Deciphering Common Traits of Breast and Ovarian Cancer Stem Cells and Possible Therapeutic Approaches
Abstract
1. Introduction
2. Cancer Development and Progression
3. Biomarkers of Breast Cancer Stem Cells (BCSC) and Ovarian Cancer Stem Cells (OCSC)
| Common Markers | Biological Function in BCSC and OCSC | Reference |
|---|---|---|
| CD44 | Transmembrane glycoprotein associated with chemoresistance. | [45,101] |
| CD24 | Cell-surface glycoprotein associated with metastasis, chemoresistance, and poor prognosis. | [49,52] |
| ALDH1 | Aldehyde dehydrogenase, an enzyme involved in oxidation of aldehydes, associated with cell proliferation, migration, poor survival, and chemoresistance. | [69,72] |
| CD133 | Transmembrane glycoprotein associated with chemoresistance, elevated migration, and invasiveness. | [75,78] |
| CD326 | Glycoprotein important for calcium-independent cell adhesion. | [80,83] |
| CD338 | Adenosine triphosphate (ATP)-binding cassette transporter associated with chemoresistance. | [90,102] |
| NANOG | Transcription factor essential for maintaining self-renewal and pluripotency. | [94,95] |
| SOX2 | Transcription factor important for cell proliferation, migration, drug resistance, and expression of stemness-related and EMT-related genes. | [96,98] |
| OCT4 | Transcription factor often co-expressed with NANOG and SOX2 associated with increased drug resistance and tumorigenicity. | [31,103] |
| ROR1 | Receptor tyrosine kinase important for proliferation, migration, and invasion. | [104,105] |
| LIN28 | RNA-binding protein important for reprogramming of somatic cells to induced pluripotent stem cells. | [106,107] |
| CD90 | Membrane glycosylphosphatidyl inositol (GPI)-anchored protein, related to tumor initiation and aggressiveness, and poorer patient prognosis. | [108,109] |
| LGR5 | Transmembrane receptor, increases the stemness of BC cells, BC recurrence, poor outcome, and high tumorigenicity. It promotes EOC proliferation, metastasis, and EMT. Yet, its high expression in HGSOC is linked with improved progression-free survival. | [110,111,112] |
| CD49f | Increases tumorsphere-formation ability, enhances tumorigenicity, drug resistance, and self-renewal properties. | [113,114] |
| SALL4 | Transcription factor, involved in cancer cell stemness, invasion, proliferation, tumor aggressiveness, and poor survival. | [115,116] |
| BMI1 | Regulator of gene expression, necessary for self-renewal properties of stem cells, plays a role in tumor aggressiveness, invasion, EMT, and drug resistance. | [117,118] |
| BCSC Markers | Biological Function in BCSC | Reference |
| CD61, ESA | Increases tumorsphere-formation ability, enhances tumorigenicity, drug resistance, and self-renewal properties. | [113] |
| Sca-1 | Enhances cancer cell tumorigenic ability. | [119] |
| CD70 | BCSC self-renewal, metastasis, and tumorgenicity. | [120] |
| CD29 | Integrin protein, enhances metastatic potential, EMT, and cell migration. | [121] |
| KLF4 | Transcription factor, maintenance of breast cancer stem cells, cell migration, and invasion. | [122] |
| OCSC Markers | Biological Function in OCSC | Reference |
| CD117 | Receptor tyrosine kinase, promotes self-renewal, differentiation, and regeneration of tumor in xenograft model, chemoresistance, and metastasis. | [123] |
| CD166 | Glycoprotein, increases sphere-forming ability, adhesion, cell migration, and chemoresistance, high tumorigenic potential. | [124] |
| CD184 | Chemokine receptor, important for migration and proliferation. | [125] |
| CD243 | ABC transporter, responsible for paclitaxel resistance in OCSC. | [126] |
| ETRA | Endothelin receptor-A, ETRA inhibition prevents chemotherapy-induced increase in CSCs, reduces the formation of tumor spheres. | [127] |
| NPRA | Atrial natriuretic peptide receptor, associated with CSCs induced tumorigenesis. | [128] |
| ZIP4 | Transmembrane zinc transporter, marker for tumor formation in vivo, self-renewal, and differentiation abilities in vitro. | [129] |
| IL-17R | Promotes self-renewal of CD133+ CSC in OC by binding IL-17 produced by the tumor microenvironment. | [130] |
| MISRs | Müllerian-inhibiting substance receptors, overexpressed in CD44+CD24+EpCAM+ cells of various OC cell lines. | [131] |
| c-MYC | Transcription factor, involved in reprogramming of OCSC through interaction with the tumor microenvironment. | [31] |
| MyD88 | Adaptor protein associated with Toll-like receptor (TLR) signaling, resistance to proapoptotic signals, and the ability to create a pro-inflammatory microenvironment. | [132] |
| SNORA72 | Highly expressed in ovarian sphere cells with CSC-like characteristics. | [133] |
4. Signaling Pathways in BCSC and OCSC
5. BCSC- and OCSC-Microenvironment Communication
5.1. The Connection between Adipocyte Populations and BCSC and OCSC
5.2. The Effect of Immune Cells on BCSC and OCSC
5.3. Cancer-Associated Fibroblasts and Mesenchymal Stem Cells
6. Cancer Immunogenicity and Evasion of Immunosurveillance of OC and BC
7. Experimental Models for Distinguishing CSC Populations
8. CSC-Targeted Therapies
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today. Available online: https://gco.iarc.fr/today (accessed on 21 March 2023).
- Reid, B.M.; Permuth, J.B.; Sellers, T.A. Epidemiology of ovarian cancer: A review. Cancer Biol. Med. 2017, 14, 9–32. [Google Scholar] [CrossRef]
- Rebbeck, T.R.; Mitra, N.; Wan, F.; Sinilnikova, O.M.; Healey, S.; McGuffog, L.; Mazoyer, S.; Chenevix-Trench, G.; Easton, D.F.; Antoniou, A.C.; et al. Association of type and location of BRCA1 and BRCA2 mutations with risk of breast and ovarian cancer. JAMA 2015, 313, 1347–1361. [Google Scholar] [CrossRef]
- Fu, X.; Tan, W.; Song, Q.; Pei, H.; Li, J. BRCA1 and Breast Cancer: Molecular Mechanisms and Therapeutic Strategies. Front. Cell Dev. Biol. 2022, 10, 813457. [Google Scholar] [CrossRef]
- Eliade, M.; Skrzypski, J.; Baurand, A.; Jacquot, C.; Bertolone, G.; Loustalot, C.; Coutant, C.; Guy, F.; Fumoleau, P.; Duffourd, Y.; et al. The transfer of multigene panel testing for hereditary breast and ovarian cancer to healthcare: What are the implications for the management of patients and families? Oncotarget 2017, 8, 1957–1971. [Google Scholar] [CrossRef]
- Kuchenbaecker, K.B.; Hopper, J.L.; Barnes, D.R.; Phillips, K.-A.; Mooij, T.M.; Roos-Blom, M.-J.; Jervis, S.; van Leeuwen, F.E.; Milne, R.L.; Andrieu, N.; et al. Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA 2017, 317, 2402–2416. [Google Scholar] [CrossRef]
- Chen, J.; Bae, E.; Zhang, L.; Hughes, K.; Parmigiani, G.; Braun, D.; Rebbeck, T.R. Penetrance of Breast and Ovarian Cancer in Women Who Carry a BRCA1/2 Mutation and Do Not Use Risk-Reducing Salpingo-Oophorectomy: An Updated Meta-Analysis. JNCI cancer Spectr. 2020, 4, pkaa029. [Google Scholar] [CrossRef]
- Hartmann, L.C.; Schaid, D.J.; Woods, J.E.; Crotty, T.P.; Myers, J.L.; Arnold, P.G.; Petty, P.M.; Sellers, T.A.; Johnson, J.L.; McDonnell, S.K.; et al. Efficacy of bilateral prophylactic mastectomy in women with a family history of breast cancer. N. Engl. J. Med. 1999, 340, 77–84. [Google Scholar] [CrossRef]
- Domchek, S.M.; Friebel, T.M.; Singer, C.F.; Evans, D.G.; Lynch, H.T.; Isaacs, C.; Garber, J.E.; Neuhausen, S.L.; Matloff, E.; Eeles, R.; et al. Association of risk-reducing surgery in BRCA1 or BRCA2 mutation carriers with cancer risk and mortality. JAMA 2010, 304, 967–975. [Google Scholar] [CrossRef]
- Rebbeck, T.R.; Friebel, T.; Lynch, H.T.; Neuhausen, S.L.; van ’t Veer, L.; Garber, J.E.; Evans, G.R.; Narod, S.A.; Isaacs, C.; Matloff, E.; et al. Bilateral prophylactic mastectomy reduces breast cancer risk in BRCA1 and BRCA2 mutation carriers: The PROSE Study Group. J. Clin. Oncol. 2004, 22, 1055–1062. [Google Scholar] [CrossRef]
- Choi, Y.-H.; Terry, M.B.; Daly, M.B.; MacInnis, R.J.; Hopper, J.L.; Colonna, S.; Buys, S.S.; Andrulis, I.L.; John, E.M.; Kurian, A.W.; et al. Association of Risk-Reducing Salpingo-Oophorectomy with Breast Cancer Risk in Women with BRCA1 and BRCA2 Pathogenic Variants. JAMA Oncol. 2021, 7, 585–592. [Google Scholar] [CrossRef]
- Perri, T.; Levin, G.; Naor-Revel, S.; Eliassi-Revivo, P.; Lifshitz, D.; Friedman, E.; Korach, J. Risk-reducing salpingo-oophorectomy and breast cancer incidence among Jewish BRCA1/BRCA2-mutation carriers-an Israeli matched-pair study. Int. J. Gynaecol. Obstet. 2022, 157, 431–436. [Google Scholar] [CrossRef]
- Conduit, C.; Milne, R.L.; Friedlander, M.L.; Phillips, K.-A. Bilateral Salpingo-oophorectomy and Breast Cancer Risk for BRCA1 and BRCA2 Mutation Carriers: Assessing the Evidence. Cancer Prev. Res. 2021, 14, 983–994. [Google Scholar] [CrossRef] [PubMed]
- Michaelson-Cohen, R.; Gabizon–Peretz, S.; Armon, S.; Srebnik-Moshe, N.; Mor, P.; Tomer, A.; Levy-Lahad, E.; Paluch-Shimon, S. Breast cancer risk and hormone replacement therapy among BRCA carriers after risk-reducing salpingo-oophorectomy. Eur. J. Cancer 2021, 148, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Prat, J. New insights into ovarian cancer pathology. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2012, 23 (Suppl. S1), x111–x117. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.S.; Conant, E.F.; Soo, M.S. Molecular Subtypes of Breast Cancer: A Review for Breast Radiologists. J. Breast Imaging 2021, 3, 12–24. [Google Scholar] [CrossRef]
- Feng, Z.; Wen, H.; Bi, R.; Ju, X.; Chen, X.; Yang, W.; Wu, X. A clinically applicable molecular classification for high-grade serous ovarian cancer based on hormone receptor expression. Sci. Rep. 2016, 6, 25408. [Google Scholar] [CrossRef]
- Lafourcade, A.; His, M.; Baglietto, L.; Boutron-Ruault, M.C.; Dossus, L.; Rondeau, V. Factors associated with breast cancer recurrences or mortality and dynamic prediction of death using history of cancer recurrences: The French E3N cohort. BMC Cancer 2018, 18, 171. [Google Scholar] [CrossRef]
- Garzon, S.; Laganà, A.S.; Casarin, J.; Raffaelli, R.; Cromi, A.; Franchi, M.; Barra, F.; Alkatout, I.; Ferrero, S.; Ghezzi, F. Secondary and tertiary ovarian cancer recurrence: What is the best management? Gland Surg. 2020, 9, 1118–1129. [Google Scholar] [CrossRef]
- Courtney, D.; Davey, M.G.; Moloney, B.M.; Barry, M.K.; Sweeney, K.; McLaughlin, R.P.; Malone, C.M.; Lowery, A.J.; Kerin, M.J. Breast cancer recurrence: Factors impacting occurrence and survival. Ir. J. Med. Sci. 2022, 191, 2501–2510. [Google Scholar] [CrossRef]
- Pedersen, R.N.; Esen, B.Ö.; Mellemkjær, L.; Christiansen, P.; Ejlertsen, B.; Lash, T.L.; Nørgaard, M.; Cronin-Fenton, D. The Incidence of Breast Cancer Recurrence 10-32 Years After Primary Diagnosis. JNCI J. Natl. Cancer Inst. 2022, 114, 391–399. [Google Scholar] [CrossRef]
- Colombo, N.; Lorusso, D.; Scollo, P. Impact of Recurrence of Ovarian Cancer on Quality of Life and Outlook for the Future. Int. J. Gynecol. Cancer 2017, 27, 1134–1140. [Google Scholar] [CrossRef]
- Bishop, A.J.; Ensor, J.; Moulder, S.L.; Shaitelman, S.F.; Edson, M.A.; Whitman, G.J.; Bishnoi, S.; Hoffman, K.E.; Stauder, M.C.; Valero, V.; et al. Prognosis for patients with metastatic breast cancer who achieve a no-evidence-of-disease status after systemic or local therapy. Cancer 2015, 121, 4324–4332. [Google Scholar] [CrossRef]
- Bilyk, O.; Coatham, M.; Jewer, M.; Postovit, L.-M. Epithelial-to-Mesenchymal Transition in the Female Reproductive Tract: From Normal Functioning to Disease Pathology. Front. Oncol. 2017, 7, 145. [Google Scholar] [CrossRef] [PubMed]
- Yamulla, R.J.; Nalubola, S.; Flesken-Nikitin, A.; Nikitin, A.Y.; Schimenti, J.C. Most Commonly Mutated Genes in High-Grade Serous Ovarian Carcinoma Are Nonessential for Ovarian Surface Epithelial Stem Cell Transformation. Cell Rep. 2020, 32, 108086. [Google Scholar] [CrossRef]
- Deng, K.; Yang, C.; Tan, Q.; Song, W.; Lu, M.; Zhao, W.; Lou, G.; Li, Z.; Li, K.; Hou, Y. Sites of distant metastases and overall survival in ovarian cancer: A study of 1481 patients. Gynecol. Oncol. 2018, 150, 460–465. [Google Scholar] [CrossRef]
