Albuminuria-Related Genetic Biomarkers: Replication and Predictive Evaluation in Individuals with and without Diabetes from the UK Biobank
Abstract
1. Introduction
2. Results
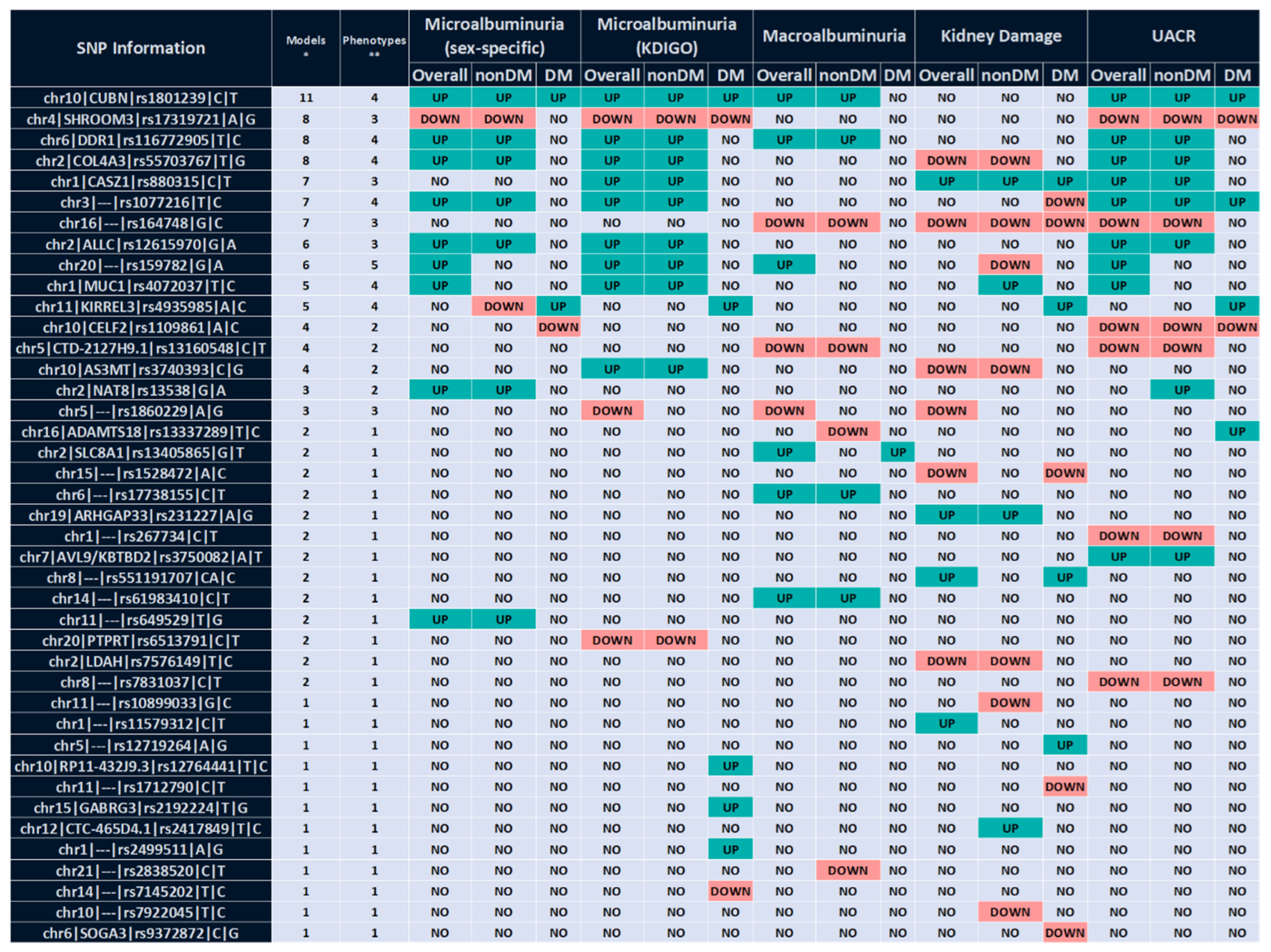
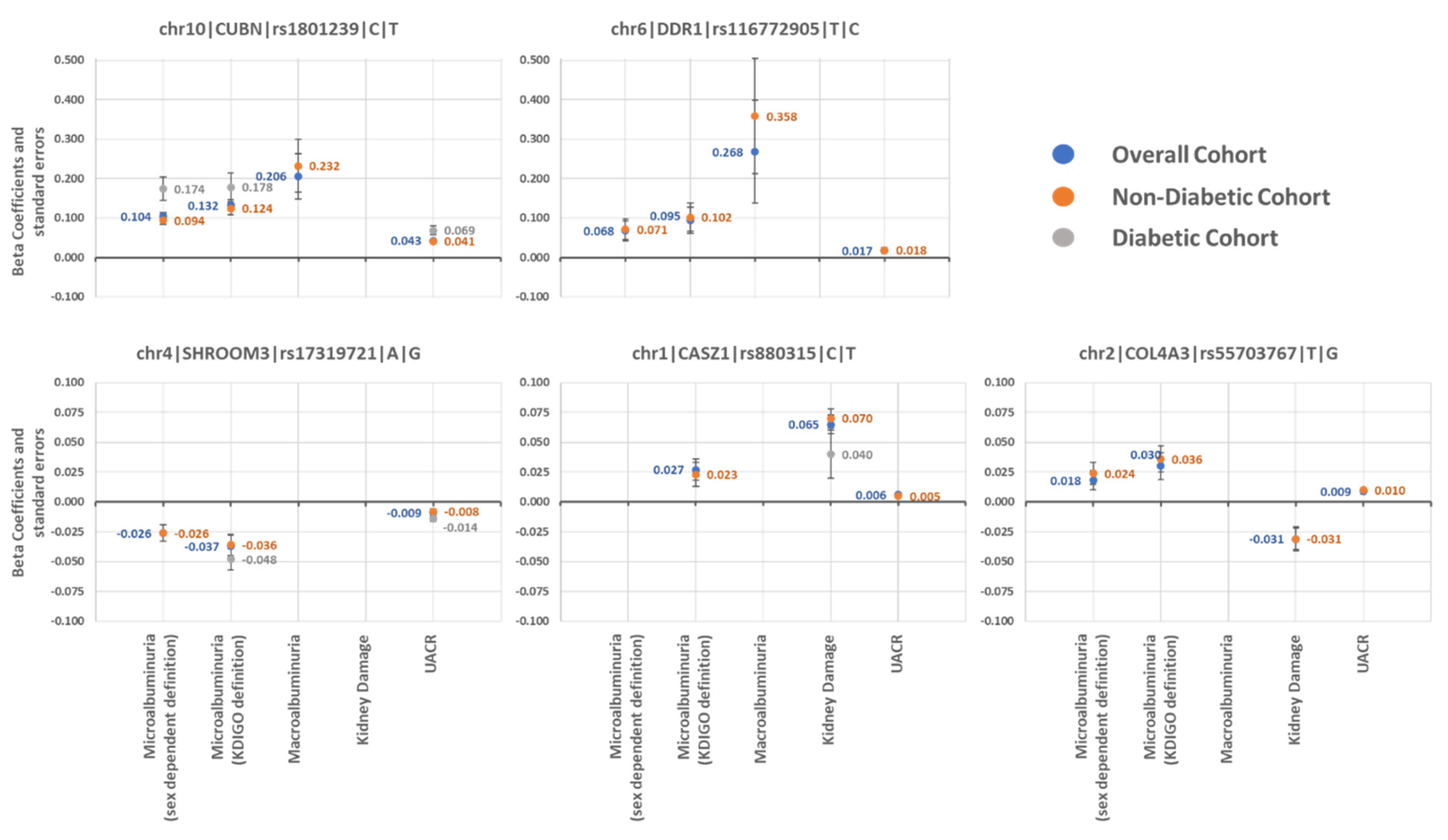
2.1. Multivariable Analysis
2.1.1. Association with UACR-Derived Phenotypes
2.1.2. Association with Kidney Damage
2.1.3. Specific Association in Patients with Diabetes
3. Discussion
4. Methods and Materials
4.1. Study Design and Population
4.2. Phenotypic Variables
4.2.1. Outcome Variables
Kidney Damage
Macroalbuminuria
Microalbuminuria
UACR
4.2.2. Independent Variables (Confounders)
Anti-Hypertensive Medication
Cholesterol Medication
Diabetes
Insulin
Other
4.3. Statistical Analysis
4.3.1. Descriptive Analysis
4.3.2. Bivariate Analysis
4.3.3. Genotyping, Imputation, and Quality Control
4.3.4. Genetic Risk Score Derivation
4.3.5. Multivariable Analysis
4.3.6. ROC Analysis
4.3.7. Power Calculations
5. Conclusions
Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACEi | angiotensin-converting enzyme inhibitors |
| AER | albumin excretion rate |
| AUROC | area under the receiver operating characteristic curve |
| BMI | body mass index |
| CKD | chronic kidney disease |
