Int. J. Cancer 2012, 130, 808–816. [Google Scholar] [CrossRef]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J.; Ciciliano, J.C.; Wells, M.N.; Shah, A.M.; et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013, 339, 580–584. [Google Scholar] [CrossRef]
- Polyak, K.; Weinberg, R.A. Transitions between epithelial and mesenchymal states: Acquisition of malignant and stem cell traits. Nat. Rev. Cancer 2009, 9, 265–273. [Google Scholar] [CrossRef]
- Ribatti, D.; Tamma, R.; Annese, T. Epithelial-Mesenchymal Transition in Cancer: A Historical Overview. Transl. Oncol. 2020, 13, 100773. [Google Scholar] [CrossRef] [PubMed]
- Jolly, M.K.; Boareto, M.; Huang, B.; Jia, D.; Lu, M.; Ben-Jacob, E.; Onuchic, J.N.; Levine, H. Implications of the Hybrid Epithelial/Mesenchymal Phenotype in Metastasis. Front. Oncol. 2015, 5, 155. [Google Scholar] [CrossRef]
- Liao, T.-T.; Yang, M.-H. Hybrid Epithelial/Mesenchymal State in Cancer Metastasis: Clinical Significance and Regulatory Mechanisms. Cells 2020, 9, 623. [Google Scholar] [CrossRef]
- Brabletz, S.; Schuhwerk, H.; Brabletz, T.; Stemmler, M.P. Dynamic EMT: A multi-tool for tumor progression. EMBO J. 2021, 40, e108647. [Google Scholar] [CrossRef]
- Zeng, Q.; Li, W.; Lu, D.; Wu, Z.; Duan, H.; Luo, Y.; Feng, J.; Yang, D.; Fu, L.; Yan, X. CD146, an epithelial-mesenchymal transition inducer, is associated with triple-negative breast cancer. Proc. Natl. Acad. Sci. USA 2012, 109, 1127–1132. [Google Scholar] [CrossRef]
- Pereira, L.; Mariadason, J.M.; Hannan, R.D.; Dhillon, A.S. Implications of Epithelial–Mesenchymal Plasticity for Heterogeneity in Colorectal Cancer. Front. Oncol. 2015, 5, 13. [Google Scholar] [CrossRef]
- Lembessis, P.; Msaouel, P.; Halapas, A.; Sourla, A.; Panteleakou, Z.; Pissimissis, N.; Milathianakis, C.; Bogdanos, J.; Papaioannou, A.; Maragoudakis, E.; et al. Combined androgen blockade therapy can convert RT-PCR detection of prostate-specific antigen (PSA) and prostate-specific membrane antigen (PSMA) transcripts from positive to negative in the peripheral blood of patients with clinically localized prostate cancer and increase biochemical failure-free survival after curative therapy. Clin. Chem. Lab. Med. 2007, 45, 1488–1494. [Google Scholar]
- Ignatiadis, M.; Kallergi, G.; Ntoulia, M.; Perraki, M.; Apostolaki, S.; Kafousi, M.; Chlouverakis, G.; Stathopoulos, E.; Lianidou, E.; Georgoulias, V.; et al. Prognostic Value of the Molecular Detection of Circulating Tumor Cells Using a Multimarker Reverse Transcription-PCR Assay for Cytokeratin 19, Mammaglobin A, and HER2 in Early Breast Cancer. Clin. Cancer Res. 2008, 14, 2593–2600. [Google Scholar] [CrossRef] [PubMed]
- Theodoropoulos, P.A.; Polioudaki, H.; Agelaki, S.; Kallergi, G.; Saridaki, Z.; Mavroudis, D.; Georgoulias, V. Circulating tumor cells with a putative stem cell phenotype in peripheral blood of patients with breast cancer. Cancer Lett. 2010, 288, 99–106. [Google Scholar] [CrossRef]