- Tan, D.S.P.; Agarwal, R.; Kaye, S.B. Mechanisms of transcoelomic metastasis in ovarian cancer. Lancet Oncol. 2006, 7, 925–934. [Google Scholar] [CrossRef] [PubMed]
- Tjhay, F.; Motohara, T.; Tayama, S.; Narantuya, D.; Fujimoto, K.; Guo, J.; Sakaguchi, I.; Honda, R.; Tashiro, H.; Katabuchi, H. CD44 variant 6 is correlated with peritoneal dissemination and poor prognosis in patients with advanced epithelial ovarian cancer. Cancer Sci. 2015, 106, 1421–1428. [Google Scholar] [CrossRef]
- Al Habyan, S.; Kalos, C.; Szymborski, J.; McCaffrey, L. Multicellular detachment generates metastatic spheroids during intra-abdominal dissemination in epithelial ovarian cancer. Oncogene 2018, 37, 5127–5135. [Google Scholar] [CrossRef]
- Yeung, T.-L.; Leung, C.S.; Yip, K.-P.; Au Yeung, C.L.; Wong, S.T.C.; Mok, S.C. Cellular and molecular processes in ovarian cancer metastasis. A Review in the Theme: Cell and Molecular Processes in Cancer Metastasis. Am. J. Physiol. Cell Physiol. 2015, 309, C444–C456. [Google Scholar] [CrossRef]
- Di, J.; Duiveman-de Boer, T.; Zusterzeel, P.L.M.; Figdor, C.G.; Massuger, L.F.A.G.; Torensma, R. The stem cell markers Oct4A, Nanog and c-Myc are expressed in ascites cells and tumor tissue of ovarian cancer patients. Cell. Oncol. 2013, 36, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Latifi, A.; Luwor, R.B.; Bilandzic, M.; Nazaretian, S.; Stenvers, K.; Pyman, J.; Zhu, H.; Thompson, E.W.; Quinn, M.A.; Findlay, J.K.; et al. Isolation and characterization of tumor cells from the ascites of ovarian cancer patients: Molecular phenotype of chemoresistant ovarian tumors. PLoS ONE 2012, 7, e46858. [Google Scholar] [CrossRef]
- Bregenzer, M.E.; Horst, E.N.; Mehta, P.; Novak, C.M.; Repetto, T.; Mehta, G. The Role of Cancer Stem Cells and Mechanical Forces in Ovarian Cancer Metastasis. Cancers 2019, 11, 1008. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Rakha, E.; Toss, M.; Quinn, C. Specific cell differentiation in breast cancer: A basis for histological classification. J. Clin. Pathol. 2022, 75, 76–84. [Google Scholar] [CrossRef]
- Feng, Y.; Spezia, M.; Huang, S.; Yuan, C.; Zeng, Z.; Zhang, L.; Ji, X.; Liu, W.; Huang, B.; Luo, W.; et al. Breast cancer development and progression: Risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis. Genes Dis. 2018, 5, 77–106. [Google Scholar] [CrossRef]
- Ng, C.K.Y.; Bidard, F.-C.; Piscuoglio, S.; Geyer, F.C.; Lim, R.S.; de Bruijn, I.; Shen, R.; Pareja, F.; Berman, S.H.; Wang, L.; et al. Genetic Heterogeneity in Therapy-Naïve Synchronous Primary Breast Cancers and Their Metastases. Clin. Cancer Res. 2017, 23, 4402–4415. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [CrossRef]
- Martins, F.C.; de Santiago, I.; Trinh, A.; Xian, J.; Guo, A.; Sayal, K.; Jimenez-Linan, M.; Deen, S.; Driver, K.; Mack, M.; et al. Combined image and genomic analysis of high-grade serous ovarian cancer reveals PTEN loss as a common driver event and prognostic classifier. Genome Biol. 2014, 15, 526. [Google Scholar] [CrossRef]
- Martins, F.C.; Couturier, D.-L.; Paterson, A.; Karnezis, A.N.; Chow, C.; Nazeran, T.M.; Odunsi, A.; Gentry-Maharaj, A.; Vrvilo, A.; Hein, A.; et al. Clinical and pathological associations of PTEN expression in ovarian cancer: A multicentre study from the Ovarian Tumour Tissue Analysis Consortium. Br. J. Cancer 2020, 123, 793–802. [Google Scholar] [CrossRef]
- Li, W.; Gu, X.; Liu, C.; Shi, Y.; Wang, P.; Zhang, N.; Wu, R.; Leng, L.; Xie, B.; Song, C.; et al. A synergetic effect of BARD1 mutations on tumorigenesis. Nat. Commun. 2021, 12, 1243. [Google Scholar] [CrossRef]
- Tegally, H.; Kensler, K.H.; Mungloo-Dilmohamud, Z.; Ghoorah, A.W.; Rebbeck, T.R.; Baichoo, S. Discovering novel driver mutations from pan-cancer analysis of mutational and gene expression profiles. PLoS ONE 2020, 15, e0242780. [Google Scholar] [CrossRef] [PubMed]
- Dick, J.E. Breast cancer stem cells revealed. Proc. Natl. Acad. Sci. USA 2003, 100, 3547–3549. [Google Scholar] [CrossRef] [PubMed]
- Chhabra, S.N.; Booth, B.W. Asymmetric cell division of mammary stem cells. Cell Div. 2021, 16, 5. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef]
- Xu, H.; Tian, Y.; Yuan, X.; Wu, H.; Liu, Q.; Pestell, R.G.; Wu, K. The role of CD44 in epithelial-mesenchymal transition and cancer development. Onco. Targets. Ther. 2015, 8, 3783–3792. [Google Scholar] [CrossRef]
- Al-Othman, N.; Alhendi, A.; Ihbaisha, M.; Barahmeh, M.; Alqaraleh, M.; Al-Momany, B.Z. Role of CD44 in breast cancer. Breast Dis. 2020, 39, 1–13. [Google Scholar] [CrossRef]
- Lim, S.-C.; Oh, S.-H. The role of CD24 in various human epithelial neoplasias. Pathol. Res. Pract. 2005, 201, 479–486. [Google Scholar] [CrossRef]
- Kristiansen, G.; Winzer, K.J.; Mayordomo, E.; Bellach, J.; Schlüns, K.; Denkert, C.; Dahl, E.; Pilarsky, C.; Altevogt, P.; Guski, H.; et al. CD24 Expression Is a New Prognostic Marker in Breast Cancer. Clin. Cancer Res. 2003, 9, 4906–4913. [Google Scholar]
- Kristiansen, G.; Denkert, C.; Schlüns, K.; Dahl, E.; Pilarsky, C.; Hauptmann, S. CD24 is expressed in ovarian cancer and is a new independent prognostic marker of patient survival. Am. J. Pathol. 2002, 161, 1215–1221. [Google Scholar] [CrossRef]
- Park, S.Y.; Lee, H.E.; Li, H.; Shipitsin, M.; Gelman, R.; Polyak, K. Heterogeneity for stem cell-related markers according to tumor subtype and histologic stage in breast cancer. Clin. Cancer Res. 2010, 16, 876–887. [Google Scholar] [CrossRef]
- Gao, M.-Q.; Choi, Y.-P.; Kang, S.; Youn, J.H.; Cho, N.-H. CD24+ cells from hierarchically organized ovarian cancer are enriched in cancer stem cells. Oncogene 2010, 29, 2672–2680. [Google Scholar] [CrossRef] [PubMed]
- Surowiak, P.; Materna, V.; Kaplenko, I.; Spaczyński, M.; Dietel, M.; Kristiansen, G.; Lage, H.; Zabel, M. Unfavorable prognostic value of CD24 expression in sections from primary and relapsed ovarian cancer tissue. Int. J. Gynecol. Cancer 2006, 16, 515–521. [Google Scholar] [CrossRef]
- Choi, Y.-L.; Kim, S.-H.; Shin, Y.K.; Hong, Y.-C.; Lee, S.-J.; Kang, S.Y.; Ahn, G. Cytoplasmic CD24 expression in advanced ovarian serous borderline tumors. Gynecol. Oncol. 2005, 97, 379–386. [Google Scholar] [CrossRef]
- Gao, Y.; Foster, R.; Yang, X.; Feng, Y.; Shen, J.K.; Mankin, H.J.; Hornicek, F.J.; Amiji, M.M.; Duan, Z. Up-regulation of CD44 in the development of metastasis, recurrence and drug resistance of ovarian cancer. Oncotarget 2015, 6, 9313–9326. [Google Scholar] [CrossRef] [PubMed]
- Ponta, H.; Sherman, L.; Herrlich, P.A. CD44: From adhesion molecules to signalling regulators. Nat. Rev. Mol. Cell Biol. 2003, 4, 33–45. [Google Scholar] [CrossRef]
- Uchino, M.; Kojima, H.; Wada, K.; Imada, M.; Onoda, F.; Satofuka, H.; Utsugi, T.; Murakami, Y. Nuclear β-catenin and CD44 upregulation characterize invasive cell populations in non-aggressive MCF-7 breast cancer cells. BMC Cancer 2010, 10, 414. [Google Scholar] [CrossRef] [PubMed]
- Börjesson, P.K.E.; Postema, E.J.; Roos, J.C.; Colnot, D.R.; Marres, H.A.M.; van Schie, M.H.; Stehle, G.; de Bree, R.; Snow, G.B.; Oyen, W.J.G.; et al. Phase I therapy study with (186)Re-labeled humanized monoclonal antibody BIWA 4 (bivatuzumab) in patients with head and neck squamous cell carcinoma. Clin. Cancer Res. 2003, 9, 3961S–3972S. [Google Scholar] [PubMed]
- Meng, E.; Long, B.; Sullivan, P.; McClellan, S.; Finan, M.A.; Reed, E.; Shevde, L.; Rocconi, R.P. CD44+/CD24− ovarian cancer cells demonstrate cancer stem cell properties and correlate to survival. Clin. Exp. Metastasis 2012, 29, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lewis, M.T.; Huang, J.; Gutierrez, C.; Osborne, C.K.; Wu, M.F.; Hilsenbeck, S.G.; Pavlick, A.; Zhang, X.; Chamness, G.C.; et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J. Natl. Cancer Inst. 2008, 100, 672–679. [Google Scholar] [CrossRef]
- Roy, R.; Willan, P.; Clarke, R.; Farnie, G. Differentiation therapy: Targeting breast cancer stem cells to reduce resistance to radiotherapy and chemotherapy. Breast Cancer Res. 2010, 12, O5. [Google Scholar] [CrossRef]
- Ricardo, S.; Vieira, A.F.; Gerhard, R.; Leitão, D.; Pinto, R.; Cameselle-Teijeiro, J.F.; Milanezi, F.; Schmitt, F.; Paredes, J. Breast cancer stem cell markers CD44, CD24 and ALDH1: Expression distribution within intrinsic molecular subtype. J. Clin. Pathol. 2011, 64, 937–944. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, Q.; Wang, Q.; Wang, Y.; Chen, J. Prognostic Significance of CD24 and CD44 in Breast Cancer: A Meta-Analysis. Int. J. Biol. Markers 2017, 32, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Friedl, P.; Alexander, S. Cancer invasion and the microenvironment: Plasticity and reciprocity. Cell 2011, 147, 992–1009. [Google Scholar] [CrossRef]
- Sheridan, C.; Kishimoto, H.; Fuchs, R.K.; Mehrotra, S.; Bhat-Nakshatri, P.; Turner, C.H.; Goulet, R.; Badve, S.; Nakshatri, H. CD44+/CD24- breast cancer cells exhibit enhanced invasive properties: An early step necessary for metastasis. Breast Cancer Res. 2006, 8, R59. [Google Scholar] [CrossRef]
- Li, W.; Ma, H.; Zhang, J.; Zhu, L.; Wang, C.; Yang, Y. Unraveling the roles of CD44/CD24 and ALDH1 as cancer stem cell markers in tumorigenesis and metastasis. Sci. Rep. 2017, 7, 13856. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Cao, M.; Liu, Y.; He, Y.; Du, Y.; Zhang, G.; Gao, F. Inducible formation of leader cells driven by CD44 switching gives rise to collective invasion and metastases in luminal breast carcinomas. Oncogene 2019, 38, 7113–7132. [Google Scholar] [CrossRef]
- Zanoni, M.; Bravaccini, S.; Fabbri, F.; Arienti, C. Emerging Roles of Aldehyde Dehydrogenase Isoforms in Anti-cancer Therapy Resistance. Front. Med. 2022, 9, 795762. [Google Scholar] [CrossRef]
- Deng, S.; Yang, X.; Lassus, H.; Liang, S.; Kaur, S.; Ye, Q.; Li, C.; Wang, L.-P.; Roby, K.F.; Orsulic, S.; et al. Distinct expression levels and patterns of stem cell marker, aldehyde dehydrogenase isoform 1 (ALDH1), in human epithelial cancers. PLoS ONE 2010, 5, e10277. [Google Scholar] [CrossRef]
- Landen, C.N.; Goodman, B.; Katre, A.A.; Steg, A.D.; Nick, A.M.; Stone, R.L.; Miller, L.D.; Mejia, P.V.; Jennings, N.B.; Gershenson, D.M.; et al. Targeting aldehyde dehydrogenase cancer stem cells in ovarian cancer. Mol. Cancer Ther. 2010, 9, 3186–3199. [Google Scholar] [CrossRef]
- Tanei, T.; Morimoto, K.; Shimazu, K.; Kim, S.J.; Tanji, Y.; Taguchi, T.; Tamaki, Y.; Noguchi, S. Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential Paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin. Cancer Res. 2009, 15, 4234–4241. [Google Scholar] [CrossRef]
- Ginestier, C.; Hur, M.H.; Charafe-Jauffret, E.; Monville, F.; Dutcher, J.; Brown, M.; Jacquemier, J.; Viens, P.; Kleer, C.G.; Liu, S.; et al. ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell Stem Cell 2007, 1, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Corbeil, D.; Röper, K.; Fargeas, C.A.; Joester, A.; Huttner, W.B. Prominin: A Story of Cholesterol, Plasma Membrane Protrusions and Human Pathology. Traffic 2001, 2, 82–91. [Google Scholar] [CrossRef]
- Anderson, L.H.; Boulanger, C.A.; Smith, G.H.; Carmeliet, P.; Watson, C.J. Stem cell marker prominin-1 regulates branching morphogenesis, but not regenerative capacity, in the mammary gland. Dev. Dyn. 2011, 240, 674–681. [Google Scholar] [CrossRef] [PubMed]
- Wright, M.H.; Calcagno, A.M.; Salcido, C.D.; Carlson, M.D.; Ambudkar, S.V.; Varticovski, L. Brca1 breast tumors contain distinct CD44+/CD24-and CD133+cells with cancer stem cell characteristics. Breast Cancer Res. 2008, 10, R10. [Google Scholar] [CrossRef]
- Brugnoli, F.; Grassilli, S.; Al-Qassab, Y.; Capitani, S.; Bertagnolo, V. CD133 in Breast Cancer Cells: More than a Stem Cell Marker. J. Oncol. 2019, 2019, 7512632. [Google Scholar] [CrossRef]
- Liu, T.J.; Sun, B.C.; Zhao, X.L.; Zhao, X.M.; Sun, T.; Gu, Q.; Yao, Z.; Dong, X.Y.; Zhao, N.; Liu, N. CD133+ cells with cancer stem cell characteristics associates with vasculogenic mimicry in triple-negative breast cancer. Oncogene 2013, 32, 544–553. [Google Scholar] [CrossRef]