| CKD-EPI | Chronic Kidney Disease Epidemiology Collaboration |
| DKD | diabetic kidney disease |
| DM | diabetes mellitus |
| eGFR | estimated glomerular filtration rate |
| ESRD | end-stage renal disease |
| GFR | glomerular filtration rate |
| GTEx | gene tissue expression |
| GWAS | genome-wide association studies |
| HWE | Hardy-Weinberg equilibrium |
| LD | linkage disequilibrium |
| KDIGO | Kidney Disease: Improving Global Outcomes |
| MAF | minor allele frequency |
| mAlb | microalbuminuria |
| PCA | principal component analysis |
| PRS | polygenic risk scores |
| ROC | receiver operating characteristic curve |
| QC | quality control |
| SNP | single nucleotide polymorphism |
| UACR | urine albumin-to-creatinine ratio |
| UKB | UK Biobank |
References
- Levey, A.S.; Atkins, R.; Coresh, J.; Cohen, E.P.; Collins, A.J.; Eckardt, K.-U.; Nahas, M.E.; Jaber, B.L.; Jadoul, M.; Levin, A.; et al. Chronic Kidney Disease as a Global Public Health Problem: Approaches and Initiatives—A Position Statement from Kidney Disease Improving Global Outcomes. Kidney Int. 2007, 72, 247–259. [Google Scholar] [CrossRef]
- CDC. Centers for Disease Control and Prevention Chronic Kidney Disease Initiative. Available online: https://www.cdc.gov/kidneydisease/index.html (accessed on 3 December 2018).