- Okabe, H.; Ishimoto, T.; Mima, K.; Nakagawa, S.; Hayashi, H.; Kuroki, H.; Imai, K.; Nitta, H.; Saito, S.; Hashimoto, D.; et al. CD44s signals the acquisition of the mesenchymal phenotype required for anchorage-independent cell survival in hepatocellular carcinoma. Br. J. Cancer 2014, 110, 958–966. [Google Scholar] [CrossRef]
- Satelli, A.; Brownlee, Z.; Mitra, A.; Meng, Q.H.; Li, S. Circulating Tumor Cell Enumeration with a Combination of Epithelial Cell Adhesion Molecule– and Cell-Surface Vimentin–Based Methods for Monitoring Breast Cancer Therapeutic Response. Clin. Chem. 2015, 61, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Jin, S.; Han, J.; Li, T.; Shi, J.; Zhong, Q.; Li, W.; Tang, W.; Huang, Q.; Zong, H. Detection and clinical significance of circulating tumor cells in colorectal cancer. Biomark. Res. 2021, 9, 85. [Google Scholar] [CrossRef]
- Cieślikowski, W.A.; Antczak, A.; Nowicki, M.; Zabel, M.; Budna-Tukan, J. Clinical Relevance of Circulating Tumor Cells in Prostate Cancer Management. Biomedicines 2021, 9, 1179. [Google Scholar] [CrossRef] [PubMed]
- Markou, A.; Lazaridou, M.; Paraskevopoulos, P.; Chen, S.; Świerczewska, M.; Budna, J.; Kuske, A.; Gorges, T.M.; Joosse, S.A.; Kroneis, T.; et al. Multiplex Gene Expression Profiling of In Vivo Isolated Circulating Tumor Cells in High-Risk Prostate Cancer Patients. Clin. Chem. 2018, 64, 297–306. [Google Scholar] [CrossRef]
- De Bono, J.S.; Scher, H.I.; Montgomery, R.B.; Parker, C.; Miller, M.C.; Tissing, H.; Doyle, G.V.; Terstappen, L.W.; Pienta, K.J.; Raghavan, D. Circulating Tumor Cells Predict Survival Benefit from Treatment in Metastatic Castration-Resistant Prostate Cancer. Clin. Cancer Res. 2008, 14, 6302–6309. [Google Scholar] [CrossRef] [PubMed]
- Larsson, A.-M.; Jansson, S.; Bendahl, P.-O.; Levin Tykjaer Jörgensen, C.; Loman, N.; Graffman, C.; Lundgren, L.; Aaltonen, K.; Rydén, L. Longitudinal enumeration and cluster evaluation of circulating tumor cells improve prognostication for patients with newly diagnosed metastatic breast cancer in a prospective observational trial. Breast Cancer Res. 2018, 20, 48. [Google Scholar] [CrossRef]
- Balakrishnan, A.; George, I.A.; Kumar, P. Circulating tumor cells as an emerging tool in cancer therapy. Front. Biosci. 2020, 25, 606–631. [Google Scholar]
- Toss, A.; Mu, Z.; Fernandez, S.; Cristofanilli, M. CTC enumeration and characterization: Moving toward personalized medicine. Ann. Transl. Med. 2014, 2, 108. [Google Scholar]
- Miyamoto, D.T.; Sequist, L.V.; Lee, R.J. Circulating tumour cells—Monitoring treatment response in prostate cancer. Nat. Rev. Clin. Oncol. 2014, 11, 401–412. [Google Scholar] [CrossRef] [PubMed]
- Polzer, B.; Medoro, G.; Pasch, S.; Fontana, F.; Zorzino, L.; Pestka, A.; Andergassen, U.; Meier-Stiegen, F.; Czyz, Z.T.; Alberter, B.; et al. Molecular profiling of single circulating tumor cells with diagnostic intention. EMBO Mol. Med. 2014, 6, 1371–1386. [Google Scholar] [CrossRef]