- Ferrandina, G.; Bonanno, G.; Pierelli, L.; Perillo, A.; Procoli, A.; Mariotti, A.; Corallo, M.; Martinelli, E.; Rutella, S.; Paglia, A.; et al. Expression of CD133-1 and CD133-2 in ovarian cancer. Int. J. Gynecol. Cancer 2008, 18, 506–514. [Google Scholar] [CrossRef]
- Long, H.; Xie, R.; Xiang, T.; Zhao, Z.; Lin, S.; Liang, Z.; Chen, Z.; Zhu, B. Autocrine CCL5 signaling promotes invasion and migration of CD133+ ovarian cancer stem-like cells via NF-κB-mediated MMP-9 upregulation. Stem Cells 2012, 30, 2309–2319. [Google Scholar] [CrossRef]
- Mal, A.; Bukhari, A.B.; Singh, R.K.; Kapoor, A.; Barai, A.; Deshpande, I.; Wadasadawala, T.; Ray, P.; Sen, S.; De, A. EpCAM-Mediated Cellular Plasticity Promotes Radiation Resistance and Metastasis in Breast Cancer. Front. Cell Dev. Biol. 2020, 8, 597673. [Google Scholar] [CrossRef]
- Hiraga, T.; Ito, S.; Nakamura, H. EpCAM expression in breast cancer cells is associated with enhanced bone metastasis formation. Int. J. Cancer 2016, 138, 1698–1708. [Google Scholar] [CrossRef]
- Walters Haygood, C.L.; Arend, R.C.; Straughn, J.M.; Buchsbaum, D.J. Ovarian cancer stem cells: Can targeted therapy lead to improved progression-free survival? World J. Stem Cells 2014, 6, 441–447. [Google Scholar] [CrossRef]
- Zheng, J.; Zhao, S.; Yu, X.; Huang, S.; Liu, H.Y. Simultaneous targeting of CD44 and EpCAM with a bispecific aptamer effectively inhibits intraperitoneal ovarian cancer growth. Theranostics 2017, 7, 1373–1388. [Google Scholar] [CrossRef]
- Münz, M.; Murr, A.; Kvesic, M.; Rau, D.; Mangold, S.; Pflanz, S.; Lumsden, J.; Volkland, J.; Fagerberg, J.; Riethmüller, G.; et al. Side-by-side analysis of five clinically tested anti-EpCAM monoclonal antibodies. Cancer Cell Int. 2010, 10, 44. [Google Scholar] [CrossRef]
- Schmidt, M.; Scheulen, M.E.; Dittrich, C.; Obrist, P.; Marschner, N.; Dirix, L.; Schmidt, M.; Rüttinger, D.; Schuler, M.; Reinhardt, C.; et al. An open-label, randomized phase II study of adecatumumab, a fully human anti-EpCAM antibody, as monotherapy in patients with metastatic breast cancer. Ann. Oncol. 2010, 21, 275–282. [Google Scholar] [CrossRef]
- Seimetz, D.; Lindhofer, H.; Bokemeyer, C. Development and approval of the trifunctional antibody catumaxomab (anti-EpCAM x anti-CD3) as a targeted cancer immunotherapy. Cancer Treat. Rev. 2010, 36, 458–467. [Google Scholar] [CrossRef]
- Szotek, P.P.; Pieretti-Vanmarcke, R.; Masiakos, P.T.; Dinulescu, D.M.; Connolly, D.; Foster, R.; Dombkowski, D.; Preffer, F.; MacLaughlin, D.T.; Donahoe, P.K. Ovarian cancer side population defines cells with stem cell-like characteristics and Mullerian Inhibiting Substance responsiveness. Proc. Natl. Acad. Sci. USA 2006, 103, 11154–11159. [Google Scholar] [CrossRef]
- Moserle, L.; Indraccolo, S.; Ghisi, M.; Frasson, C.; Fortunato, E.; Canevari, S.; Miotti, S.; Tosello, V.; Zamarchi, R.; Corradin, A.; et al. The Side Population of Ovarian Cancer Cells Is a Primary Target of IFN-α Antitumor Effects. Cancer Res. 2008, 68, 5658–5668. [Google Scholar] [CrossRef]
- Hu, L.; McArthur, C.; Jaffe, R.B. Ovarian cancer stem-like side-population cells are tumourigenic and chemoresistant. Br. J. Cancer 2010, 102, 1276–1283. [Google Scholar] [CrossRef]
- Leccia, F.; Del Vecchio, L.; Mariotti, E.; Di Noto, R.; Morel, A.P.; Puisieux, A.; Salvatore, F.; Ansieau, S. ABCG2, a novel antigen to sort luminal progenitors of BRCA1- breast cancer cells. Mol. Cancer 2014, 13, 213. [Google Scholar] [CrossRef]
- Honeth, G.; Bendahl, P.O.; Ringnér, M.; Saal, L.H.; Gruvberger-Saal, S.K.; Lövgren, K.; Grabau, D.; Fernö, M.; Borg, Å.; Hegardt, C. The CD44+/CD24− phenotype is enriched in basal-like breast tumors. Breast Cancer Res. 2008, 10, R53. [Google Scholar] [CrossRef]
- Croker, A.K.; Goodale, D.; Chu, J.; Postenka, C.; Hedley, B.D.; Hess, D.A.; Allan, A.L. High aldehyde dehydrogenase and expression of cancer stem cell markers selects for breast cancer cells with enhanced malignant and metastatic ability. J. Cell. Mol. Med. 2009, 13, 2236–2252. [Google Scholar] [CrossRef]
- Kryczek, I.; Liu, S.; Roh, M.; Vatan, L.; Szeliga, W.; Wei, S.; Banerjee, M.; Mao, Y.; Kotarski, J.; Wicha, M.S.; et al. Expression of aldehyde dehydrogenase and CD133 defines ovarian cancer stem cells. Int. J. Cancer 2012, 130, 29–39. [Google Scholar] [CrossRef]
- Siu, M.K.Y.; Wong, E.S.Y.; Kong, D.S.H.; Chan, H.Y.; Jiang, L.; Wong, O.G.W.; Lam, E.W.-F.; Chan, K.K.L.; Ngan, H.Y.S.; Le, X.-F.; et al. Stem cell transcription factor NANOG controls cell migration and invasion via dysregulation of E-cadherin and FoxJ1 and contributes to adverse clinical outcome in ovarian cancers. Oncogene 2013, 32, 3500–3509. [Google Scholar] [CrossRef]
- Han, J.; Zhang, F.; Yu, M.; Zhao, P.; Ji, W.; Zhang, H.; Wu, B.; Wang, Y.; Niu, R. RNA interference-mediated silencing of NANOG reduces cell proliferation and induces G0/G1 cell cycle arrest in breast cancer cells. Cancer Lett. 2012, 321, 80–88. [Google Scholar] [CrossRef]
- Liu, K.; Xie, F.; Gao, A.; Zhang, R.; Zhang, L.; Xiao, Z.; Hu, Q.; Huang, W.; Huang, Q.; Lin, B.; et al. SOX2 regulates multiple malignant processes of breast cancer development through the SOX2/miR-181a-5p, miR-30e-5p/TUSC3 axis. Mol. Cancer 2017, 16, 62. [Google Scholar] [CrossRef]
- Leis, O.; Eguiara, A.; Lopez-Arribillaga, E.; Alberdi, M.J.; Hernandez-Garcia, S.; Elorriaga, K.; Pandiella, A.; Rezola, R.; Martin, A.G. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene 2012, 31, 1354–1365. [Google Scholar] [CrossRef]
- Wen, Y.; Hou, Y.; Huang, Z.; Cai, J.; Wang, Z. SOX2 is required to maintain cancer stem cells in ovarian cancer. Cancer Sci. 2017, 108, 719–731. [Google Scholar] [CrossRef]
- Mohiuddin, I.S.; Wei, S.J.; Kang, M.H. Role of OCT4 in cancer stem-like cells and chemotherapy resistance. Biochim. Biophys. Acta—Mol. Basis Dis. 2020, 1866, 165432. [Google Scholar] [CrossRef]
- Yan, H.C.; Fang, L.S.; Xu, J.; Qiu, Y.Y.; Lin, X.M.; Huang, H.X.; Han, Q.Y. The identification of the biological characteristics of human ovarian cancer stem cells. Eur. Rev. Med. Pharmacol. Sci. 2014, 18, 3497–3503. [Google Scholar]
- Zhang, S.; Balch, C.; Chan, M.W.W.; Lai, H.-C.; Matei, D.; Schilder, J.M.M.; Yan, P.S.S.; Huang, T.H.-M.H.-M.; Nephew, K.P.P. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008, 68, 4311–4320. [Google Scholar] [CrossRef]
- Boesch, M.; Zeimet, A.G.; Reimer, D.; Schmidt, S.; Gastl, G.; Parson, W.; Spoeck, F.; Hatina, J.; Wolf, D.; Sopper, S. The side population of ovarian cancer cells defines a heterogeneous compartment exhibiting stem cell characteristics. Oncotarget 2014, 5, 7027–7039. [Google Scholar] [CrossRef]
- Zhang, Q.; Han, Z.; Zhu, Y.; Chen, J.; Li, W. The Role and Specific Mechanism of OCT4 in Cancer Stem Cells: A Review. Int. J. Stem Cells 2020, 13, 312. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, H.; Ghia, E.M.; Huang, J.; Wu, L.; Zhang, J.; Lam, S.; Lei, Y.; He, J.; Cui, B.; et al. Inhibition of chemotherapy resistant breast cancer stem cells by a ROR1 specific antibody. Proc. Natl. Acad. Sci. USA 2019, 116, 1370–1377. [Google Scholar] [CrossRef]
- Henry, C.; Llamosas, E.; Knipprath-Meszaros, A.; Schoetzau, A.; Obermann, E.; Fuenfschilling, M.; Caduff, R.; Fink, D.; Hacker, N.; Ward, R.; et al. Targeting the ROR1 and ROR2 receptors in epithelial ovarian cancer inhibits cell migration and invasion. Oncotarget 2015, 6, 40310–40326. [Google Scholar] [CrossRef]
- Zou, H.; Luo, J.; Guo, Y.; Liu, Y.; Wang, Y.; Deng, L.; Li, P. RNA-binding protein complex LIN28/MSI2 enhances cancer stem cell-like properties by modulating Hippo-YAP1 signaling and independently of Let-7. Oncogene 2022, 41, 1657–1672. [Google Scholar] [CrossRef]
- Peng, S.; Maihle, N.J.; Huang, Y. Pluripotency factors Lin28 and Oct4 identify a sub-population of stem cell-like cells in ovarian cancer. Oncogene 2010, 29, 2153–2159. [Google Scholar] [CrossRef]
- Lu, H.; Clauser, K.R.; Tam, W.L.; Fröse, J.; Ye, X.; Eaton, E.N.; Reinhardt, F.; Donnenberg, V.S.; Bhargava, R.; Carr, S.A.; et al. A breast cancer stem cell niche supported by juxtacrine signalling from monocytes and macrophages. Nat. Cell Biol. 2014, 16, 1105–1117. [Google Scholar] [CrossRef]
- Connor, E.V.; Saygin, C.; Braley, C.; Wiechert, A.C.; Karunanithi, S.; Crean-Tate, K.; Abdul-Karim, F.W.; Michener, C.M.; Rose, P.G.; Lathia, J.D.; et al. Thy-1 predicts poor prognosis and is associated with self-renewal in ovarian cancer. J. Ovarian Res. 2019, 12, 112. [Google Scholar] [CrossRef]
- Yang, L.; Tang, H.; Kong, Y.; Xie, X.; Chen, J.; Song, C.; Liu, X.; Ye, F.; Li, N.; Wang, N.; et al. LGR5 Promotes Breast Cancer Progression and Maintains Stem-Like Cells Through Activation of Wnt/β-Catenin Signaling. Stem Cells 2015, 33, 2913–2924. [Google Scholar] [CrossRef]
- Kim, H.; Lee, D.H.; Park, E.; Myung, J.K.; Park, J.H.; Kim, D.I.; Kim, S.I.; Lee, M.; Kim, Y.; Park, C.M.; et al. Differential epithelial and stromal LGR5 expression in ovarian carcinogenesis. Sci. Rep. 2022, 12, 11200. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, J.; Gan, X.; Shen, F.; Yang, X.; Du, N.; Xia, D.; Liu, L.; Qiao, L.; Pan, J.; et al. LGR5 promotes epithelial ovarian cancer proliferation, metastasis, and epithelial–mesenchymal transition through the Notch1 signaling pathway. Cancer Med. 2018, 7, 3132–3142. [Google Scholar] [CrossRef]
- Lo, P.K.; Kanojia, D.; Liu, X.; Singh, U.P.; Berger, F.G.; Wang, Q.; Chen, H. CD49f and CD61 identify Her2/neu-induced mammary tumor-initiating cells that are potentially derived from luminal progenitors and maintained by the integrin-TGFΒ signaling. Oncogene 2012, 31, 2614–2626. [Google Scholar] [CrossRef] [PubMed]
- Wiechert, A.; Saygin, C.; Thiagarajan, P.S.; Rao, V.S.; Hale, J.S.; Gupta, N.; Hitomi, M.; Nagaraj, A.B.; DiFeo, A.; Lathia, J.D.; et al. Cisplatin induces stemness in ovarian cancer. Oncotarget 2016, 7, 30511–30522. [Google Scholar] [CrossRef] [PubMed]
- Moein, S.; Tenen, D.G.; Amabile, G.; Chai, L. SALL4: An Intriguing Therapeutic Target in Cancer Treatment. Cells 2022, 11, 2601. [Google Scholar] [CrossRef] [PubMed]
- Sharbatoghli, M.; Shamshiripour, P.; Fattahi, F.; Kalantari, E.; Habibi Shams, Z.; Panahi, M.; Totonchi, M.; Asadi-Lari, Z.; Madjd, Z.; Saeednejad Zanjani, L. Co-expression of cancer stem cell markers, SALL4/ALDH1A1, is associated with tumor aggressiveness and poor survival in patients with serous ovarian carcinoma. J. Ovarian Res. 2022, 15, 17. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, M.; Bharali, D.J.; Sudha, T.; Khedr, M.; Guest, I.; Sell, S.; Glinsky, G.V.; Mousa, S.A. Downregulation of Bmi1 in breast cancer stem cells suppresses tumor growth and proliferation. Oncotarget 2017, 8, 38731–38742. [Google Scholar] [CrossRef]
- Zhao, Q.; Qian, Q.; Cao, D.; Yang, J.; Gui, T.; Shen, K. Role of BMI1 in epithelial ovarian cancer: Investigated via the CRISPR/Cas9 system and RNA sequencing. J. Ovarian Res. 2018, 11, 31. [Google Scholar] [CrossRef]
- Grange, C.; Lanzardo, S.; Cavallo, F.; Camussi, G.; Bussolati, B. SCA-1 identifies the tumor-initiating cells in mammary tumors of BALB-neuT trangenic mice. Neoplasia 2008, 10, 1433–1443. [Google Scholar] [CrossRef]
- Liu, L.; Yin, B.; Yi, Z.; Liu, X.J.; Hu, Z.Q.; Gao, W.C.; Yu, H.W.; Li, Q.Q. Breast cancer stem cells characterized by CD70 expression preferentially metastasize to the lungs. Breast Cancer 2018, 25, 706–716. [Google Scholar] [CrossRef]
- Vassilopoulos, A.; Chisholm, C.; Lahusen, T.; Zheng, H.; Deng, C.-X. A critical role of CD29 and CD49f in mediating metastasis for cancer-initiating cells isolated from a Brca1-associated mouse model of breast cancer. Oncogene 2014, 33, 5477–5482. [Google Scholar] [CrossRef]