- Bash, L.D.; Coresh, J.; Köttgen, A.; Parekh, R.S.; Fulop, T.; Wang, Y.; Astor, B.C. Defining Incident Chronic Kidney Disease in the Research Setting: The ARIC Study. Am. J. Epidemiol. 2009, 170, 414–424. [Google Scholar] [CrossRef]
- GBD 2017 Disease and Injury Incidence and Prevalence Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1789–1858. [Google Scholar] [CrossRef]
- GBD 2017 Causes of Death Collaborators. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: A systematic analysis for the Global Burden of Disease Study 2017; GBD 2017 Causes of Death Collaborators. Lancet 2018, 392, 1736–1788. [Google Scholar] [CrossRef] [PubMed]
- Foreman, K.J.; Marquez, N.; Dolgert, A.; Fukutaki, K.; Fullman, N.; McGaughey, M.; Pletcher, M.A.; Smith, A.E.; Tang, K.; Yuan, C.-W.; et al. Forecasting Life Expectancy, Years of Life Lost, and All-Cause and Cause-Specific Mortality for 250 Causes of Death: Reference and Alternative Scenarios for 2016-40 for 195 Countries and Territories. Lancet 2018, 392, 2052–2090. [Google Scholar] [CrossRef]
- Kazancioğlu, R. Risk Factors for Chronic Kidney Disease: An Update. Kidney Int. Suppl. 2013, 3, 368–371. [Google Scholar] [CrossRef] [PubMed]
- The Kidney Disease Outcomes Quality Initiative (KDOQI). KDIGO 2012 Clinical Practice Guideline for the Evaluation and Management of Chronic Kidney Disease. Kidney Int. 2021, 99, S1–S87. [Google Scholar]
- Levey, A.S.; De Jong, P.E.; Coresh, J.; Nahas, M.E.; Astor, B.C.; Matsushita, K.; Gansevoort, R.T.; Kasiske, B.L.; Eckardt, K.U. The Definition, Classification, and Prognosis of Chronic Kidney Disease: A KDIGO Controversies Conference Report. Kidney Int. 2011, 80, 17–28. [Google Scholar] [CrossRef]
- Hallan, S.I.; Ritz, E.; Lydersen, S.; Romundstad, S.; Kvenild, K.; Orth, S.R. Combining GFR and Albuminuria to Classify CKD Improves Prediction of ESRD. J. Am. Soc. Nephrol. 2009, 20, 1069–1077. [Google Scholar] [CrossRef]
- Brantsma, A.H.; Bakker, S.J.L.; Hillege, H.L.; De Zeeuw, D.; De Jong, P.E.; Gansevoort, R.T. Cardiovascular and Renal Outcome in Subjects with K/DOQI Stage 1–3 Chronic Kidney Disease: The Importance of Urinary Albumin Excretion. Nephrol. Dial. Transplant. 2008, 23, 3851–3858. [Google Scholar] [CrossRef]
- Astor, B.C.; Matsushita, K.; Gansevoort, R.T.; Van Der Velde, M.; Woodward, M.; Levey, A.S.; Jong, P.E.D.; Coresh, J.; El-Nahas, M.; Eckardt, K.U.; et al. Lower Estimated Glomerular Filtration Rate and Higher Albuminuria Are Associated with Mortality and End-Stage Renal Disease. A Collaborative Meta-Analysis of Kidney Disease Population Cohorts. Kidney Int. 2011, 79, 1331–1340. [Google Scholar] [CrossRef] [PubMed]
- Gansevoort, R.T.; Matsushita, K.; Van Der Velde, M.; Astor, B.C.; Woodward, M.; Levey, A.S.; Jong, P.E.D.; Coresh, J. Lower Estimated GFR and Higher Albuminuria Are Associated with Adverse Kidney Outcomes. A Collaborative Meta-Analysis of General and High-Risk Population Cohorts. Kidney Int. 2011, 80, 93–104. [Google Scholar] [CrossRef]
- Matsushita, K.; van der Velde, M.; Astor, B.C.; Woodward, M.; Levey, A.S.; de Jong, P.E.; Coresh, J.; Gansevoort, R.T.; El-Nahas, M.; Eckardt, K.U.; et al. Association of Estimated Glomerular Filtration Rate and Albuminuria with All-Cause and Cardiovascular Mortality in General Population Cohorts: A Collaborative Meta-Analysis. Lancet 2010, 375, 2073–2081. [Google Scholar] [CrossRef]
- Van Der Velde, M.; Matsushita, K.; Coresh, J.; Astor, B.C.; Woodward, M.; Levey, A.; De Jong, P.; Gansevoort, R.T.; El-Nahas, M.; Eckardt, K.U.; et al. Lower Estimated Glomerular Filtration Rate and Higher Albuminuria Are Associated with All-Cause and Cardiovascular Mortality. A Collaborative Meta-Analysis of High-Risk Population Cohorts. Kidney Int. 2011, 79, 1341–1352. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, K.; Coresh, J.; Sang, Y.; Chalmers, J.; Fox, C.; Guallar, E.; Jafar, T.; Jassal, S.K.; Landman, G.W.D.; Muntner, P.; et al. Estimated Glomerular Filtration Rate and Albuminuria for Prediction of Cardiovascular Outcomes: A Collaborative Meta-Analysis of Individual Participant Data. Lancet Diabetes Endocrinol. 2015, 3, 514–525. [Google Scholar] [CrossRef]