- Keller, L.; Pantel, K. Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells. Nat. Rev. Cancer 2019, 19, 553–567. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef]
- Hensler, M.; Vančurová, I.; Becht, E.; Palata, O.; Strnad, P.; Tesařová, P.; Čabiňaková, M.; Švec, D.; Kubista, M.; Bartůňková, J.; et al. Gene expression profiling of circulating tumor cells and peripheral blood mononuclear cells from breast cancer patients. Oncoimmunology 2016, 5, e1102827. [Google Scholar] [CrossRef]
- Yan, W.-T.; Cui, X.; Chen, Q.; Li, Y.-F.; Cui, Y.-H.; Wang, Y.; Jiang, J. Circulating tumor cell status monitors the treatment responses in breast cancer patients: A meta-analysis. Sci. Rep. 2017, 7, 43464. [Google Scholar] [CrossRef]
- Cani, A.K.; Dolce, E.M.; Darga, E.P.; Hu, K.; Liu, C.-J.; Pierce, J.; Bradbury, K.; Kilgour, E.; Aung, K.; Schiavon, G.; et al. Serial monitoring of genomic alterations in circulating tumor cells of ER-positive/HER2-negative advanced breast cancer: Feasibility of precision oncology biomarker detection. Mol. Oncol. 2022, 16, 1969–1985. [Google Scholar] [CrossRef]
- Franken, A.; Kraemer, A.; Sicking, A.; Watolla, M.; Rivandi, M.; Yang, L.; Warfsmann, J.; Polzer, B.M.; Friedl, T.W.P.; Meier-Stiegen, F.; et al. Comparative analysis of EpCAM high-expressing and low-expressing circulating tumour cells with regard to their clonal relationship and clinical value. Br. J. Cancer 2023, 128, 1742–1752. [Google Scholar] [CrossRef] [PubMed]
- Bankó, P.; Lee, S.Y.; Nagygyörgy, V.; Zrínyi, M.; Chae, C.H.; Cho, D.H.; Telekes, A. Technologies for circulating tumor cell separation from whole blood. J. Hematol. Oncol. 2019, 12, 48. [Google Scholar] [CrossRef]
- Grover, P.K.; Cummins, A.G.; Price, T.J.; Roberts-Thomson, I.C.; Hardingham, J.E. Circulating tumour cells: The evolving concept and the inadequacy of their enrichment by EpCAM-based methodology for basic and clinical cancer research. Ann. Oncol. 2014, 25, 1506–1516. [Google Scholar] [CrossRef] [PubMed]
- Königsberg, R.; Obermayr, E.; Bises, G.; Pfeiler, G.; Gneist, M.; Wrba, F.; de Santis, M.; Zeillinger, R.; Hudec, M.; Dittrich, C. Detection of EpCAM positive and negative circulating tumor cells in metastatic breast cancer patients. Acta. Oncol. 2011, 50, 700–710. [Google Scholar] [CrossRef]
- Gorges, T.M.; Tinhofer, I.; Drosch, M.; Röse, L.; Zollner, T.M.; Krahn, T.; von Ahsen, O. Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer 2012, 12, 178. [Google Scholar] [CrossRef] [PubMed]
- Jie, X.-X.; Zhang, X.-Y.; Xu, C.-J. Epithelial-to-mesenchymal transition, circulating tumor cells and cancer metastasis: Mechanisms and clinical applications. Oncotarget 2017, 8, 81558–81571. [Google Scholar] [CrossRef]
- Zhang, X.; Wei, L.; Li, J.; Zheng, J.; Zhang, S.; Zhou, J. Epithelial-mesenchymal transition phenotype of circulating tumor cells is associated with distant metastasis in patients with NSCLC. Mol. Med. Rep. 2019, 19, 601–608. [Google Scholar] [CrossRef]