- Yu, F.; Li, J.; Chen, H.; Fu, J.; Ray, S.; Huang, S.; Zheng, H.; Ai, W. Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene 2011, 30, 2161–2172. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Zeng, J.; Liang, B.; Zhao, Z.; Sun, L.; Cao, D.; Yang, J.; Shen, K. Ovarian cancer cells with the CD117 phenotype are highly tumorigenic and are related to chemotherapy outcome. Exp. Mol. Pathol. 2011, 91, 596–602. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.K.; Ham, M.H.; Lee, S.Y.; Shin, M.J.; Kim, Y.E.; Song, P.; Suh, D.-S.; Kim, J.H. CD166 promotes the cancer stem-like properties of primary epithelial ovarian cancer cells. BMB Rep. 2020, 53, 622–627. [Google Scholar] [CrossRef] [PubMed]
- Mitsui, H.; Shibata, K.; Suzuki, S.; Umezu, T.; Mizuno, M.; Kajiyama, H.; Kikkawa, F. Functional interaction between peritoneal mesothelial cells and stem cells of ovarian yolk sac tumor (SC-OYST) in peritoneal dissemination. Gynecol. Oncol. 2012, 124, 303–310. [Google Scholar] [CrossRef]
- Eyre, R.; Harvey, I.; Stemke-Hale, K.; Lennard, T.W.J.; Tyson-Capper, A.; Meeson, A.P. Reversing paclitaxel resistance in ovarian cancer cells via inhibition of the ABCB1 expressing side population. Tumour Biol. 2014, 35, 9879–9892. [Google Scholar] [CrossRef]
- Coffman, L.; Mooney, C.; Lim, J.; Bai, S.; Silva, I.; Gong, Y.; Yang, K.; Buckanovich, R.J. Endothelin receptor-A is required for the recruitment of antitumor T cells and modulates chemotherapy induction of cancer stem cells. Cancer Biol. Ther. 2013, 14, 184–192. [Google Scholar] [CrossRef]
- Kong, X.; Wang, X.; Xu, W.; Behera, S.; Hellermann, G.; Kumar, A.; Lockey, R.F.; Mohapatra, S.; Mohapatra, S.S. Natriuretic peptide receptor a as a novel anticancer target. Cancer Res. 2008, 68, 249–256. [Google Scholar] [CrossRef]
- Fan, Q.; Zhang, W.; Emerson, R.E.; Xu, Y. ZIP4 Is a Novel Cancer Stem Cell Marker in High-Grade Serous Ovarian Cancer. Cancers 2020, 12, 3692. [Google Scholar] [CrossRef]
- Xiang, T.; Long, H.; He, L.; Han, X.; Lin, K.; Liang, Z.; Zhuo, W.; Xie, R.; Zhu, B. Interleukin-17 produced by tumor microenvironment promotes self-renewal of CD133+ cancer stem-like cells in ovarian cancer. Oncogene 2015, 34, 165–176. [Google Scholar] [CrossRef]
- Wei, X.; Dombkowski, D.; Meirelles, K.; Pieretti-Vanmarcke, R.; Szotek, P.P.; Chang, H.L.; Preffer, F.I.; Mueller, P.R.; Teixeira, J.; MacLaughlin, D.T.; et al. Mullerian inhibiting substance preferentially inhibits stem/progenitors in human ovarian cancer cell lines compared with chemotherapeutics. Proc. Natl. Acad. Sci. USA 2010, 107, 18874–18879. [Google Scholar] [CrossRef] [PubMed]
- d’Adhemar, C.J.; Spillane, C.D.; Gallagher, M.F.; Bates, M.; Costello, K.M.; Barry-O’Crowley, J.; Haley, K.; Kernan, N.; Murphy, C.; Smyth, P.C.; et al. The MyD88+ phenotype is an adverse prognostic factor in epithelial ovarian cancer. PLoS ONE 2014, 9, e100816. [Google Scholar] [CrossRef]
- Zhang, L.; Ma, R.; Gao, M.; Zhao, Y.; Lv, X.; Zhu, W.; Han, L.; Su, P.; Fan, Y.; Yan, Y.; et al. SNORA72 Activates the Notch1/c-Myc Pathway to Promote Stemness Transformation of Ovarian Cancer Cells. Front. Cell Dev. Biol. 2020, 8, 583087. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.A.; Nusse, R. Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell Stem Cell 2010, 6, 568–577. [Google Scholar] [CrossRef]
- Ng, A.; Tan, S.; Singh, G.; Rizk, P.; Swathi, Y.; Tan, T.Z.; Huang, R.Y.-J.; Leushacke, M.; Barker, N. Lgr5 marks stem/progenitor cells in ovary and tubal epithelia. Nat. Cell Biol. 2014, 16, 745–757. [Google Scholar] [CrossRef]
- Logan, C.Y.; Nusse, R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004, 20, 781–810. [Google Scholar] [CrossRef]
- Hojo, N.; Huisken, A.L.; Wang, H.; Chirshev, E.; Kim, N.S.; Nguyen, S.M.; Campos, H.; Glackin, C.A.; Ioffe, Y.J.; Unternaehrer, J.J. Snail knockdown reverses stemness and inhibits tumour growth in ovarian cancer. Sci. Rep. 2018, 8, 8704. [Google Scholar] [CrossRef]
- Chau, W.K.; Ip, C.K.; Mak, A.S.C.; Lai, H.-C.; Wong, A.S.T. c-Kit mediates chemoresistance and tumor-initiating capacity of ovarian cancer cells through activation of Wnt/β-catenin-ATP-binding cassette G2 signaling. Oncogene 2013, 32, 2767–2781. [Google Scholar] [CrossRef]
- Raghavan, S.; Mehta, P.; Xie, Y.; Lei, Y.L.; Mehta, G. Ovarian cancer stem cells and macrophages reciprocally interact through the WNT pathway to promote pro-tumoral and malignant phenotypes in 3D engineered microenvironments. J. Immunother. Cancer 2019, 7, 190. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.Y.; Xia, W.; Wang, J.C.; Kwong, K.Y.; Spohn, B.; Wen, Y.; Pestell, R.G.; Hung, M.C. β-catenin, a novel prognostic marker for breast cancer: Its roles in cyclin D1 expression and cancer progression. Proc. Natl. Acad. Sci. USA 2000, 97, 4262–4266. [Google Scholar] [CrossRef]
- Monteiro, J.; Gaspar, C.; Richer, W.; Franken, P.F.; Sacchetti, A.; Joosten, R.; Idali, A.; Brandao, J.; Decraene, C.; Fodde, R. Cancer stemness in Wnt-driven mammary tumorigenesis. Carcinogenesis 2014, 35, 2–13. [Google Scholar] [CrossRef]
- Wu, Y.; Ginther, C.; Kim, J.; Mosher, N.; Chung, S.; Slamon, D.; Vadgama, J.V. Expression of Wnt3 activates Wnt/β-catenin pathway and promotes EMT-like phenotype in trastuzumab-resistant HER2-overexpressing breast cancer cells. Mol. Cancer Res. 2012, 10, 1597–1606. [Google Scholar] [CrossRef] [PubMed]
- Jang, G.B.; Kim, J.Y.; Cho, S.D.; Park, K.S.; Jung, J.Y.; Lee, H.Y.; Hong, I.S.; Nam, J.S. Blockade of Wnt/β-catenin signaling suppresses breast cancer metastasis by inhibiting CSC-like phenotype. Sci. Rep. 2015, 5, 12465. [Google Scholar] [CrossRef] [PubMed]
- Shan, N.L.; Shin, Y.; Yang, G.; Furmanski, P.; Suh, N. Breast Cancer Stem Cells: A Review of Their Characteristics and The Agents That Affect Them. Mol. Carcinog. 2021, 60, 73. [Google Scholar] [CrossRef]
- Xu, J.; Prosperi, J.R.; Choudhury, N.; Olopade, O.I.; Goss, K.H. β-Catenin Is Required for the Tumorigenic Behavior of Triple-Negative Breast Cancer Cells. PLoS ONE 2015, 10, e0117097. [Google Scholar] [CrossRef]
- Yan, Y.; Liu, F.; Han, L.; Zhao, L.; Chen, J.; Olopade, O.I.; He, M.; Wei, M. HIF-2α promotes conversion to a stem cell phenotype and induces chemoresistance in breast cancer cells by activating Wnt and Notch pathways. J. Exp. Clin. Cancer Res. 2018, 37, 256. [Google Scholar] [CrossRef]
- Rascio, F.; Spadaccino, F.; Rocchetti, M.T.; Castellano, G.; Stallone, G.; Netti, G.S.; Ranieri, E. The Pathogenic Role of PI3K/AKT Pathway in Cancer Onset and Drug Resistance: An Updated Review. Cancers 2021, 13, 3949. [Google Scholar] [CrossRef]
- Stemke-Hale, K.; Gonzalez-Angulo, A.M.; Lluch, A.; Neve, R.M.; Kuo, W.L.; Davies, M.; Carey, M.; Hu, Z.; Guan, Y.; Sahin, A.; et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008, 68, 6084–6091. [Google Scholar] [CrossRef]
- Guo, T.; Dong, X.; Xie, S.; Zhang, L.; Zeng, P.; Zhang, L. Cellular Mechanism of Gene Mutations and Potential Therapeutic Targets in Ovarian Cancer. Cancer Manag. Res. 2021, 13, 3081–3100. [Google Scholar] [CrossRef]
- Li, H.; Prever, L.; Hirsch, E.; Gulluni, F. Targeting PI3K/AKT/mTOR Signaling Pathway in Breast Cancer. Cancers (Basel). 2021, 13, 3517. [Google Scholar] [CrossRef]
- Kinross, K.M.; Montgomery, K.G.; Kleinschmidt, M.; Waring, P.; Ivetac, I.; Tikoo, A.; Saad, M.; Hare, L.; Roh, V.; Mantamadiotis, T.; et al. An activating Pik3ca mutation coupled with Pten loss is sufficient to initiate ovarian tumorigenesis in mice. J. Clin. Invest. 2012, 122, 553–557. [Google Scholar] [CrossRef]
- du Rusquec, P.; Blonz, C.; Frenel, J.S.; Campone, M. Targeting the PI3K/Akt/mTOR pathway in estrogen-receptor positive HER2 negative advanced breast cancer. Ther. Adv. Med. Oncol. 2020, 12, 1758835920940939. [Google Scholar] [CrossRef]
- Li, H.X.; Zeng, J.F.; Shen, K. PI3K/AKT/mTOR signaling pathway as a therapeutic target for ovarian cancer. Arch. Gynecol. Obstet. 2014, 290, 1067–1078. [Google Scholar] [CrossRef]
- Miricescu, D.; Totan, A.; Stanescu-Spinu, I.I.; Badoiu, S.C.; Stefani, C.; Greabu, M. PI3K/AKT/mTOR Signaling Pathway in Breast Cancer: From Molecular Landscape to Clinical Aspects. Int. J. Mol. Sci. 2021, 22, 173. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Bai, X.; Feng, X.; Ni, J.; Beretov, J.; Graham, P.; Li, Y. Inhibition of PI3K/Akt/mTOR signaling pathway alleviates ovarian cancer chemoresistance through reversing epithelial-mesenchymal transition and decreasing cancer stem cell marker expression. BMC Cancer 2019, 19, 618. [Google Scholar] [CrossRef] [PubMed]
- Shayesteh, L.; Lu, Y.; Kuo, W.L.; Baldocchi, R.; Godfrey, T.; Collins, C.; Pinkel, D.; Powell, B.; Mills, G.B.; Gray, J.W. PlK3CA is implicated as an oncogene in ovarian cancer. Nat. Genet. 1999, 21, 99–102. [Google Scholar] [CrossRef]
- Madsen, R.R.; Erickson, E.C.; Rueda, O.M.; Robin, X.; Caldas, C.; Toker, A.; Semple, R.K.; Vanhaesebroeck, B. Positive correlation between transcriptomic stemness and PI3K/AKT/mTOR signaling scores in breast cancer, and a counterintuitive relationship with PIK3CA genotype. PLoS Genet. 2021, 17, e1009876. [Google Scholar] [CrossRef]
- Zhou, J.; Wulfkuhle, J.; Zhang, H.; Gu, P.; Yang, Y.; Deng, J.; Margolick, J.B.; Liotta, L.A.; Petricoin, E.; Zhang, Y. Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc. Natl. Acad. Sci. USA 2007, 104, 16158–16163. [Google Scholar] [CrossRef]
- Russo, A.; Colina, J.A.; Moy, J.; Baligod, S.; Czarnecki, A.A.; Varughese, P.; Lantvit, D.D.; Dean, M.J.; Burdette, J.E. Silencing PTEN in the fallopian tube promotes enrichment of cancer stem cell-like function through loss of PAX2. Cell Death Dis. 2021, 12, 375. [Google Scholar] [CrossRef]
- Rivas, S.; Gómez-Oro, C.; Antón, I.M.; Wandosell, F. Role of Akt isoforms controlling cancer stem cell survival, phenotype and self-renewal. Biomedicines 2018, 6, 29. [Google Scholar] [CrossRef] [PubMed]
- Douville, J.; Beaulieu, R.; Balicki, D. ALDH1 as a Functional Marker of Cancer Stem and Progenitor Cells. Stem Cells Dev. 2009, 18, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Park, H.J.; Park, C.S.; Oh, E.T.; Choi, B.H.; Williams, B.; Lee, C.K.; Song, C.W. Response of breast cancer cells and cancer stem cells to metformin and hyperthermia alone or combined. PLoS ONE 2014, 9, e87979. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhang, P.; Wang, H.; Hou, D.; Li, W.; Xiao, G.; Li, C. Inhibitory effects of metformin at low concentration on epithelial-mesenchymal transition of CD44(+)CD117(+) ovarian cancer stem cells. Stem Cell Res. Ther. 2015, 6, 262. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Luo, X.; Wu, N.; Liao, Q.; Wang, J. SRC-3/TRAF4 facilitates ovarian cancer development by activating the PI3K/AKT signaling pathway. Med. Oncol. 2023, 40, 76. [Google Scholar] [CrossRef]
- Gu, Y.; Gao, H.; Zhang, H.; John, A.; Zhu, X.; Shivaram, S.; Yu, J.; Weinshilboum, R.M.; Wang, L. TRAF4 hyperactivates HER2 signaling and contributes to Trastuzumab resistance in HER2-positive breast cancer. Oncogene 2022, 41, 4119–4129. [Google Scholar] [CrossRef]
- Li, L.; Deng, C.-X.; Chen, Q. SRC-3, a Steroid Receptor Coactivator: Implication in Cancer. Int. J. Mol. Sci. 2021, 22, 4760. [Google Scholar] [CrossRef]
- Jiang, N.; Hu, Y.; Wang, M.; Zhao, Z.; Li, M. The Notch Signaling Pathway Contributes to Angiogenesis and Tumor Immunity in Breast Cancer. Breast Cancer Targets Ther. 2022, 14, 291–309. [Google Scholar] [CrossRef]
- Lobry, C.; Oh, P.; Mansour, M.R.; Look, A.T.; Aifantis, I. Notch signaling: Switching an oncogene to a tumor suppressor. Blood 2014, 123, 2451–2459. [Google Scholar] [CrossRef]
- Hopfer, O.; Zwahlen, D.; Fey, M.F.; Aebi, S. The Notch pathway in ovarian carcinomas and adenomas. Br. J. Cancer 2005, 93, 709–718. [Google Scholar] [CrossRef]
- Giuli, M.V.; Mancusi, A.; Giuliani, E.; Screpanti, I.; Checquolo, S. Notch signaling in female cancers: A multifaceted node to overcome drug resistance. Cancer drug Resist. 2021, 4, 805–836. [Google Scholar] [CrossRef]