- Tuttle, K.R.; Bakris, G.L.; Bilous, R.W.; Chiang, J.L.; de Boer, I.H.; Goldstein-Fuchs, J.; Hirsch, I.B.; Kalantar-Zadeh, K.; Narva, A.S.; Navaneethan, S.D.; et al. Diabetic Kidney Disease: A Report from an ADA Consensus Conference. Am. J. Kidney Dis. 2014, 64, 510–533. [Google Scholar] [CrossRef]
- Hillege, H.L.; Fidler, V.; Diercks, G.F.H.; van Gilst, W.H.; de Zeeuw, D.; van Veldhuisen, D.J.; Gans, R.O.B.; Janssen, W.M.T.; Grobbee, D.E.; de Jong, P.E.; et al. Urinary Albumin Excretion Predicts Cardiovascular and Noncardiovascular Mortality in General Population. Circulation 2002, 106, 1777–1782. [Google Scholar] [CrossRef] [PubMed]
- Arnlöv, J.; Evans, J.C.; Meigs, J.B.; Wang, T.J.; Fox, C.S.; Levy, D.; Benjamin, E.J.; D’Agostino, R.B.; Vasan, R.S. Low-Grade Albuminuria and Incidence of Cardiovascular Disease Events in Nonhypertensive and Nondiabetic Individuals: The Framingham Heart Study. Circulation 2005, 112, 969–975. [Google Scholar] [CrossRef]
- Klausen, K.; Borch-Johnsen, K.; Feldt-Rasmussen, B.; Jensen, G.; Clausen, P.; Scharling, H.; Appleyard, M.; Jensen, J.S. Very Low Levels of Microalbuminuria Are Associated with Increased Risk of Coronary Heart Disease and Death Independently of Renal Function, Hypertension, and Diabetes. Circulation 2004, 110, 32–35. [Google Scholar] [CrossRef]
- Parving, H.H.; Oxenbøll, B.; Svendsen, P.A.; Christiansen, J.S.; Andersen, A.R. Early Detection of Patients at Risk of Developing Diabetic Nephropathy. A Longitudinal Study of Urinary Albumin Excretion. Acta Endocrinol. 1982, 100, 550–555. [Google Scholar] [CrossRef]
- Mogensen, C.E.; Christensen, C.K.; Vittinghus, E. The Stages in Diabetic Renal Disease: With Emphasis on the Stage of Incipient Diabetic Nephropathy. Diabetes 1983, 32, 64–78. [Google Scholar] [CrossRef]
- Canadas-Garre, M.; Anderson, K.; McGoldrick, J.; Maxwell, A.P.; McKnight, A.J. Genomic Approaches in the Search for Molecular Biomarkers in Chronic Kidney Disease. J. Transl. Med. 2018, 16, 292. [Google Scholar] [CrossRef]
- Canadas-Garre, M.; Anderson, K.; Cappa, R.; Skelly, R.; Smyth, L.J.; McKnight, A.J.; Maxwell, A.P.; Cañadas-Garre, M.; Anderson, K.; Cappa, R.; et al. Genetic Susceptibility to Chronic Kidney Disease—Some More Pieces for the Heritability Puzzle. Front. Genet. 2019, 10, 453. [Google Scholar] [CrossRef]
- van Zuydam, N.R.; Ahlqvist, E.; Sandholm, N.; Deshmukh, H.; Rayner, N.W.; Abdalla, M.; Ladenvall, C.; Ziemek, D.; Fauman, E.; Robertson, N.R.; et al. A Genome-Wide Association Study of Diabetic Kidney Disease in Subjects with Type 2 Diabetes. Diabetes 2018, 67, 1414–1427. [Google Scholar] [CrossRef]
- Teumer, A.; Tin, A.; Sorice, R.; Gorski, M.; Yeo, N.C.; Chu, A.Y.; Li, M.; Li, Y.; Mijatovic, V.; Ko, Y.-A.; et al. Genome-Wide Association Studies Identify Genetic Loci Associated with Albuminuria in Diabetes. Diabetes 2016, 65, 803–817. [Google Scholar] [CrossRef]
- Böger, C.A.; Chen, M.-H.; Tin, A.; Olden, M.; Köttgen, A.; de Boer, I.H.; Fuchsberger, C.; O’Seaghdha, C.M.; Pattaro, C.; Teumer, A.; et al. CUBN Is a Gene Locus for Albuminuria. J. Am. Soc. Nephrol. 2011, 22, 555–570. [Google Scholar] [CrossRef] [PubMed]
- Salem, R.M.; Todd, J.N.; Sandholm, N.; Cole, J.B.; Chen, W.-M.; Andrews, D.; Pezzolesi, M.G.; McKeigue, P.M.; Hiraki, L.T.; Qiu, C.; et al. Genome-Wide Association Study of Diabetic Kidney Disease Highlights Biology Involved in Glomerular Basement Membrane Collagen. J. Am. Soc. Nephrol. 2019, 30, 2000–2016. [Google Scholar] [CrossRef] [PubMed]