- Fang, J.; Wang, W.; Fang, J.; Wang, H.; Lin, L.; Li, F.; Sun, Q.; Li, F.; Qi, J.; Sun, X.; et al. Epithelial-mesenchymal transition classification of circulating tumor cells in lung and colon cancer patients: Potential role in clinical practice. Transl. Cancer Res. 2020, 9, 6639–6651. [Google Scholar] [CrossRef]
- Papadaki, M.A.; Aggouraki, D.; Vetsika, E.-K.; Xenidis, N.; Kallergi, G.; Kotsakis, A.; Georgoulias, V. Epithelial-to-mesenchymal Transition Heterogeneity of Circulating Tumor Cells and Their Correlation with MDSCs and Tregs in HER2-negative Metastatic Breast Cancer Patients. Anticancer Res. 2021, 41, 661–670. [Google Scholar] [CrossRef]
- Ridge, K.M.; Eriksson, J.E.; Pekny, M.; Goldman, R.D. Roles of vimentin in health and disease. Genes Dev. 2022, 36, 391–407. [Google Scholar] [CrossRef]
- Ratnayake, W.S.; Breedy, S.; Apostolatos, C.; Hill, R.; Dennison, C.; Acevedo-Duncan, M. Abstract 2857: Vimentin is a novel PKC-ι target facilitating migration and invasion in melanoma. Cancer Res. 2019, 79, 2857. [Google Scholar] [CrossRef]
- Zhu, Q.-Q.; Ma, C.; Wang, Q.; Song, Y.; Lv, T. The role of TWIST1 in epithelial-mesenchymal transition and cancers. Tumour Biol. 2016, 37, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Loh, C.Y.; Chai, J.Y.; Tang, T.F.; Wong, W.F.; Sethi, G.; Shanmugam, M.K.; Chong, P.P.; Looi, C.Y. The E-Cadherin and N-Cadherin Switch in Epithelial-to-Mesenchymal Transition: Signaling, Therapeutic Implications, and Challenges. Cells 2019, 8, 1118. [Google Scholar] [CrossRef] [PubMed]
- Sinha, D.; Saha, P.; Samanta, A.; Bishayee, A. Emerging Concepts of Hybrid Epithelial-to-Mesenchymal Transition in Cancer Progression. Biomolecules 2020, 10, 1561. [Google Scholar] [CrossRef] [PubMed]
- Simeonov, K.P.; Byrns, C.N.; Clark, M.L.; Norgard, R.J.; Martin, B.; Stanger, B.Z.; Shendure, J.; McKenna, A.; Lengner, C.J. Single-cell lineage tracing of metastatic cancer reveals selection of hybrid EMT states. Cancer Cell 2021, 39, 1150–1162.e9. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.S.; Abdollahi, B.; Wilkins, O.M.; Lu, H.; Chakraborty, P.; Ognjenovic, N.B.; Muller, K.E.; Jolly, M.K.; Christensen, B.C.; Hassanpour, S.; et al. Phenotypic heterogeneity driven by plasticity of the intermediate EMT state governs disease progression and metastasis in breast cancer. Sci. Adv. 2022, 8, eabj8002. [Google Scholar] [CrossRef]
- Caramel, J.; Papadogeorgakis, E.; Hill, L.; Browne, G.J.; Richard, G.; Wierinckx, A.; Saldanha, G.; Osborne, J.; Hutchinson, P.; Tse, G. A Switch in the Expression of Embryonic EMT-Inducers Drives the Development of Malignant Melanoma. Cancer Cell 2013, 24, 466. Available online: https://www.sciencedirect.com/science/article/pii/S1535610813003644 (accessed on 25 June 2023). [CrossRef]
- Richard, G.; Dalle, S.; Monet, M.-A.; Ligier, M.; Boespflug, A.; Pommier, R.M.; de la Fouchardière, A.; Perier-Muzet, M.; Depaepe, L.; Barnault, R.; et al. ZEB1-mediated melanoma cell plasticity enhances resistance to MAPK inhibitors. EMBO Mol. Med. 2016, 8, 1143–1161. Available online: https://www.embopress.org/doi/abs/10.15252/emmm.201505971 (accessed on 25 June 2023). [CrossRef]