- Liu, Z.; Zhu, Y.; Li, F.; Xie, Y. GATA1-regulated JAG1 promotes ovarian cancer progression by activating Notch signal pathway. Protoplasma 2020, 257, 901–910. [Google Scholar] [CrossRef]
- Kim, M.J.; Kim, A.-R.; Jeong, J.-Y.; Kim, K.; Kim, T.-H.; Lee, C.; Chung, K.; Ko, Y.-H.; An, H.-J. Correlation of ALDH1 and Notch3 Expression: Clinical implication in Ovarian Carcinomas. J. Cancer 2017, 8, 3331–3342. [Google Scholar] [CrossRef] [PubMed]
- Edwards, A.; Brennan, K. Notch Signalling in Breast Development and Cancer. Front. cell Dev. Biol. 2021, 9, 692173. [Google Scholar] [CrossRef] [PubMed]
- Ibragimova, M.; Tsyganov, M.; Litviakov, N. Tumour Stem Cells in Breast Cancer. Int. J. Mol. Sci. 2022, 23, 5058. [Google Scholar] [CrossRef]
- Seo, E.J.; Kim, D.K.; Jang, I.H.; Choi, E.J.; Shin, S.H.; Lee, S.I.; Kwon, S.-M.; Kim, K.-H.; Suh, D.-S.; Kim, J.H. Hypoxia-NOTCH1-SOX2 signaling is important for maintaining cancer stem cells in ovarian cancer. Oncotarget 2016, 7, 55624–55638. [Google Scholar] [CrossRef]
- Weijzen, S.; Rizzo, P.; Braid, M.; Vaishnav, R.; Jonkheer, S.M.; Zlobin, A.; Osborne, B.A.; Gottipati, S.; Aster, J.C.; Hahn, W.C.; et al. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat. Med. 2002, 8, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Nigam, A. Breast Cancer Stem Cells, Pathways and Therapeutic Perspectives 2011. Indian J. Surg. 2013, 75, 170–180. [Google Scholar] [CrossRef][Green Version]
- McGowan, P.M.; Simedrea, C.; Ribot, E.J.; Foster, P.J.; Palmieri, D.; Steeg, P.S.; Allan, A.L.; Chambers, A.F. Notch1 inhibition alters the CD44 hi/CD24 lo population and reduces the formation of brain metastases from breast cancer. Mol. Cancer Res. 2011, 9, 834–844. [Google Scholar] [CrossRef]
- Qiu, M.; Peng, Q.; Jiang, I.; Carroll, C.; Han, G.; Rymer, I.; Lippincott, J.; Zachwieja, J.; Gajiwala, K.; Kraynov, E.; et al. Specific inhibition of Notch1 signaling enhances the antitumor efficacy of chemotherapy in triple negative breast cancer through reduction of cancer stem cells. Cancer Lett. 2013, 328, 261–270. [Google Scholar] [CrossRef]
- Ma, H.; Tian, T.; Cui, Z. Targeting ovarian cancer stem cells: A new way out. Stem Cell Res. Ther. 2023, 14, 28. [Google Scholar] [CrossRef]
- Grudzien, P.; Lo, S.; Albain, K.S.; Robinson, P.; Rajan, P.; Strack, P.R.; Golde, T.E.; Miele, L.; Foreman, K.E. Inhibition of notch signaling reduces the stem-like population of breast cancer cells and prevents mammosphere formation. Anticancer Res. 2010, 30, 3853–3867. [Google Scholar] [CrossRef]
- Muñoz-Galván, S.; Carnero, A. Targeting Cancer Stem Cells to Overcome Therapy Resistance in Ovarian Cancer. Cells 2020, 9, 1402. [Google Scholar] [CrossRef] [PubMed]
- Varjosalo, M.; Taipale, J. Hedgehog: Functions and mechanisms. Genes Dev. 2008, 22, 2454–2472. [Google Scholar] [CrossRef] [PubMed]
- Cochrane, C.R.; Szczepny, A.; Watkins, D.N.; Cain, J.E. Hedgehog Signaling in the Maintenance of Cancer Stem Cells. Cancers 2015, 7, 1554–1585. [Google Scholar] [CrossRef]
- Sari, I.N.; Phi, L.T.H.; Jun, N.; Wijaya, Y.T.; Lee, S.; Kwon, H.Y. Hedgehog Signaling in Cancer: A Prospective Therapeutic Target for Eradicating Cancer Stem Cells. Cells 2018, 7, 208. [Google Scholar] [CrossRef]
- Pietrobono, S.; Gagliardi, S.; Stecca, B. Non-canonical Hedgehog Signaling Pathway in Cancer: Activation of GLI Transcription Factors Beyond Smoothened. Front. Genet. 2019, 10, 556. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Siu, M.K.Y.; Au, C.W.H.; Wong, E.S.Y.; Chan, H.Y.; Ip, P.P.C.; Ngan, H.Y.S.; Cheung, A.N.Y. Aberrant activation of hedgehog signaling pathway in ovarian cancers: Effect on prognosis, cell invasion and differentiation. Carcinogenesis 2009, 30, 131–140. [Google Scholar] [CrossRef]
- Clement, V.; Sanchez, P.; de Tribolet, N.; Radovanovic, I.; Ruiz i Altaba, A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr. Biol. 2007, 17, 165–172. [Google Scholar] [CrossRef]
- Zhao, H.; Li, N.; Pang, Y.; Zhao, J.; Wu, X. Gli affects the stemness and prognosis of epithelial ovarian cancer via homeobox protein NANOG. Mol. Med. Rep. 2021, 23, 128. [Google Scholar] [CrossRef]
- Ray, A.; Meng, E.; Reed, E.; Shevde, L.A.; Rocconi, R.P. Hedgehog signaling pathway regulates the growth of ovarian cancer spheroid forming cells. Int. J. Oncol. 2011, 39, 797–804. [Google Scholar] [CrossRef]
- Liu, S.; Dontu, G.; Mantle, I.D.; Patel, S.; Ahn, N.S.; Jackson, K.W.; Suri, P.; Wicha, M.S. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006, 66, 6063–6071. [Google Scholar] [CrossRef]
- He, M.; Fu, Y.; Yan, Y.; Xiao, Q.; Wu, H.; Yao, W.; Zhaov, H.; Zhao, L.; Jiang, Q.; Yu, Z.; et al. The Hedgehog signalling pathway mediates drug response of MCF-7 mammosphere cells in breast cancer patients. Clin. Sci. 2015, 129, 809–822. [Google Scholar] [CrossRef] [PubMed]
- Riaz, S.K.; Khan, J.S.; Shah, S.T.A.; Wang, F.; Ye, L.; Jiang, W.G.; Malik, M.F.A. Involvement of hedgehog pathway in early onset, aggressive molecular subtypes and metastatic potential of breast cancer. Cell Commun. Signal. 2018, 16, 3. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Qu, Y.; Jin, Y.; Yu, Y.; Deng, N.; Wawrowsky, K.; Zhang, X.; Li, N.; Bose, S.; Wang, Q.; et al. FOXC1 Activates Smoothened-Independent Hedgehog Signaling in Basal-like Breast Cancer. Cell Rep. 2015, 13, 1046–1058. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Gao, G.; Luo, S. Hedgehog signaling pathway and ovarian cancer. Chin. J. Cancer Res. 2013, 25, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Xu, R.; Zeng, C.; Lu, Q.; Huang, D.; Shi, C.; Zhang, W.; Deng, L.; Yan, R.; Rao, H.; et al. Down-Regulation of Gli Transcription Factor Leads to the Inhibition of Migration and Invasion of Ovarian Cancer Cells via Integrin β4-Mediated FAK Signaling. PLoS ONE 2014, 9, e88386. [Google Scholar] [CrossRef]
- Bhateja, P.; Cherian, M.; Majumder, S.; Ramaswamy, B. The Hedgehog Signaling Pathway: A Viable Target in Breast Cancer? Cancers 2019, 11, 1126. [Google Scholar] [CrossRef] [PubMed]
- Levy, D.E.; Darnell, J.E. Stats: Transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 2002, 3, 651–662. [Google Scholar] [CrossRef]
- Chung, S.S.; Aroh, C.; Vadgama, J.V. Constitutive activation of STAT3 signaling regulates hTERT and promotes stem cell-like traits in human breast cancer cells. PLoS ONE 2013, 8, e83971. [Google Scholar] [CrossRef]
- Hu, X.; Li, J.; Fu, M.; Zhao, X.; Wang, W. The JAK/STAT signaling pathway: From bench to clinic. Signal Transduct. Target. Ther. 2021, 6, 402. [Google Scholar] [CrossRef]
- Abubaker, K.; Luwor, R.B.; Zhu, H.; McNally, O.; Quinn, M.A.; Burns, C.J.; Thompson, E.W.; Findlay, J.K.; Ahmed, N. Inhibition of the JAK2/STAT3 pathway in ovarian cancer results in the loss of cancer stem cell-like characteristics and a reduced tumor burden. BMC Cancer 2014, 14, 317. [Google Scholar] [CrossRef]
- Burgos-Ojeda, D.; Wu, R.; McLean, K.; Chen, Y.-C.; Talpaz, M.; Yoon, E.; Cho, K.R.; Buckanovich, R.J. CD24+ Ovarian Cancer Cells Are Enriched for Cancer-Initiating Cells and Dependent on JAK2 Signaling for Growth and Metastasis. Mol. Cancer Ther. 2015, 14, 1717–1727. [Google Scholar] [CrossRef]
- Ruan, Z.; Yang, X.; Cheng, W. OCT4 accelerates tumorigenesis through activating JAK/STAT signaling in ovarian cancer side population cells. Cancer Manag. Res. 2019, 11, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Xiang, T.; Zhao, Z.; Lin, K.; Yin, P.; Jiang, L.; Liang, Z.; Zhu, B. Autocrine interleukin-23 promotes self-renewal of CD133+ ovarian cancer stem-like cells. Oncotarget 2016, 7, 76006–76020. [Google Scholar] [CrossRef] [PubMed]
- To, S.Q.; Dmello, R.S.; Richards, A.K.; Ernst, M.; Chand, A.L. STAT3 Signaling in Breast Cancer: Multicellular Actions and Therapeutic Potential. Cancers 2022, 14, 429. [Google Scholar] [CrossRef]
- Wei, W.; Tweardy, D.J.; Zhang, M.; Zhang, X.; Landua, J.; Petrovic, I.; Bu, W.; Roarty, K.; Hilsenbeck, S.G.; Rosen, J.M.; et al. STAT3 Signaling Is Activated Preferentially in Tumor-Initiating Cells in Claudin-Low Models of Human Breast Cancer. Stem Cells 2014, 32, 2571–2582. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Hutzen, B.; Lee, H.F.; Peng, Z.; Wang, W.; Zhao, C.; Lin, H.J.; Sun, D.; Li, P.K.; Li, C.; et al. Evaluation of STAT3 signaling in ALDH+ and ALDH+/CD44+/CD24- subpopulations of breast cancer cells. PLoS ONE 2013, 8, e82821. [Google Scholar] [CrossRef]
- Weng, Y.-S.; Tseng, H.-Y.; Chen, Y.-A.; Shen, P.-C.; Al Haq, A.T.; Chen, L.-M.; Tung, Y.-C.; Hsu, H.-L. MCT-1/miR-34a/IL-6/IL-6R signaling axis promotes EMT progression, cancer stemness and M2 macrophage polarization in triple-negative breast cancer. Mol. Cancer 2019, 18, 42. [Google Scholar] [CrossRef]
- Deng, X.; Zhang, P.; Liang, T.; Deng, S.; Chen, X.; Zhu, L. Ovarian cancer stem cells induce the M2 polarization of macrophages through the PPARγ and NF-κB pathways. Int. J. Mol. Med. 2015, 36, 449–454. [Google Scholar] [CrossRef]
- Tzavlaki, K.; Moustakas, A. TGF-β Signaling. Biomolecules 2020, 10, 487. [Google Scholar] [CrossRef]
- Bruna, A.; Greenwood, W.; Le Quesne, J.; Teschendorff, A.; Miranda-Saavedra, D.; Rueda, O.M.; Sandoval, J.L.; Vidakovic, A.T.; Saadi, A.; Pharoah, P.; et al. TGFβ induces the formation of tumour-initiating cells in claudin low breast cancer. Nat. Commun. 2012, 3, 1055. [Google Scholar] [CrossRef]
- Rafehi, S.; Ramos Valdes, Y.; Bertrand, M.; McGee, J.; Préfontaine, M.; Sugimoto, A.; DiMattia, G.E.; Shepherd, T.G. TGFβ signaling regulates epithelial-mesenchymal plasticity in ovarian cancer ascites-derived spheroids. Endocr. Relat. Cancer 2016, 23, 147–159. [Google Scholar] [CrossRef]
- Asiedu, M.K.; Ingle, J.N.; Behrens, M.D.; Radisky, D.C.; Knutson, K.L. TGFβ/TNFα-Mediated Epithelial–Mesenchymal Transition Generates Breast Cancer Stem Cells with a Claudin-Low Phenotype. Cancer Res. 2011, 71, 4707–4719. [Google Scholar] [CrossRef] [PubMed]
- Bellomo, C.; Caja, L.; Moustakas, A. Transforming growth factor β as regulator of cancer stemness and metastasis. Br. J. Cancer 2016, 115, 761–769. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, L.; He, X.; Zhang, P.; Sun, C.; Xu, X.; Lu, Y.; Li, F. TGF-β plays a vital role in triple-negative breast cancer (TNBC) drug-resistance through regulating stemness, EMT and apoptosis. Biochem. Biophys. Res. Commun. 2018, 502, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Chihara, Y.; Shimoda, M.; Hori, A.; Ohara, A.; Naoi, Y.; Ikeda, J.; Kagara, N.; Tanei, T.; Shimomura, A.; Shimazu, K.; et al. A small-molecule inhibitor of SMAD3 attenuates resistance to anti-HER2 drugs in HER2-positive breast cancer cells. Breast Cancer Res. Treat. 2017, 166, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Shao, M.; Schilder, J.; Guise, T.; Mohammad, K.S.; Matei, D. Tissue transglutaminase links TGF-β, epithelial to mesenchymal transition and a stem cell phenotype in ovarian cancer. Oncogene 2012, 31, 2521–2534. [Google Scholar] [CrossRef]
- Wang, C.-W.; Lee, B.-H.; Tai, C.-J. The inhibition of cordycepin on cancer stemness in TGF-beta induced chemo-resistant ovarian cancer cell. Oncotarget 2017, 8, 111912–111921. [Google Scholar] [CrossRef]
- Bhola, N.E.; Balko, J.M.; Dugger, T.C.; Kuba, M.G.; Sánchez, V.; Sanders, M.; Stanford, J.; Cook, R.S.; Arteaga, C.L. TGF-β inhibition enhances chemotherapy action against triple-negative breast cancer. J. Clin. Investig. 2013, 123, 1348–1358. [Google Scholar] [CrossRef]
- Guido, C.; Whitaker-Menezes, D.; Capparelli, C.; Balliet, R.; Lin, Z.; Pestell, R.G.; Howell, A.; Aquila, S.; Andò, S.; Martinez-Outschoorn, U.; et al. Metabolic reprogramming of cancer-associated fibroblasts by TGF-β drives tumor growth: Connecting TGF-β signaling with “Warburg-like” cancer metabolism and L-lactate production. Cell Cycle 2012, 11, 3019–3035. [Google Scholar] [CrossRef]
- Yeung, T.-L.; Leung, C.S.; Wong, K.-K.; Samimi, G.; Thompson, M.S.; Liu, J.; Zaid, T.M.; Ghosh, S.; Birrer, M.J.; Mok, S.C. TGF-β modulates ovarian cancer invasion by upregulating CAF-derived versican in the tumor microenvironment. Cancer Res. 2013, 73, 5016–5028. [Google Scholar] [CrossRef]