- Shiffman, D.; Pare, G.; Oberbauer, R.; Louie, J.Z.; Rowland, C.M.; Devlin, J.J.; Mann, J.F.; McQueen, M.J. A Gene Variant in CERS2 Is Associated with Rate of Increase in Albuminuria in Patients with Diabetes from ONTARGET and TRANSCEND. PLoS ONE 2014, 9, e106631. [Google Scholar] [CrossRef]
- Brown, L.A.; Sofer, T.; Stilp, A.M.; Baier, L.J.; Kramer, H.J.; Masindova, I.; Levy, D.; Hanson, R.L.; Moncrieft, A.E.; Redline, S.; et al. Admixture Mapping Identifies an Amerindian Ancestry Locus Associated with Albuminuria in Hispanics in the United States. J. Am. Soc. Nephrol. 2017, 28, 2211–2220. [Google Scholar] [CrossRef]
- Salem, R.M.; Todd, J.N.; Sandholm, N.; Cole, J.B.; Chen, W.-M.; Andrews, D.; Pezzolesi, M.G.; McKeigue, P.M.; Hiraki, L.T.; Qiu, C.; et al. Genome-Wide Association Study of Diabetic Kidney Disease Highlights Biology Involved in Renal Basement Membrane Collagen. bioRxiv 2018, 43, 499616. [Google Scholar] [CrossRef]
- Sandholm, N.; Forsblom, C.; Mäkinen, V.-P.; McKnight, A.J.; Österholm, A.-M.; He, B.; Harjutsalo, V.; Lithovius, R.; Gordin, D.; Parkkonen, M.; et al. Genome-Wide Association Study of Urinary Albumin Excretion Rate in Patients with Type 1 Diabetes. Diabetologia 2014, 57, 1143–1153. [Google Scholar] [CrossRef]
- Pattaro, C.; Teumer, A.; Gorski, M.; Chu, A.Y.; Li, M.; Mijatovic, V.; Garnaas, M.; Tin, A.; Sorice, R.; Li, Y.; et al. Genetic Associations at 53 Loci Highlight Cell Types and Biological Pathways Relevant for Kidney Function. Nat. Commun. 2016, 7, 10023. [Google Scholar] [CrossRef]
- Hwang, S.-J.; Yang, Q.; Meigs, J.B.; Pearce, E.N.; Fox, C.S. A Genome-Wide Association for Kidney Function and Endocrine-Related Traits in the NHLBI’s Framingham Heart Study. BMC Med. Genet. 2007, 8 (Suppl. S1), S10. [Google Scholar] [CrossRef] [PubMed]
- Böger, C.A.; Heid, I.M. Chronic Kidney Disease: Novel Insights from Genome-Wide Association Studies. Kidney Blood Press. Res. 2011, 34, 225–234. [Google Scholar] [CrossRef]
- Pattaro, C. Genome-Wide Association Studies of Albuminuria: Towards Genetic Stratification in Diabetes? J. Nephrol. 2018, 31, 475–487. [Google Scholar] [CrossRef] [PubMed]
- Haas, M.E.; Aragam, K.G.; Emdin, C.A.; Bick, A.G.; Hemani, G.; Davey Smith, G.; Kathiresan, S. Genetic Association of Albuminuria with Cardiometabolic Disease and Blood Pressure. Am. J. Hum. Genet. 2018, 103, 461–473. [Google Scholar] [CrossRef]
- Birn, H.; Fyfe, J.C.; Jacobsen, C.; Mounier, F.; Verroust, P.J.; Orskov, H.; Willnow, T.E.; Moestrup, S.K.; Christensen, E.I. Cubilin Is an Albumin Binding Protein Important for Renal Tubular Albumin Reabsorption. J. Clin. Investig. 2000, 105, 1353–1361. [Google Scholar] [CrossRef] [PubMed]
- Yanes, T.; McInerney-Leo, A.M.; Law, M.; Cummings, S. The Emerging Field of Polygenic Risk Scores and Perspective for Use in Clinical Care. Hum. Mol. Genet. 2020, 29, R165–R176. [Google Scholar] [CrossRef]
- Lambert, S.A.; Abraham, G.; Inouye, M. Towards Clinical Utility of Polygenic Risk Scores. Hum. Mol. Genet. 2019, 28, R133–R142. [Google Scholar] [CrossRef]
- Torkamani, A.; Wineinger, N.E.; Topol, E.J. The Personal and Clinical Utility of Polygenic Risk Scores. Nat. Rev. Genet. 2018, 19, 581–590. [Google Scholar] [CrossRef]
- Zanetti, D.; Rao, A.; Gustafsson, S.; Assimes, T.L.; Montgomery, S.B.; Ingelsson, E. Identification of 22 Novel Loci Associated with Urinary Biomarkers of Albumin, Sodium, and Potassium Excretion. Kidney Int. 2019, 95, 1197–1208. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, A.; Rodan, A.R.; Le, T.H.; Gaulton, K.J.; Haessler, J.; Stilp, A.M.; Kamatani, Y.; Zhu, G.; Sofer, T.; Puri, S.; et al. Trans-Ethnic Fine Mapping Highlights Kidney-Function Genes Linked to Salt Sensitivity. Am. J. Hum. Genet. 2016, 99, 636–646. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, J.; Haloui, M.; Attaoua, R.; Tahir, R.; Hishmih, C.; Harvey, F.; Marois-Blanchet, F.-C.; Long, C.; Simon, P.; Santucci, L.; et al. Polygenic Risk Scores Predict Diabetes Complications and Their Response to Intensive Blood Pressure and Glucose Control. Diabetologia 2021, 64, 2012–2025. [Google Scholar] [CrossRef] [PubMed]