- Fenouille, N.; Tichet, M.; Dufies, M.; Pottier, A.; Mogha, A.; Soo, J.K.; Rocchi, S.; Mallavialle, A.; Galibert, M.D.; Khammari, A.; et al. The Epithelial-Mesenchymal Transition (EMT) Regulatory Factor SLUG (SNAI2) Is a Downstream Target of SPARC and AKT in Promoting Melanoma Cell Invasion. PLoS ONE 2012, 7, e40378. [Google Scholar] [CrossRef]
- Shirley, S.H.; Greene, V.R.; Duncan, L.M.; Torres Cabala, C.A.; Grimm, E.A.; Kusewitt, D.F. Slug Expression during Melanoma Progression. Am. J. Pathol. 2012, 180, 2479–2489. Available online: https://www.sciencedirect.com/science/article/pii/S0002944012002350 (accessed on 25 June 2023). [CrossRef] [PubMed]
- Srivastava, K.; Pickard, A.; Craig, S.G.; Quinn, G.P.; Lambe, S.M.; James, J.A.; McDade, S.S.; McCance, D.J. ΔNp63γ/SRC/Slug Signaling Axis Promotes Epithelial-to-Mesenchymal Transition in Squamous Cancers. Clin. Cancer Res. 2018, 24, 3917–3927. [Google Scholar] [CrossRef] [PubMed]
- Visciano, C.; Liotti, F.; Prevete, N.; Cali’, G.; Franco, R.; Collina, F.; Collina, F.; de Paulis, A.; Marone, G.; Santoro, M.; et al. Mast cells induce epithelial-to-mesenchymal transition and stem cell features in human thyroid cancer cells through an IL-8–Akt–Slug pathway. Oncogene 2015, 34, 5175–5186. [Google Scholar] [CrossRef] [PubMed]
- Beck, B.; Lapouge, G.; Rorive, S.; Drogat, B.; Desaedelaere, K.; Delafaille, S.; Dubois, C.; Salmon, I.; Willekens, K.; Marine, J.C.; et al. Different levels of Twist1 regulate skin tumor initiation, stemness, and progression. Cell Stem Cell 2015, 16, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Weiss, M.B.; Abel, E.V.; Mayberry, M.M.; Basile, K.J.; Berger, A.C.; Aplin, A.E. TWIST1 is an ERK1/2 effector that promotes invasion and regulates MMP-1 expression in human melanoma cells. Cancer Res. 2012, 72, 6382–6392. [Google Scholar] [CrossRef] [PubMed]
- Gray, E.S.; Reid, A.L.; Bowyer, S.; Calapre, L.; Siew, K.; Pearce, R.; Cowell, L.; Frank, M.H.; Millward, M.; Ziman, M. Circulating Melanoma Cell Subpopulations: Their Heterogeneity and Differential Responses to Treatment. J. Investig. Dermatol. 2015, 135, 2040–2048. [Google Scholar] [CrossRef]
- Aya-Bonilla, C.A.; Morici, M.; Hong, X.; McEvoy, A.C.; Sullivan, R.J.; Freeman, J.; Calapre, L.; Khattak, M.A.; Meniawy, T.; Millward, M.; et al. Detection and prognostic role of heterogeneous populations of melanoma circulating tumour cells. Br. J. Cancer 2020, 122, 1059–1067. [Google Scholar] [CrossRef]
- Setia, N.; Abbas, O.; Sousa, Y.; Garb, J.L.; Mahalingam, M. Profiling of ABC transporters ABCB5, ABCF2 and nestin-positive stem cells in nevi, in situ and invasive melanoma. Mod. Pathol. 2012, 25, 1169–1175. [Google Scholar] [CrossRef]
- Huaman, J.; Ogunwobi, O.O. Circulating Tumor Cell Migration Requires Fibronectin Acting through Integrin B1 or SLUG. Cells 2020, 9, 1594. [Google Scholar] [CrossRef]