- Labelle, M.; Begum, S.; Hynes, R.O. Direct Signaling between Platelets and Cancer Cells Induces an Epithelial-Mesenchymal-Like Transition and Promotes Metastasis. Cancer Cell 2011, 20, 576–590. [Google Scholar] [CrossRef] [PubMed]
- Wen, H.; Qian, M.; He, J.; Li, M.; Yu, Q.; Leng, Z. Inhibiting of self-renewal, migration and invasion of ovarian cancer stem cells by blocking TGF-β pathway. PLoS ONE 2020, 15, e0230230. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Santillan, K.; Melendez-Zajgla, J.; Jimenez-Hernandez, L.E.; Gaytan-Cervantes, J.; Munõz-Galindo, L.; Pinã-Sanchez, P.; Martinez-Ruiz, G.; Torres, J.; Garcia-Lopez, P.; Gonzalez-Torres, C.; et al. NF-kappaΒ-inducing kinase regulates stem cell phenotype in breast cancer. Sci. Reports 2016, 6, 37340. [Google Scholar] [CrossRef]
- Sarkar, F.H.; Li, Y.; Wang, Z.; Kong, D. NF-kappaB signaling pathway and its therapeutic implications in human diseases. Int. Rev. Immunol. 2008, 27, 293–319. [Google Scholar] [CrossRef]
- Rinkenbaugh, A.L.; Baldwin, A.S. The NF-κB Pathway and Cancer Stem Cells. Cells 2016, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Taguchi, Y.; Ito-Kureha, T.; Semba, K.; Yamaguchi, N.; Inoue, J.I. NF-κB non-cell-autonomously regulates cancer stem cell populations in the basal-like breast cancer subtype. Nat. Commun. 2013, 4, 2299. [Google Scholar] [CrossRef]
- Smith, S.M.; Lyu, Y.L.; Cai, L. NF-κB affects proliferation and invasiveness of breast cancer cells by regulating CD44 expression. PLoS ONE 2014, 9, e106966. [Google Scholar] [CrossRef]
- Alvero, A.B.; Chen, R.; Fu, H.-H.; Montagna, M.; Schwartz, P.E.; Rutherford, T.; Silasi, D.-A.; Steffensen, K.D.; Waldstrom, M.; Visintin, I.; et al. Molecular phenotyping of human ovarian cancer stem cells unravel the mechanisms for repair and chemo-resistance. Cell Cycle 2009, 8, 158–166. [Google Scholar] [CrossRef]
- House, C.D.; Jordan, E.; Hernandez, L.; Ozaki, M.; James, J.M.; Kim, M.; Kruhlak, M.J.; Batchelor, E.; Elloumi, F.; Cam, M.C.; et al. NFκB Promotes Ovarian Tumorigenesis via Classical Pathways That Support Proliferative Cancer Cells and Alternative Pathways That Support ALDH+ Cancer Stem-like Cells. Cancer Res. 2017, 77, 6927–6940. [Google Scholar] [CrossRef]
- Gonzalez-Torres, C.; Gaytan-Cervantes, J.; Vazquez-Santillan, K.; Mandujano-Tinoco, E.A.; Ceballos-Cancino, G.; Garcia-Venzor, A.; Zampedri, C.; Sanchez-Maldonado, P.; Mojica-Espinosa, R.; Jimenez-Hernandez, L.E.; et al. NF-κB Participates in the Stem Cell Phenotype of Ovarian Cancer Cells. Arch. Med. Res. 2017, 48, 343–351. [Google Scholar] [CrossRef]
- Alvero, A.B.; Fu, H.-H.; Holmberg, J.; Visintin, I.; Mor, L.; Marquina, C.C.; Oidtman, J.; Silasi, D.-A.; Mor, G. Stem-like ovarian cancer cells can serve as tumor vascular progenitors. Stem Cells 2009, 27, 2405–2413. [Google Scholar] [CrossRef] [PubMed]
- Hallis, S.P.; Kim, S.K.; Lee, J.-H.; Kwak, M.-K. Association of NRF2 with HIF-2α-induced cancer stem cell phenotypes in chronic hypoxic condition. Redox Biol. 2023, 60, 102632. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhang, H.; Gu, P.; Bai, J.; Margolick, J.B.; Zhang, Y. NF-κB pathway inhibitors preferentially inhibit breast cancer stem-like cells. Breast Cancer Res. Treat. 2008, 111, 419–427. [Google Scholar] [CrossRef] [PubMed]
- Calses, P.C.; Crawford, J.J.; Lill, J.R.; Dey, A. Hippo Pathway in Cancer: Aberrant Regulation and Therapeutic Opportunities. Trends in cancer 2019, 5, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Hu, Y.; Lan, T.; Guan, K.-L.; Luo, T.; Luo, M. The Hippo signalling pathway and its implications in human health and diseases. Signal Transduct. Target. Ther. 2022, 7, 376. [Google Scholar] [CrossRef]
- Hall, C.A.; Wang, R.; Miao, J.; Oliva, E.; Shen, X.; Wheeler, T.; Hilsenbeck, S.G.; Orsulic, S.; Goode, S. Hippo pathway effector Yap is an ovarian cancer oncogene. Cancer Res. 2010, 70, 8517–8525. [Google Scholar] [CrossRef]
- Muñoz-Galván, S.; Felipe-Abrio, B.; Verdugo-Sivianes, E.M.; Perez, M.; Jiménez-García, M.P.; Suarez-Martinez, E.; Estevez-Garcia, P.; Carnero, A. Downregulation of MYPT1 increases tumor resistance in ovarian cancer by targeting the Hippo pathway and increasing the stemness. Mol. Cancer 2020, 19, 7. [Google Scholar] [CrossRef]
- Quinn, H.M.; Vogel, R.; Popp, O.; Mertins, P.; Lan, L.; Messerschmidt, C.; Landshammer, A.; Lisek, K.; Château-Joubert, S.; Marangoni, E.; et al. YAP and β-Catenin Cooperate to Drive Oncogenesis in Basal Breast Cancer. Cancer Res. 2021, 81, 2116–2127. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Zhang, Y.-L.; Yu, C.; Chang, T.; Fan, H.-Y. YAP/TEAD co-activator regulated pluripotency and chemoresistance in ovarian cancer initiated cells. PLoS ONE 2014, 9, e109575. [Google Scholar] [CrossRef]
- Park, J.H.; Shin, J.E.; Park, H.W. The Role of Hippo Pathway in Cancer Stem Cell Biology. Mol. Cells 2018, 41, 83–92. [Google Scholar] [CrossRef]
- Tian, Q.; Gao, H.; Zhou, Y.; Zhu, L.; Yang, J.; Wang, B.; Liu, P.; Yang, J. RICH1 inhibits breast cancer stem cell traits through activating kinases cascade of Hippo signaling by competing with Merlin for binding to Amot-p80. Cell Death Dis. 2022, 13, 71. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Feng, J.; Gou, J.; Jia, J.; Yi, T.; Cui, T. Verteporfin, a suppressor of YAP–TEAD complex, presents promising antitumor properties on ovarian cancer. Onco. Targets. Ther. 2016, 9, 5371–5381. [Google Scholar] [CrossRef]
- Jaganjac, M.; Milkovic, L.; Sunjic, S.B.; Zarkovic, N. The NRF2, Thioredoxin, and Glutathione System in Tumorigenesis and Anticancer Therapies. Antioxidants 2020, 9, 1151. [Google Scholar] [CrossRef] [PubMed]
- Croker, A.K.; Allan, A.L. Inhibition of aldehyde dehydrogenase (ALDH) activity reduces chemotherapy and radiation resistance of stem-like ALDHhiCD44+ human breast cancer cells. Breast Cancer Res. Treat. 2012, 133, 75–87. [Google Scholar] [CrossRef]
- Ayub, T.H.; Keyver-Paik, M.-D.; Debald, M.; Rostamzadeh, B.; Thiesler, T.; Schröder, L.; Barchet, W.; Abramian, A.; Kaiser, C.; Kristiansen, G.; et al. Accumulation of ALDH1-positive cells after neoadjuvant chemotherapy predicts treatment resistance and prognosticates poor outcome in ovarian cancer. Oncotarget 2015, 6, 16437–16448. [Google Scholar] [CrossRef]
- Ryoo, I.-G.; Choi, B.-H.; Kwak, M.-K. Activation of NRF2 by p62 and proteasome reduction in sphere-forming breast carcinoma cells. Oncotarget 2015, 6, 8167–8184. [Google Scholar] [CrossRef]
- Kim, D.; Choi, B.-H.; Ryoo, I.-G.; Kwak, M.-K. High NRF2 level mediates cancer stem cell-like properties of aldehyde dehydrogenase (ALDH)-high ovarian cancer cells: Inhibitory role of all-trans retinoic acid in ALDH/NRF2 signaling. Cell Death Dis. 2018, 9, 896. [Google Scholar] [CrossRef]
- Ryoo, I.; Choi, B.; Ku, S.-K.; Kwak, M.-K. High CD44 expression mediates p62-associated NFE2L2/NRF2 activation in breast cancer stem cell-like cells: Implications for cancer stem cell resistance. Redox Biol. 2018, 17, 246–258. [Google Scholar] [CrossRef]
- Qin, S.; He, X.; Lin, H.; Schulte, B.A.; Zhao, M.; Tew, K.D.; Wang, G.Y. Nrf2 inhibition sensitizes breast cancer stem cells to ionizing radiation via suppressing DNA repair. Free Radic. Biol. Med. 2021, 169, 238–247. [Google Scholar] [CrossRef]
- Kamble, D.; Mahajan, M.; Dhat, R.; Sitasawad, S. Keap1-Nrf2 Pathway Regulates ALDH and Contributes to Radioresistance in Breast Cancer Stem Cells. Cells 2021, 10, 83. [Google Scholar] [CrossRef]
- Zhao, H.; Gao, Y.; Miao, J.; Chen, S.; Li, J.; Li, Z.; Yin, C.; Yue, W. Single-cell RNA-seq highlights a specific carcinoembryonic cluster in ovarian cancer. Cell Death Dis. 2021, 12, 1082. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.V.; Patel, K.S.; Teras, L.R. Excess body fatness and cancer risk: A summary of the epidemiologic evidence. Surg. Obes. Relat. Dis. 2023, 19, 742–745. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Dannenberg, A.J. The obese adipose tissue microenvironment in cancer development and progression. Nat. Rev. Endocrinol. 2019, 15, 139–154. [Google Scholar] [CrossRef] [PubMed]
- Massier, L.; Jalkanen, J.; Elmastas, M.; Zhong, J.; Wang, T.; Nono Nankam, P.A.; Frendo-Cumbo, S.; Bäckdahl, J.; Subramanian, N.; Sekine, T.; et al. An integrated single cell and spatial transcriptomic map of human white adipose tissue. Nat. Commun. 2023, 14, 1438. [Google Scholar] [CrossRef] [PubMed]
- Ham, S.; Choi, J.H.; Shin, S.G.; Lee, E.-J. High visceral fat-to-muscle ratio is an independent factor that predicts worse overall survival in patients with primary epithelial ovarian, fallopian tube, and peritoneal cancer. J. Ovarian Res. 2023, 16, 19. [Google Scholar] [CrossRef] [PubMed]
- Parafiniuk, K.; Skiba, W.; Pawłowska, A.; Suszczyk, D.; Maciejczyk, A.; Wertel, I. The Role of the Adipokine Resistin in the Pathogenesis and Progression of Epithelial Ovarian Cancer. Biomedicines 2022, 10, 920. [Google Scholar] [CrossRef]
- Brock, C.K.; Hebert, K.L.; Artiles, M.; Wright, M.K.; Cheng, T.; Windsor, G.O.; Nguyen, K.; Alzoubi, M.S.; Collins-Burow, B.M.; Martin, E.C.; et al. A Role for Adipocytes and Adipose Stem Cells in the Breast Tumor Microenvironment and Regenerative Medicine. Front. Physiol. 2021, 12, 751239. [Google Scholar] [CrossRef]
- Mukherjee, A.; Chiang, C.-Y.; Daifotis, H.A.; Nieman, K.M.; Fahrmann, J.F.; Lastra, R.R.; Romero, I.L.; Fiehn, O.; Lengyel, E. Adipocyte-Induced FABP4 Expression in Ovarian Cancer Cells Promotes Metastasis and Mediates Carboplatin Resistance. Cancer Res. 2020, 80, 1748–1761. [Google Scholar] [CrossRef]
- Nieman, K.M.; Kenny, H.A.; Penicka, C.V.; Ladanyi, A.; Buell-Gutbrod, R.; Zillhardt, M.R.; Romero, I.L.; Carey, M.S.; Mills, G.B.; Hotamisligil, G.S.; et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med. 2011, 17, 1498–1503. [Google Scholar] [CrossRef]
- Salimian Rizi, B.; Caneba, C.; Nowicka, A.; Nabiyar, A.W.; Liu, X.; Chen, K.; Klopp, A.; Nagrath, D. Nitric Oxide Mediates Metabolic Coupling of Omentum-Derived Adipose Stroma to Ovarian and Endometrial Cancer Cells. Cancer Res. 2015, 75, 456–471. [Google Scholar] [CrossRef]
- Bochet, L.; Lehuédé, C.; Dauvillier, S.; Wang, Y.Y.; Dirat, B.; Laurent, V.; Dray, C.; Guiet, R.; Maridonneau-Parini, I.; Le Gonidec, S.; et al. Adipocyte-Derived Fibroblasts Promote Tumor Progression and Contribute to the Desmoplastic Reaction in Breast Cancer. Cancer Res. 2013, 73, 5657–5668. [Google Scholar] [CrossRef]
- Ritter, A.; Kreis, N.-N.; Roth, S.; Friemel, A.; Safdar, B.K.; Hoock, S.C.; Wildner, J.M.; Allert, R.; Louwen, F.; Solbach, C.; et al. Cancer-educated mammary adipose tissue-derived stromal/stem cells in obesity and breast cancer: Spatial regulation and function. J. Exp. Clin. Cancer Res. 2023, 42, 35. [Google Scholar] [CrossRef] [PubMed]
- Ritter, A.; Kreis, N.-N.; Hoock, S.C.; Solbach, C.; Louwen, F.; Yuan, J. Adipose Tissue-Derived Mesenchymal Stromal/Stem Cells, Obesity and the Tumor Microenvironment of Breast Cancer. Cancers 2022, 14, 3908. [Google Scholar] [CrossRef] [PubMed]
- Iyoshi, S.; Yoshihara, M.; Nakamura, K.; Sugiyama, M.; Koya, Y.; Kitami, K.; Uno, K.; Mogi, K.; Tano, S.; Tomita, H.; et al. Pro-tumoral behavior of omental adipocyte-derived fibroblasts in tumor microenvironment at the metastatic site of ovarian cancer. Int. J. Cancer 2021, 149, 1961–1972. [Google Scholar] [CrossRef]
- Ladanyi, A.; Mukherjee, A.; Kenny, H.A.; Johnson, A.; Mitra, A.K.; Sundaresan, S.; Nieman, K.M.; Pascual, G.; Benitah, S.A.; Montag, A.; et al. Adipocyte-induced CD36 expression drives ovarian cancer progression and metastasis. Oncogene 2018, 37, 2285–2301. [Google Scholar] [CrossRef] [PubMed]