- Major, R.W.; Shepherd, D.; Medcalf, J.F.; Xu, G.; Gray, L.J.; Brunskill, N.J. The Kidney Failure Risk Equation for Prediction of End Stage Renal Disease in UK Primary Care: An External Validation and Clinical Impact Projection Cohort Study. PLoS Med. 2019, 16, e1002955. [Google Scholar] [CrossRef]
- Tangri, N.; Grams, M.E.; Levey, A.S.; Coresh, J.; Appel, L.J.; Astor, B.C.; Chodick, G.; Collins, A.J.; Djurdjev, O.; Raina Elley, C.; et al. Multinational Assessment of Accuracy of Equations for Predicting Risk of Kidney Failure Ameta-Analysis. JAMA-J. Am. Med. Assoc. 2016, 315, 164–174. [Google Scholar] [CrossRef]
- Stapleton, C.P.; Heinzel, A.; Guan, W.; van der Most, P.J.; van Setten, J.; Lord, G.M.; Keating, B.J.; Israni, A.K.; de Borst, M.H.; Bakker, S.J.L.; et al. The Impact of Donor and Recipient Common Clinical and Genetic Variation on Estimated Glomerular Filtration Rate in a European Renal Transplant Population. Am. J. Transplant. 2019, 19, 2262–2273. [Google Scholar] [CrossRef]
- Kunzmann, A.T.; Canadas Garre, M.; Thrift, A.P.; McMenamin, U.C.; Johnston, B.T.; Cardwell, C.R.; Anderson, L.A.; Spence, A.D.; Lagergren, J.; Xie, S.-H.; et al. Information on Genetic Variants Does Not Increase Identification of Individuals at Risk of Esophageal Adenocarcinoma Compared to Clinical Risk Factors. Gastroenterology 2019, 156, 43–45. [Google Scholar] [CrossRef]
- Canadas-Garre, M.; Anderson, K.; McGoldrick, J.; Maxwell, A.P.; McKnight, A.J. Proteomic and Metabolomic Approaches in the Search for Biomarkers in Chronic Kidney Disease. J. Proteom. 2019, 193, 93–122. [Google Scholar] [CrossRef]
- Sirugo, G.; Williams, S.M.; Tishkoff, S.A. The Missing Diversity in Human Genetic Studies. Cell 2019, 177, 26–31. [Google Scholar] [CrossRef]
- Cooke Bailey, J.N.; Bush, W.S.; Crawford, D.C. Editorial: The Importance of Diversity in Precision Medicine Research. Front. Genet. 2020, 11, 875. [Google Scholar] [CrossRef]
- Keyes, K.M.; Westreich, D. UK Biobank, Big Data, and the Consequences of Non-Representativeness. Lancet 2019, 393, 1297. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Turchin, M.C.; Patki, A.; Srinivasasainagendra, V.; Shang, N.; Nadukuru, R.; Jones, A.C.; Malolepsza, E.; Dikilitas, O.; Kullo, I.J.; et al. Genome-Wide Polygenic Score to Predict Chronic Kidney Disease across Ancestries. Nat. Med. 2022, 28, 1412–1420. [Google Scholar] [CrossRef] [PubMed]
- Duncan, L.; Shen, H.; Gelaye, B.; Meijsen, J.; Ressler, K.; Feldman, M.; Peterson, R.; Domingue, B. Analysis of Polygenic Risk Score Usage and Performance in Diverse Human Populations. Nat. Commun. 2019, 10, 3328. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Patel, K.P.; Teng, A.K.; Berens, A.J.; Lachance, J. Genetic Disease Risks Can Be Misestimated across Global Populations. Genome Biol. 2018, 19, 179. [Google Scholar] [CrossRef] [PubMed]
- Grinde, K.E.; Qi, Q.; Thornton, T.A.; Liu, S.; Shadyab, A.H.; Chan, K.H.K.; Reiner, A.P.; Sofer, T. Generalizing Polygenic Risk Scores from Europeans to Hispanics/Latinos. Genet. Epidemiol. 2019, 43, 50–62. [Google Scholar] [CrossRef] [PubMed]
- Ware, E.B.; Schmitz, L.L.; Faul, J.; Gard, A.; Mitchell, C.; Smith, J.A.; Zhao, W.; Weir, D.; Kardia, S.L. Heterogeneity in Polygenic Scores for Common Human Traits. bioRxiv 2017, 106062. [Google Scholar] [CrossRef]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’Connell, J.; et al. The UK Biobank Resource with Deep Phenotyping and Genomic Data. Nature 2018, 562, 203–209. [Google Scholar] [CrossRef]