- Guo, W.; Wang, H.; Li, C. Signal pathways of melanoma and targeted therapy. Signal Transduct. Target Ther. 2021, 6, 424. [Google Scholar] [CrossRef]
- Gu, Y.; Zhang, Z.; Ten Dijke, P. Harnessing epithelial-mesenchymal plasticity to boost cancer immunotherapy. Cell Mol. Immunol. 2023, 20, 318–340. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Durand, S.; Pommier, R.M.; Benboubker, V.; Grimont, M.; Cumunel, E.; Boivin, F.; Dupeuble, F.; Barbollat-Boutrand, L.; Eberhardt, A.; et al. ZEB1 Controls a Lineage-Specific Transcriptional Program Essential for Melanoma Cell State Transitions; Cold Spring Harbor Laboratory: Laurel Hollow, NY, USA, 2023. [Google Scholar]
- Yang, J.; Antin, P.; Berx, G.; Blanpain, C.; Brabletz, T.; Bronner, M.; Campbell, K.; Cano, A.; Casanova, J.; Christofori, G.; et al. Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020, 21, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Xu, Q.; Xiao, C.; Li, H.; Wu, W.; Du, W.; Zhao, J.; Liu, H.; Wang, H.; Liu, C. SYBR Green real-time qPCR method: Diagnose drowning more rapidly and accurately. Forensic. Sci. Int. 2021, 321, 110720. [Google Scholar] [CrossRef] [PubMed]
| Control Cell Line | Gene | Ct Value | Anneal | #Cycles RTqPCR | Thr | Report Standard Curve and Efficiency Curve (90–110%) | Melting Curve (°C) | |
|---|---|---|---|---|---|---|---|---|
| V126 | SNAI2 | 25.1 | 60 °C | 48 | 0.2 | Slope: −3.19 R2: 0.993 | 105.4% | 82 |
| hFN1 | 22 | 60 °C | 48 | 0.2 | Slope: −3.09 R2: 0.99 | 110% | 78.5 | |
| A375 | n-CADH | 26.3 | 60 °C | 48 | 0.3 | Slope: −3.37 R2: 0.98 | 97% | 78 |
| TWIST1 | 26 | 60 °C | 48 | 0.3 | Slope: −3.4 R2: 0.979 | 94% | 89 | |
| VIMENTIN | 18.9 | 60 °C | 48 | 0.3 | Slope: −3.47 R2: 0.97 | 94% | 88 | |
| ZEB1 | 24.6 | 60 °C | 48 | 0.3 | Slope: −3.33 R2: 0.97 | 99% | 81 | |
| β-ACT | 19.07 | 60 °C | 48 | 0.3 | Slope: −3.40 R2: 0.96 | 96% | 88 | |
| Hela | MCAM | 23.88 | 60 °C | 48 | 0.2 | Slope: −3.33 R2: 0.998 | 99% | 82 |
| HaCaT | e-CADH | 22.0 | 60 °C | 48 | 0.2 | Slope: −3.00 R2: 0.99 | 99% | 80.4 |
| Gene | Frequency | Percentage | χ2 p |
|---|---|---|---|
| TWIST1 | Onset: 1/7 | 14% | 0.0063 |
| Follow-up: 3/10 | 30% | ||
| SNAI2 | Onset: 1/7 | 14% | ns |
| Follow-up: 2/10 | 20% | ||
| ZEB1 | Onset: 6/7 | 86% | 0.0063 |
| Follow-up: 7/10 | 70% | ||
| CD146/MCAM | Onset: 2/7 | 29% | 0.0007 |
| Follow-up: 0/10 | 0% | ||
| CDH2 | Onset: 5/7 | 71% | 0.0024 |
| Follow-up: 5/10 | 50% | ||
| CDH1 | Onset: 1/7 | 14% | 0.0063 |
| Follow-up: 3/10 | 30% | ||
| VIM | Onset: 7/7 | 100% | <0.0001 |
| Follow-up: 7/10 | 70% |
| Gene | Frequency | Percentage | χ2 p |
|---|---|---|---|
| TWIST1 | Responder: 0/5 | 0% | <0.0001 |
| Non-Responder: 3/5 | 60% | ||
| SNAI2 | Responder: 0/5 | 0% | <0.0001 |
| Non-Responder: 1/5 | 20% | ||
| ZEB1 | Responder: 3/5 | 60% | 0.002 |
| Non-Responder: 4/5 | 80% | ||
| CD146M/CAM | Responder: 1/5 | 20% | <0.0001 |
| Non-Responder: 0/5 | 0% | ||
| CDH2 | Responder: 1/5 | 20% | <0.0001 |