- John, B.; Naczki, C.; Patel, C.; Ghoneum, A.; Qasem, S.; Salih, Z.; Said, N. Regulation of the bi-directional cross-talk between ovarian cancer cells and adipocytes by SPARC. Oncogene 2019, 38, 4366–4383. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, G.; Huo, X.; Wang, Y.; Tigyi, G.; Zhu, B.-M.; Yue, J.; Zhang, W. Adipose-Derived Stem Cells Facilitate Ovarian Tumor Growth and Metastasis by Promoting Epithelial to Mesenchymal Transition Through Activating the TGF-β Pathway. Front. Oncol. 2021, 11, 756011. [Google Scholar] [CrossRef]
- Nowicka, A.; Marini, F.C.; Solley, T.N.; Elizondo, P.B.; Zhang, Y.; Sharp, H.J.; Broaddus, R.; Kolonin, M.; Mok, S.C.; Thompson, M.S.; et al. Human omental-derived adipose stem cells increase ovarian cancer proliferation, migration, and chemoresistance. PLoS ONE 2013, 8, e81859. [Google Scholar] [CrossRef]
- Wei, H.-J.; Zeng, R.; Lu, J.-H.; Lai, W.-F.T.; Chen, W.-H.; Liu, H.-Y.; Chang, Y.-T.; Deng, W.-P. Adipose-derived stem cells promote tumor initiation and accelerate tumor growth by interleukin-6 production. Oncotarget 2015, 6, 7713–7726. [Google Scholar] [CrossRef]
- Cho, J.A.; Park, H.; Lim, E.H.; Kim, K.H.; Choi, J.S.; Lee, J.H.; Shin, J.W.; Lee, K.W. Exosomes from ovarian cancer cells induce adipose tissue-derived mesenchymal stem cells to acquire the physical and functional characteristics of tumor-supporting myofibroblasts. Gynecol. Oncol. 2011, 123, 379–386. [Google Scholar] [CrossRef]
- Cho, J.A.; Park, H.; Lim, E.H.; Lee, K.W. Exosomes from breast cancer cells can convert adipose tissue-derived mesenchymal stem cells into myofibroblast-like cells. Int. J. Oncol. 2012, 40, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Divella, R.; De Luca, R.; Abbate, I.; Naglieri, E.; Daniele, A. Obesity and cancer: The role of adipose tissue and adipo-cytokines-induced chronic inflammation. J. Cancer 2016, 7, 2346–2359. [Google Scholar] [CrossRef] [PubMed]
- Rausch, L.K.; Netzer, N.C.; Hoegel, J.; Pramsohler, S. The Linkage between Breast Cancer, Hypoxia, and Adipose Tissue. Front. Oncol. 2017, 7, 211. [Google Scholar] [CrossRef] [PubMed]
- Bachelot, T.; Ray-Coquard, I.; Menetrier-Caux, C.; Rastkha, M.; Duc, A.; Blay, J.-Y. Prognostic value of serum levels of interleukin 6 and of serum and plasma levels of vascular endothelial growth factor in hormone-refractory metastatic breast cancer patients. Br. J. Cancer 2003, 88, 1721–1726. [Google Scholar] [CrossRef]
- Marotta, L.L.C.; Almendro, V.; Marusyk, A.; Shipitsin, M.; Schemme, J.; Walker, S.R.; Bloushtain-Qimron, N.; Kim, J.J.; Choudhury, S.A.; Maruyama, R.; et al. The JAK2/STAT3 signaling pathway is required for growth of CD44+CD24– stem cell–like breast cancer cells in human tumors. J. Clin. Investig. 2011, 121, 2723–2735. [Google Scholar] [CrossRef]
- Liu, S.; Lee, J.S.; Jie, C.; Park, M.H.; Iwakura, Y.; Patel, Y.; Soni, M.; Reisman, D.; Chen, H. HER2 Overexpression Triggers an IL1α Proinflammatory Circuit to Drive Tumorigenesis and Promote Chemotherapy Resistance. Cancer Res. 2018, 78, 2040–2051. [Google Scholar] [CrossRef]
- Browning, L.; Patel, M.; Bring Horvath, E.; Tawara, K.; Jorcyk, C.L. IL-6 and ovarian cancer: Inflammatory cytokines in promotion of metastasis. Cancer Manag. Res. 2018, 10, 6685–6693. [Google Scholar] [CrossRef]
- Sansone, P.; Storci, G.; Tavolari, S.; Guarnieri, T.; Giovannini, C.; Taffurelli, M.; Ceccarelli, C.; Santini, D.; Paterini, P.; Marcu, K.B.; et al. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. J. Clin. Investig. 2007, 117, 3988–4002. [Google Scholar] [CrossRef]
- Zhao, P.; Sun, J.; Huang, X.; Zhang, X.; Liu, X.; Liu, R.; Du, G.; Gan, W.; Yang, C.; Tang, Y.; et al. Targeting the KLF5-EphA2 axis can restrain cancer stemness and overcome chemoresistance in basal-like breast cancer. Int. J. Biol. Sci. 2023, 19, 1861–1874. [Google Scholar] [CrossRef]
- Nickel, A.; Blücher, C.; Al Kadri, O.; Schwagarus, N.; Müller, S.; Schaab, M.; Thiery, J.; Burkhardt, R.; Stadler, S.C. Adipocytes induce distinct gene expression profiles in mammary tumor cells and enhance inflammatory signaling in invasive breast cancer cells. Sci. Rep. 2018, 8, 9482. [Google Scholar] [CrossRef]
- Muthukumaran, N.; Miletti-González, K.E.; Ravindranath, A.K.; Rodríguez-Rodríguez, L. Tumor Necrosis Factor-α Differentially Modulates CD44 Expression in Ovarian Cancer Cells. Mol. Cancer Res. 2006, 4, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Kulbe, H.; Thompson, R.; Wilson, J.L.; Robinson, S.; Hagemann, T.; Fatah, R.; Gould, D.; Ayhan, A.; Balkwill, F. The Inflammatory Cytokine Tumor Necrosis Factor-α Generates an Autocrine Tumor-Promoting Network in Epithelial Ovarian Cancer Cells. Cancer Res. 2007, 67, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Rickard, B.P.; Conrad, C.; Sorrin, A.J.; Ruhi, M.K.; Reader, J.C.; Huang, S.A.; Franco, W.; Scarcelli, G.; Polacheck, W.J.; Roque, D.M.; et al. Malignant Ascites in Ovarian Cancer: Cellular, Acellular, and Biophysical Determinants of Molecular Characteristics and Therapy Response. Cancers 2021, 13, 4318. [Google Scholar] [CrossRef] [PubMed]
- Salemme, V.; Centonze, G.; Cavallo, F.; Defilippi, P.; Conti, L. The Crosstalk Between Tumor Cells and the Immune Microenvironment in Breast Cancer: Implications for Immunotherapy. Front. Oncol. 2021, 11, 610303. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Tsang, J.Y.; Tse, G.M. Tumor Microenvironment in Breast Cancer-Updates on Therapeutic Implications and Pathologic Assessment. Cancers 2021, 13, 4233. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Dean, D.C.; Hornicek, F.J.; Shi, H.; Duan, Z. Exosomes promote pre-metastatic niche formation in ovarian cancer. Mol. Cancer 2019, 18, 124. [Google Scholar] [CrossRef] [PubMed]
- González-Callejo, P.; Gener, P.; Díaz-Riascos, Z.V.; Conti, S.; Cámara-Sánchez, P.; Riera, R.; Mancilla, S.; García-Gabilondo, M.; Peg, V.; Arango, D.; et al. Extracellular vesicles secreted by triple-negative breast cancer stem cells trigger premetastatic niche remodeling and metastatic growth in the lungs. Int. J. Cancer 2023, 152, 2153–2165. [Google Scholar] [CrossRef]
- Kogure, A.; Yoshioka, Y.; Ochiya, T. Extracellular vesicles in cancer metastasis: Potential as therapeutic targets and materials. Int. J. Mol. Sci. 2020, 21, 4463. [Google Scholar] [CrossRef]
- Bobrie, A.; Krumeich, S.; Reyal, F.; Recchi, C.; Moita, L.F.; Seabra, M.C.; Ostrowski, M.; Théry, C. Rab27a Supports Exosome-Dependent and -Independent Mechanisms That Modify the Tumor Microenvironment and Can Promote Tumor Progression. Cancer Res. 2012, 72, 4920–4930. [Google Scholar] [CrossRef]
- Fico, F.; Santamaria-Martínez, A. The Tumor Microenvironment as a Driving Force of Breast Cancer Stem Cell Plasticity. Cancers 2020, 12, 3863. [Google Scholar] [CrossRef]
- Radharani, N.N.V.; Yadav, A.S.; Nimma, R.; Kumar, T.V.S.; Bulbule, A.; Chanukuppa, V.; Kumar, D.; Patnaik, S.; Rapole, S.; Kundu, G.C. Tumor-associated macrophage derived IL-6 enriches cancer stem cell population and promotes breast tumor progression via Stat-3 pathway. Cancer Cell Int. 2022, 22, 122. [Google Scholar] [CrossRef]
- El-Arabey, A.A.; Alkhalil, S.S.; Al-Shouli, S.T.; Awadalla, M.E.; Alhamdi, H.W.; Almanaa, T.N.; Mohamed, S.S.E.M.; Abdalla, M. Revisiting macrophages in ovarian cancer microenvironment: Development, function and interaction. Med. Oncol. 2023, 40, 142. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.R.; Provenzano, E.; Dawson, S.J.; Blows, F.M.; Liu, B.; Shah, M.; Earl, H.M.; Poole, C.J.; Hiller, L.; Dunn, J.A.; et al. Association between CD8+ T-cell infiltration and breast cancer survival in 12 439 patients. Ann. Oncol. 2014, 25, 1536–1543. [Google Scholar] [CrossRef] [PubMed]
- Shipitsin, M.; Campbell, L.L.; Argani, P.; Weremowicz, S.; Bloushtain-Qimron, N.; Yao, J.; Nikolskaya, T.; Serebryiskaya, T.; Beroukhim, R.; Hu, M.; et al. Molecular Definition of Breast Tumor Heterogeneity. Cancer Cell 2007, 11, 259–273. [Google Scholar] [CrossRef] [PubMed]
- Wilczyński, J.R.; Wilczyński, M.; Paradowska, E. Cancer Stem Cells in Ovarian Cancer—A Source of Tumor Success and a Challenging Target for Novel Therapies. Int. J. Mol. Sci. 2022, 23, 2496. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Chen, M.; Wu, P.; Chen, C.; Xu, Z.P.; Gu, W. Increased PD-L1 expression in breast and colon cancer stem cells. Clin. Exp. Pharmacol. Physiol. 2017, 44, 602–604. [Google Scholar] [CrossRef]
- Matsuzaki, J.; Gnjatic, S.; Mhawech-Fauceglia, P.; Beck, A.; Miller, A.; Tsuji, T.; Eppolito, C.; Qian, F.; Lele, S.; Shrikant, P.; et al. Tumor-infiltrating NY-ESO-1–specific CD8 + T cells are negatively regulated by LAG-3 and PD-1 in human ovarian cancer. Proc. Natl. Acad. Sci. USA 2010, 107, 7875–7880. [Google Scholar] [CrossRef]
- Castagnoli, L.; Cancila, V.; Cordoba-Romero, S.L.; Faraci, S.; Talarico, G.; Belmonte, B.; Iorio, M.V.; Milani, M.; Volpari, T.; Chiodoni, C.; et al. WNT signaling modulates PD-L1 expression in the stem cell compartment of triple-negative breast cancer. Oncogene 2019, 38, 4047–4060. [Google Scholar] [CrossRef]
- Stein, R.G.; Ebert, S.; Schlahsa, L.; Scholz, C.J.; Braun, M.; Hauck, P.; Horn, E.; Monoranu, C.M.; Thiemann, V.J.; Wustrow, M.P.; et al. Cognate nonlytic interactions between CD8+ T cells and breast cancer cells induce cancer stem cell-like properties. Cancer Res. 2019, 79, 1507–1519. [Google Scholar] [CrossRef]
- Sato, E.; Olson, S.H.; Ahn, J.; Bundy, B.; Nishikawa, H.; Qian, F.; Jungbluth, A.A.; Frosina, D.; Gnjatic, S.; Ambrosone, C.; et al. Intraepithelial CD8 + tumor-infiltrating lymphocytes and a high CD8 +/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc. Natl. Acad. Sci. USA 2005, 102, 18538–18543. [Google Scholar] [CrossRef]
- Xu, Y.; Dong, X.; Qi, P.; Ye, Y.; Shen, W.; Leng, L.; Wang, L.; Li, X.; Luo, X.; Chen, Y.; et al. Sox2 communicates with Tregs through CCL1 to promote the stemness property of breast cancer cells. Stem Cells 2017, 35, 2351. [Google Scholar] [CrossRef]
- Loh, J.J.; Ma, S. The Role of Cancer-Associated Fibroblast as a Dynamic Player in Mediating Cancer Stemness in the Tumor Microenvironment. Front. Cell Dev. Biol. 2021, 9, 727640. [Google Scholar] [CrossRef] [PubMed]
- Valenti, G.; Quinn, H.M.; Heynen, G.J.J.E.; Lan, L.; Holland, J.D.; Vogel, R.; Wulf-Goldenberg, A.; Birchmeier, W. Cancer stem cells regulate cancer-associated fibroblasts via activation of hedgehog signaling in mammary gland tumors. Cancer Res. 2017, 77, 2134–2147. [Google Scholar] [CrossRef] [PubMed]
- Elwakeel, E.; Weigert, A. Breast Cancer CAFs: Spectrum of Phenotypes and Promising Targeting Avenues. Int. J. Mol. Sci. 2021, 22, 11636. [Google Scholar] [CrossRef] [PubMed]
- Ji, Z.; Tian, W.; Gao, W.; Zang, R.; Wang, H.; Yang, G. Cancer-Associated Fibroblast-Derived Interleukin-8 Promotes Ovarian Cancer Cell Stemness and Malignancy Through the Notch3-Mediated Signaling. Front. Cell Dev. Biol. 2021, 9, 684505. [Google Scholar] [CrossRef]
- Kalluri, R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer 2016, 16, 582–598. [Google Scholar] [CrossRef]
- Fang, Y.; El Aziz, M.A.A.; Tiwari, K.; Mitra, A.K. Abstract 4685: Cancer associated fibroblasts promote ovarian cancer chemoresistance by inducing cancer stem cells through Wnt signaling. Cancer Res. 2019, 79, 4685. [Google Scholar] [CrossRef]
- Sommerfeld, L.; Finkernagel, F.; Jansen, J.M.; Wagner, U.; Nist, A.; Stiewe, T.; Müller-Brüsselbach, S.; Sokol, A.M.; Graumann, J.; Reinartz, S.; et al. The multicellular signalling network of ovarian cancer metastases. Clin. Transl. Med. 2021, 11, e633. [Google Scholar] [CrossRef]
- McLean, K.; Gong, Y.; Choi, Y.; Deng, N.; Yang, K.; Bai, S.; Cabrera, L.; Keller, E.; McCauley, L.; Cho, K.R.; et al. Human ovarian carcinoma–associated mesenchymal stem cells regulate cancer stem cells and tumorigenesis via altered {BMP} production. J. Clin. Investig. 2011, 121, 3206–3219. [Google Scholar] [CrossRef]
- Coffman, L.G.; Pearson, A.T.; Frisbie, L.G.; Freeman, Z.; Christie, E.; Bowtell, D.D.; Buckanovich, R.J. Ovarian Carcinoma-Associated Mesenchymal Stem Cells Arise from Tissue-Specific Normal Stroma. Stem Cells 2019, 37, 257–269. [Google Scholar] [CrossRef]