- Mills, M.C.; Rahal, C. A Scientometric Review of Genome-Wide Association Studies. Commun. Biol. 2019, 2, 9. [Google Scholar] [CrossRef]
- Mostafavi, H.; Harpak, A.; Conley, D.; Pritchard, J.K.; Przeworski, M. Variable Prediction Accuracy of Polygenic Scores within an Ancestry Group. bioRxiv 2019, 9, 629949. [Google Scholar] [CrossRef]
- Conte, C.; Antonelli, G.; Melica, M.E.; Tarocchi, M.; Romagnani, P.; Peired, A.J. Role of Sex Hormones in Prevalent Kidney Diseases. Int. J. Mol. Sci. 2023, 24, 8244. [Google Scholar] [CrossRef]
- Gall, M.-A.; Hougaard, P.; Borch-Johnsen, K.; Parving, H.-H. Risk Factors for Development of Incipient and Overt Diabetic Nephropathy in Patients with Non-Insulin Dependent Diabetes Mellitus: Prospective, Observational Study. BMJ 1997, 314, 783. [Google Scholar] [CrossRef] [PubMed]
- Ricardo, A.C.; Yang, W.; Sha, D.; Appel, L.J.; Chen, J.; Krousel-Wood, M.; Manoharan, A.; Steigerwalt, S.; Wright, J.; Rahman, M.; et al. Sex-Related Disparities in CKD Progression. J. Am. Soc. Nephrol. 2018, 30, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Neugarten, J.; Acharya, A.; Silbiger, S.R. Effect of Gender on the Progression of Nondiabetic Renal Disease. J. Am. Soc. Nephrol. 2000, 11, 319–329. [Google Scholar] [CrossRef]
- Maric-Bilkan, C. Sex Differences in Diabetic Kidney Disease. Mayo Clin. Proc. 2020, 95, 587–599. [Google Scholar] [CrossRef] [PubMed]
- Shepard, B.D. Sex Differences in Diabetes and Kidney Disease: Mechanisms and Consequences. Am. J. Physiol. Physiol. 2019, 317, F456–F462. [Google Scholar] [CrossRef]
- Wang, Y.; Namba, S.; Lopera, E.; Kerminen, S.; Tsuo, K.; Läll, K.; Kanai, M.; Zhou, W.; Wu, K.H.; Favé, M.J.; et al. Global Biobank Analyses Provide Lessons for Developing Polygenic Risk Scores across Diverse Cohorts. Cell Genom. 2023, 3, 100241. [Google Scholar] [CrossRef]
- Sonnega, A.; Faul, J.D.; Ofstedal, M.B.; Langa, K.M.; Phillips, J.W.R.; Weir, D.R. Cohort Profile: The Health and Retirement Study (HRS). Int. J. Epidemiol. 2014, 43, 576–585. [Google Scholar] [CrossRef]
- Wang, X.; Cheng, Z. Cross-Sectional Studies: Strengths, Weaknesses, and Recommendations. Chest 2020, 158, S65–S71. [Google Scholar] [CrossRef]
- Rusticus, S.A.; Lovato, C.Y. Impact of Sample Size and Variability on the Power and Type I Error Rates of Equivalence Tests: A Simulation Study. Pract. Assess. Res. Eval. 2014, 19, 11. [Google Scholar]
- Abdelmalek, J.A.; Gansevoort, R.T.; Lambers Heerspink, H.J.; Ix, J.H.; Rifkin, D.E. Estimated Albumin Excretion Rate Versus Urine Albumin-Creatinine Ratio for the Assessment of Albuminuria: A Diagnostic Test Study from the Prevention of Renal and Vascular Endstage Disease (PREVEND) Study. Am. J. Kidney Dis. 2014, 63, 415–421. [Google Scholar] [CrossRef]
- Warram, J.H.; Gearin, G.; Laffel, L.; Krolewski, A.S. Effect of Duration of Type I Diabetes on the Prevalence of Stages of Diabetic Nephropathy Defined by Urinary Albumin/Creatinine Ratio. J. Am. Soc. Nephrol. 1996, 7, 930–937. [Google Scholar] [CrossRef] [PubMed]
- R Core Team R: A Language and Environment for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 25 June 2023).
- Chang, C.C.; Chow, C.C.; Tellier, L.C.A.M.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-Generation PLINK: Rising to the Challenge of Larger and Richer Datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.; GRAIL, I.; Human Longevity, I.; Department of Biomedical Data Science. PLINK 2.00 Alpha. Available online: https://www.cog-genomics.org/plink/2.0/ (accessed on 25 June 2023).
- StataCorp Stata Statistical Software: Release 16 2019; StataCorp LLC: College Station, TX, USA, 2019.