| Non-Responder: 4/5 | 80% | ||
| CDH1 | Responder: 1/5 | 20% | 0.002 |
| Non-Responder: 2/5 | 40% | ||
| VIM | Responder: 2/5 | 20% | <0.0001 |
| Non-Responder: 5/5 | 100% |
| TARGET GENE (ΔCT×1000) UPN | MCAM//CD146 | CDH1 | CDH2 | TWIST1 | SNAI2 | ZEB1 | VIM | hHFN1 | Therapy | Clinical Status |
|---|---|---|---|---|---|---|---|---|---|---|
| UPN1-AuV * | ||||||||||
| 1st checkpoint | 0 | 0 | 0.102 | 0.664 | 0 | 2.309 | 216.352 | 0 | In treatment | Disease Progression |
| 2nd check point | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | Targeted Therapy | Clinical Remission |
| UPN2-FuM | ||||||||||
| Onset | 0 | 0 | 7.36 | 0 | 0 | 67.485 | 171.043 | 0 | Naive | |
| 1st checkpoint | 0 | 0 | 0 | 0 | 0 | 15.441 | 0 | 0 | Checkpoint Inhibitors | Clinical Remission |
| UPN3-PaN | ||||||||||
| Onset | 0 | 0 | 0 | 0 | 0 | 38.751 | 347.245 | 0 | Naïve | |
| 1st checkpoint | 0 | 0 | 0 | 0 | 0 | 0 | 113.113 | 0 | Targeted Therapy | Disease Progression |
| UPN4-CaA | ||||||||||
| Onset | 2.795 | 0 | 4.74 | 22.581 | 0 | 10.298 | 485.60 | 0 | Naïve | |
| 1st checkpoint | 0 | 0 | 1.066 | 0 | 0.88 | 4.743 | 377.357 | 0 | Checkpoint Inhibitors | Disease Progression |
| UPN5-ZuF | ||||||||||
| Onset | 0 | 0 | 2.721 | 0 | 0 | 30.675 | 370.746 | 0 | Naïve | |
| 1st checkpoint | 0 | 3.057 | 0.247 | 0 | 0 | 4.662 | 260.04 | 0 | Checkpoint Inhibitors | Clinical Remission |
| UPN6-GiD | ||||||||||
| Onset | 2.7 | 0 | 0.156 | 0 | 0 | 2.377 | 222.476 | 0 | Naïve | |
| 1st checkpoint | 0 | 0 | 0 | 0 | 0 | 42.73 | 158.108 | 0 | Targeted Therapy | Clinical Remission |
| UPN7-PeME * | ||||||||||
| 1st checkpoint | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | In treatment | Clinical Remission |
| 2nd check point | 0.126 | 0.366 | 0.207 | 0.613 | 0 | 3.197 | 354.12 | 0 | Checkpoint Inhbitors | Disease Progression |
| UPN8-StD | ||||||||||
| Onset | 0 | 0.3874 | 0.04 | 0 | 0 | 1.742 | 287.527 | 0 | Naive | |
| 1st checkpoint | 0 | 0.146 | 0.018 | 1.775 | 0 | 5.263 | 431.553 | 0 | Checkpoint Inhibitors | Disease Progression |
| UPN9-ZaF | ||||||||||
| Onset | 0 | 0 | 0 | 0 | 0 | 0 | 79.167 | 0 | Naive | Started to Targeted Therapy |
| UPN Cod. | Sex | Age at First Observation | Primary Tumor-Site | Histology | Breslow Grade (mm) | AJCC Status at First Observation | Baseline Sample | Therapy | Clinical Status at Follow-Up |
|---|---|---|---|---|---|---|---|---|---|
| UPN1-AuV* | f | 80 | Unknown | / | / | IV | NO (+2 years) | Targeted Therapy (Anti-BRAF and anti-MEK) | At +12 months: Disease Progression At +18 months: Clinical Remission |
| UPN2-FuM | m | 40 | Trunk | SSM | 1.25 | IIB | YES | Checkpoint Inhibitors (antiPD1-PD1L) | At +6 months: Clinical Remission |
| UPN3-PaN | m | 64 | Trunk | NM | 4.0 | IV | YES | Targeted Therapy (Anti-BRAF and anti-MEK) | At +6 months: Disease Progression |
| UPN4-CaA | f | 82 | Extremity | NM | 2.4 | IV | YES | Checkpoint Inhibitors (anti-PD1-PD1L) | At +6 months: Disease Progression |