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.C.; Müller, M. PROC: An Open-Source Package for R and S+ to Analyze and Compare ROC Curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- Pencina, M.J.; D’Agostino, R.B.; D’Agostino, R.B.; Vasan, R.S. Evaluating the Added Predictive Ability of a New Marker: From Area under the ROC Curve to Reclassification and Beyond. Stat. Med. 2008, 27, 157–172; discussion 207–212. [Google Scholar] [CrossRef] [PubMed]
- Harrell, F.E., Jr. Hmisc: Harrell Miscellaneous 2022. Available online: https://hbiostat.org/R/Hmisc/ (accessed on 25 June 2023).
- Sud, A.; Horton, R.H.; Hingorani, A.D.; Tzoulaki, I.; Turnbull, C.; Houlston, R.S.; Lucassen, A. Realistic Expectations Are Key to Realising the Benefits of Polygenic Scores. BMJ 2023, 380, e073149. [Google Scholar] [CrossRef]

| Overall Cohort | Non-Diabetic Cohort | Diabetic Cohort | ||||
|---|---|---|---|---|---|---|
| Variable | N | n (%) Mean ± sd|P50 [P25-P75] | N | n (%) Mean ± sd|P50 [P25-P75] | N | n (%) Mean ± sd|P50 [P25-P75] |
| Age (years) | 389,206 | 56.9 ± 8.0 | 353,400 | 56.7 ± 8.0 | 35,806 | 58.6 ± 7.6 |
| Sex (male) | 389,206 | 179,240 (46.1) | 353,400 | 159,874 (45.2) | 35,806 | 19,366 (54.1) |
| Body Mass Index (kg/m2) | 389,206 | 27.4 ± 4.7 | 353,400 | 27.2 ± 4.6 | 35,806 | 29.4 ± 5.7 |
| Diabetes (yes) | 389,206 | 35,806 (9.2) | 353,400 | 0 (0) | 35,806 | 35,806 (100) |
| Ever Smoker (yes) | 389,206 | 176,499 (45.4) | 353,400 | 158,263 (44.8) | 35,806 | 18,236 (50.9) |
| Hypertensive Medication (yes) | 389,172 | 91,381 (23.5) | 353,374 | 74,674 (21.1) | 35,798 | 16,707 (46.7) |
| Cholesterol Medication (yes) | 389,206 | 69,617 (17.9) | 353,400 | 52,285 (14.8) | 35,806 | 17,332 (48.4) |
| Insulin (yes) | 389,206 | 3931 (1.0) | 351,942 | 0 (0) | 35,522 | 3931 (11.1) |
| Microalbuminuria | sex-specific (yes) | 389,206 | 56,682 (14.6) | 353,400 | 49,639 (14.1) | 35,806 | 7043 (19.7) |
| Microalbuminuria | KDIGO (yes) | 389,206 | 29,260 (7.5) | 353,400 | 25,161 (7.1) | 35,806 | 4099 (11.5) |
| Macroalbuminuria (yes) | 389,206 | 1429 (0.4) | 353,400 | 1017 (0.3) | 35,806 | 412 (1.2) |
| Kidney Damage (yes) | 368,108 | 56,279 (15.3) | 333,316 | 43,763 (13.1) | 34,792 | 12,516 (36.0) |
| UACR (mg/g) | 389,206 | 9.8 [6.1–16.5] | 353,400 | 9.7 [6.1–16.3] | 35,806 | 10.3 [6.4–17.9] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cañadas-Garre, M.; Kunzmann, A.T.; Anderson, K.; Brennan, E.P.; Doyle, R.; Patterson, C.C.; Godson, C.; Maxwell, A.P.; McKnight, A.J. Albuminuria-Related Genetic Biomarkers: Replication and Predictive Evaluation in Individuals with and without Diabetes from the UK Biobank. Int. J. Mol. Sci. 2023, 24, 11209. https://doi.org/10.3390/ijms241311209
Cañadas-Garre M, Kunzmann AT, Anderson K, Brennan EP, Doyle R, Patterson CC, Godson C, Maxwell AP, McKnight AJ. Albuminuria-Related Genetic Biomarkers: Replication and Predictive Evaluation in Individuals with and without Diabetes from the UK Biobank. International Journal of Molecular Sciences. 2023; 24(13):11209. https://doi.org/10.3390/ijms241311209
Chicago/Turabian StyleCañadas-Garre, Marisa, Andrew T. Kunzmann, Kerry Anderson, Eoin P. Brennan, Ross Doyle, Christopher C. Patterson, Catherine Godson, Alexander P. Maxwell, and Amy Jayne McKnight. 2023. "Albuminuria-Related Genetic Biomarkers: Replication and Predictive Evaluation in Individuals with and without Diabetes from the UK Biobank" International Journal of Molecular Sciences 24, no. 13: 11209. https://doi.org/10.3390/ijms241311209
APA StyleCañadas-Garre, M., Kunzmann, A. T., Anderson, K., Brennan, E. P., Doyle, R., Patterson, C. C., Godson, C., Maxwell, A. P., & McKnight, A. J. (2023). Albuminuria-Related Genetic Biomarkers: Replication and Predictive Evaluation in Individuals with and without Diabetes from the UK Biobank. International Journal of Molecular Sciences, 24(13), 11209. https://doi.org/10.3390/ijms241311209