| UPN5-ZuF | m | 42 | Head | NM | 2.2 | IB | YES | Checkpoint Inhibitors (anti-PD1-PD1L) | At +6 months: Clinical Remission |
| UPN6-GiD | m | 35 | Extremity | SSM | 2.2 | IIIB | YES | Targeted Therapy (Anti-BRAF and anti-MEK) | At +6 months: Clinical Remission |
| UPN7-PeME* | f | 75 | Arm | Mucous M | 2.2 | IIB | NO (+1 year) | Checkpoint Inhibitors (anti-PD1-PD1L) | At +12 months: Disease Progression At +18 months: Clinical Remission |
| UPN8-StD | f | 41 | Trunk | NM | 7.0 | IIB | YES | Checkpoint Inhibitors (anti-PD1-PD1L) | At +6 months: Disease Progression |
| UPN9-ZaF | m | 42 | Extremity | NM | 6.0 | IVres | YES | Not yet started | Not yet reached |
| Gene | Primers |
|---|---|
| CD146/MCAM | F: 5′-AGCTCCGCGTCTACAAAGC-3′ |
| R: 5′-CTACACAGGTAGCGACCTCC-3′ | |
| CDH1 | F: 5′-AAAGGCCCATTTCCTAAAAACCT-3′ |
| R: 5′-TGCGTTCTCTATCCAGAGGCT-3′ | |
| CDH2 | F: 5′-CTCCTATGA GTGGAA CAG GAA CG-3′ |
| R: 5′ -TTG GAT CAA TGT CAT AAT CAA GTG CTGTA-3′ | |
| HFN1 | F: 5′-AGCCGAGGTTTTAACTGCGA-3′ |
| R: 5′-CCC ACT CGGTAAGTGTTCCC-3′ | |
| VIM | R: 5′-GACGCCATCAACACCGAGTT-3′ |
| F: 5′-CTTTGTCGTTGGTTAGCTGGT-3′ | |
| SLUG | R: 5′-CCAAGCTTTCAGACCCCCAT-3′ |
| F: 5′-GAAAAAGGCTTCTCCCCCGT-3′ | |
| TWIST1 | R: 5′- GCTTGAGGGTCTGAATCTTGCT-3′ |
| F: 5′-GTCCGCAGTCTTACGAGGAG-3′ | |
| ZEB1 | R: 5′-CAGCTTGATACCTGTGAATGGG-3′ |
| F: 5′-TATCTGTGGTCGTGTGGGACT-3′ | |
| ACTB | R: 5′-GAGACCTTCAACACCCCAGCC-3 |
| F: 5′-AATGTCACGCACGATTTCCC-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rapanotti, M.C.; Cugini, E.; Campione, E.; Di Raimondo, C.; Costanza, G.; Rossi, P.; Ferlosio, A.; Bernardini, S.; Orlandi, A.; De Luca, A.; et al. Epithelial-to-Mesenchymal Transition Gene Signature in Circulating Melanoma Cells: Biological and Clinical Relevance. Int. J. Mol. Sci. 2023, 24, 11792. https://doi.org/10.3390/ijms241411792
Rapanotti MC, Cugini E, Campione E, Di Raimondo C, Costanza G, Rossi P, Ferlosio A, Bernardini S, Orlandi A, De Luca A, et al. Epithelial-to-Mesenchymal Transition Gene Signature in Circulating Melanoma Cells: Biological and Clinical Relevance. International Journal of Molecular Sciences. 2023; 24(14):11792. https://doi.org/10.3390/ijms241411792
Chicago/Turabian StyleRapanotti, Maria Cristina, Elisa Cugini, Elena Campione, Cosimo Di Raimondo, Gaetana Costanza, Piero Rossi, Amedeo Ferlosio, Sergio Bernardini, Augusto Orlandi, Anastasia De Luca, and et al. 2023. "Epithelial-to-Mesenchymal Transition Gene Signature in Circulating Melanoma Cells: Biological and Clinical Relevance" International Journal of Molecular Sciences 24, no. 14: 11792. https://doi.org/10.3390/ijms241411792
APA StyleRapanotti, M. C., Cugini, E., Campione, E., Di Raimondo, C., Costanza, G., Rossi, P., Ferlosio, A., Bernardini, S., Orlandi, A., De Luca, A., & Bianchi, L. (2023). Epithelial-to-Mesenchymal Transition Gene Signature in Circulating Melanoma Cells: Biological and Clinical Relevance. International Journal of Molecular Sciences, 24(14), 11792. https://doi.org/10.3390/ijms241411792