- Liang, D.; Ma, Y.; Liu, J.; Trope, C.G.; Holm, R.; Nesland, J.M.; Suo, Z. The hypoxic microenvironment upgrades stem-like properties of ovarian cancer cells. BMC Cancer 2012, 12, 201. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, S.; Lee, K.L.; Hikasa, H.; Sun, W.; Huang, R.Y.-J.; Yang, H.; Matsunaga, S.; Yamaguchi, T.; Araki, M.; Kato, H.; et al. Hypoxia-inducible factor-1α promotes cell survival during ammonia stress response in ovarian cancer stem-like cells. Oncotarget 2017, 8, 114481–114494. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Imanaka, N.; Chen, J.; Griffin, J.D. Hypoxia potentiates Notch signaling in breast cancer leading to decreased E-cadherin expression and increased cell migration and invasion. Br. J. Cancer 2010, 102, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Liu, Y.; Lu, Y.; Liu, M.; Li, M.; Li, J.; Wu, L. Hypoxia-inducible factor 1 alpha promotes cancer stem cells-like properties in human ovarian cancer cells by upregulating SIRT1 expression. Sci. Rep. 2017, 7, 10592. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Wei, Y.; Xu, F.; Zhao, M.; Dai, K.; Shen, R.; Yang, S.; Zhang, N. SIRT1 promotes formation of breast cancer through modulating Akt activity. J. Cancer 2018, 9, 2012–2023. [Google Scholar] [CrossRef]
- Shi, L.; Tang, X.; Qian, M.; Liu, Z.; Meng, F.; Fu, L.; Wang, Z.; Zhu, W.G.; Huang, J.D.; Zhou, Z.; et al. A SIRT1-centered circuitry regulates breast cancer stemness and metastasis. Oncogene 2018, 37, 6299–6315. [Google Scholar] [CrossRef]
- Lasek, W. Cancer immunoediting hypothesis: History, clinical implications and controversies. Cent. J. Immunol. 2022, 47, 168–174. [Google Scholar] [CrossRef]
- Ding, J.; Zhang, Y.; Che, Y. Ovarian cancer stem cells: Critical roles in anti-tumor immunity. Front. Genet. 2022, 13, 998220. [Google Scholar] [CrossRef]
- Gatti-Mays, M.E.; Balko, J.M.; Gameiro, S.R.; Bear, H.D.; Prabhakaran, S.; Fukui, J.; Disis, M.L.; Nanda, R.; Gulley, J.L.; Kalinsky, K.; et al. If we build it they will come: Targeting the immune response to breast cancer. NPJ Breast Cancer 2019, 5, 37. [Google Scholar] [CrossRef]
- Luo, H.; Xu, X.; Ye, M.; Sheng, B.; Zhu, X. The prognostic value of HER2 in ovarian cancer: A meta-analysis of observational studies. PLoS ONE 2018, 13, e0191972. [Google Scholar] [CrossRef]
- Iqbal, N.; Iqbal, N. Human Epidermal Growth Factor Receptor 2 (HER2) in Cancers: Overexpression and Therapeutic Implications. Mol. Biol. Int. 2014, 2014, 852748. [Google Scholar] [CrossRef] [PubMed]
- Peoples, G.E.; Goedegebuure, P.S.; Smith, R.; Linehan, D.C.; Yoshino, I.; Eberlein, T.J. Breast and ovarian cancer-specific cytotoxic T lymphocytes recognize the same HER2/neu-derived peptide. Proc. Natl. Acad. Sci. USA 1995, 92, 432–436. [Google Scholar] [CrossRef] [PubMed]
- Koh, J.; Lee, S.; Park, H.; Lee, H.J.; Cho, N.H.; Kim, J. Susceptibility of CD24+ ovarian cancer cells to anti-cancer drugs and natural killer cells. Biochem. Biophys. Res. Commun. 2012, 427, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Chovatiya, N.; Kaur, K.; Huerta-Yepez, S.; Chen, P.-C.; Neal, A.; DiBernardo, G.; Gumrukcu, S.; Memarzadeh, S.; Jewett, A. Inability of ovarian cancers to upregulate their MHC-class I surface expression marks their aggressiveness and increased susceptibility to NK cell-mediated cytotoxicity. Cancer Immunol. Immunother. 2022, 71, 2929–2941. [Google Scholar] [CrossRef]
- Uppendahl, L.D.; Dahl, C.M.; Miller, J.S.; Felices, M.; Geller, M.A. Natural Killer Cell-Based Immunotherapy in Gynecologic Malignancy: A Review. Front. Immunol. 2017, 8, 1825. [Google Scholar] [CrossRef]
- Dewan, M.Z.; Terunuma, H.; Takada, M.; Tanaka, Y.; Abe, H.; Sata, T.; Toi, M.; Yamamoto, N. Role of natural killer cells in hormone-independent rapid tumor formation and spontaneous metastasis of breast cancer cells in vivo. Breast Cancer Res. Treat. 2007, 104, 267–275. [Google Scholar] [CrossRef]
- Yin, T.; Wang, G.; He, S.; Liu, Q.; Sun, J.; Wang, Y. Human cancer cells with stem cell-like phenotype exhibit enhanced sensitivity to the cytotoxicity of IL-2 and IL-15 activated natural killer cells. Cell. Immunol. 2016, 300, 41–45. [Google Scholar] [CrossRef]
- Jin, H.; Kim, H.J. NK Cells Lose Their Cytotoxicity Function against Cancer Stem Cell-Rich Radiotherapy-Resistant Breast Cancer Cell Populations. Int. J. Mol. Sci. 2021, 22, 9639. [Google Scholar] [CrossRef]
- Wang, B.; Wang, Q.; Wang, Z.; Jiang, J.; Yu, S.-C.; Ping, Y.-F.; Yang, J.; Xu, S.-L.; Ye, X.-Z.; Xu, C.; et al. Metastatic Consequences of Immune Escape from NK Cell Cytotoxicity by Human Breast Cancer Stem Cells. Cancer Res. 2014, 74, 5746–5757. [Google Scholar] [CrossRef]
- Geller, M.A.; Cooley, S.; Judson, P.L.; Ghebre, R.; Carson, L.F.; Argenta, P.A.; Jonson, A.L.; Panoskaltsis-Mortari, A.; Curtsinger, J.; McKenna, D.; et al. A phase II study of allogeneic natural killer cell therapy to treat patients with recurrent ovarian and breast cancer. Cytotherapy 2011, 13, 98–107. [Google Scholar] [CrossRef]
- Workel, H.H.; Lubbers, J.M.; Arnold, R.; Prins, T.M.; van der Vlies, P.; de Lange, K.; Bosse, T.; van Gool, I.C.; Eggink, F.A.; Wouters, M.C.A.; et al. A Transcriptionally Distinct CXCL13+CD103+CD8+ T-cell Population Is Associated with B-cell Recruitment and Neoantigen Load in Human Cancer. Cancer Immunol. Res. 2019, 7, 784–796. [Google Scholar] [CrossRef]
- Garg, A.D.; De Ruysscher, D.; Agostinis, P. Immunological metagene signatures derived from immunogenic cancer cell death associate with improved survival of patients with lung, breast or ovarian malignancies: A large-scale meta-analysis. Oncoimmunology 2016, 5, e1069938. [Google Scholar] [CrossRef] [PubMed]
- Alwosaibai, K.; Aalmri, S.; Mashhour, M.; Ghandorah, S.; Alshangiti, A.; Azam, F.; Selwi, W.; Gharaibeh, L.; Alatawi, Y.; Alruwaii, Z.; et al. PD-L1 is highly expressed in ovarian cancer and associated with cancer stem cells populations expressing CD44 and other stem cell markers. BMC Cancer 2023, 23, 13. [Google Scholar] [CrossRef]
- Mansour, F.A.; Al-Mazrou, A.; Al-Mohanna, F.; Al-Alwan, M.; Ghebeh, H. PD-L1 is overexpressed on breast cancer stem cells through notch3/mTOR axis. Oncoimmunology 2020, 9, 1729299. [Google Scholar] [CrossRef] [PubMed]
- Darvin, P.; Sasidharan Nair, V.; Elkord, E. PD-L1 Expression in Human Breast Cancer Stem Cells Is Epigenetically Regulated through Posttranslational Histone Modifications. J. Oncol. 2019, 2019, 3958908. [Google Scholar] [CrossRef] [PubMed]
- Sobral-Leite, M.; Van de Vijver, K.; Michaut, M.; van der Linden, R.; Hooijer, G.K.J.; Horlings, H.M.; Severson, T.M.; Mulligan, A.M.; Weerasooriya, N.; Sanders, J.; et al. Assessment of PD-L1 expression across breast cancer molecular subtypes, in relation to mutation rate, BRCA1-like status, tumor-infiltrating immune cells and survival. Oncoimmunology 2018, 7, e1509820. [Google Scholar] [CrossRef]
- Pawłowska, A.; Kwiatkowska, A.; Suszczyk, D.; Chudzik, A.; Tarkowski, R.; Barczyński, B.; Kotarski, J.; Wertel, I. Clinical and Prognostic Value of Antigen-Presenting Cells with PD-L1/PD-L2 Expression in Ovarian Cancer Patients. Int. J. Mol. Sci. 2021, 22, 11563. [Google Scholar] [CrossRef]
- Wang, L. Prognostic effect of programmed death-ligand 1 (PD-L1) in ovarian cancer: A systematic review, meta-analysis and bioinformatics study. J. Ovarian Res. 2019, 12, 37. [Google Scholar] [CrossRef]
- Li, X.; Li, M.; Lian, Z.; Zhu, H.; Kong, L.; Wang, P.; Yu, J. Prognostic Role of Programmed Death Ligand-1 Expression in Breast Cancer: A Systematic Review and Meta-Analysis. Target. Oncol. 2016, 11, 753–761. [Google Scholar] [CrossRef]
- Jang, Y.S.; Kim, T.W.; Ryu, J.S.; Kong, H.J.; Jang, S.H.; Nam, G.H.; Kim, J.H.; Jeon, S. Upregulation of programmed death ligand-1 in tumor-associated macrophages affects chemotherapeutic response in ovarian cancer cells. PLoS ONE 2023, 18, e0277285. [Google Scholar] [CrossRef]
- Barkal, A.A.; Brewer, R.E.; Markovic, M.; Kowarsky, M.; Barkal, S.A.; Zaro, B.W.; Krishnan, V.; Hatakeyama, J.; Dorigo, O.; Barkal, L.J.; et al. CD24 signalling through macrophage Siglec-10 is a target for cancer immunotherapy. Nature 2019, 572, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, S.; Hersey, J.M.; Mellor, P.; Dai, W.; Santos-Silva, A.; Liber, D.; Luk, L.; Titley, I.; Carden, C.P.; Box, G.; et al. Ovarian Cancer Stem Cell–Like Side Populations Are Enriched Following Chemotherapy and Overexpress EZH2. Mol. Cancer Ther. 2011, 10, 325–335. [Google Scholar] [CrossRef]
- Steg, A.D.; Bevis, K.S.; Katre, A.A.; Ziebarth, A.; Dobbin, Z.C.; Alvarez, R.D.; Zhang, K.; Conner, M.; Landen, C.N. Stem cell pathways contribute to clinical chemoresistance in ovarian cancer. Clin. Cancer Res. 2012, 18, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Montalbán Del Barrio, I.; Penski, C.; Schlahsa, L.; Stein, R.G.; Diessner, J.; Wöckel, A.; Dietl, J.; Lutz, M.B.; Mittelbronn, M.; Wischhusen, J.; et al. Adenosine-generating ovarian cancer cells attract myeloid cells which differentiate into adenosine-generating tumor associated macrophages—A self-amplifying, CD39- and CD73-dependent mechanism for tumor immune escape. J. Immunother. Cancer 2016, 4, 49. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, J.; Liu, N.; Wu, W.; Li, H.; Chen, J.; Guo, X. Molecular mechanism of CD163+ tumor-associated macrophage (TAM)-derived exosome-induced cisplatin resistance in ovarian cancer ascites. Ann. Transl. Med. 2022, 10, 1014. [Google Scholar] [CrossRef] [PubMed]
- Allavena, P.; Digifico, E.; Belgiovine, C. Macrophages and cancer stem cells: A malevolent alliance. Mol. Med. 2021, 27, 121. [Google Scholar] [CrossRef]
- Curiel, T.J.; Coukos, G.; Zou, L.; Alvarez, X.; Cheng, P.; Mottram, P.; Evdemon-Hogan, M.; Conejo-Garcia, J.R.; Zhang, L.; Burow, M.; et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat. Med. 2004, 10, 942–949. [Google Scholar] [CrossRef] [PubMed]
- Harper, E.; Sheedy, E.; Stack, M. With Great Age Comes Great Metastatic Ability: Ovarian Cancer and the Appeal of the Aging Peritoneal Microenvironment. Cancers 2018, 10, 230. [Google Scholar] [CrossRef]
- Wefers, C.; Duiveman-de Boer, T.; Zusterzeel, P.; Massuger, L.; Fuchs, D.; Torensma, R.; Wheelock, C.; de Vries, I. Different Lipid Regulation in Ovarian Cancer: Inhibition of the Immune System. Int. J. Mol. Sci. 2018, 19, 273. [Google Scholar] [CrossRef]
- Holen, I.; Speirs, V.; Morrissey, B.; Blyth, K. In vivo models in breast cancer research: Progress, challenges and future directions. Dis. Model. Mech. 2017, 10, 359–371. [Google Scholar] [CrossRef]
- Sztankovics, D.; Moldvai, D.; Petővári, G.; Gelencsér, R.; Krencz, I.; Raffay, R.; Dankó, T.; Sebestyén, A. 3D bioprinting and the revolution in experimental cancer model systems-A review of developing new models and experiences with in vitro 3D bioprinted breast cancer tissue-mimetic structures. Pathol. Oncol. Res. 2023, 29, 1610996. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Reyes, M.; Aguilar-Rojas, A. Three-dimensional models to study breast cancer (Review). Int. J. Oncol. 2021, 58, 331–343. [Google Scholar] [CrossRef] [PubMed]
- Fröhlich, E. The Variety of 3D Breast Cancer Models for the Study of Tumor Physiology and Drug Screening. Int. J. Mol. Sci. 2023, 24, 7116. [Google Scholar] [CrossRef] [PubMed]
- Salinas-Vera, Y.M.; Valdés, J.; Pérez-Navarro, Y.; Mandujano-Lazaro, G.; Marchat, L.A.; Ramos-Payán, R.; Nuñez-Olvera, S.I.; Pérez-Plascencia, C.; López-Camarillo, C. Three-Dimensional 3D Culture Models in Gynecological and Breast Cancer Research. Front. Oncol. 2022, 12, 826113. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.F.; Rao, S.S. Three-dimensional culture models to study drug resistance in breast cancer. Biotechnol. Bioeng. 2020, 117, 2262–2278. [Google Scholar] [CrossRef]
- Polonio-Alcalá, E.; Rabionet, M.; Ruiz-Martínez, S.; Ciurana, J.; Puig, T. Three-Dimensional Manufactured Supports for Breast Cancer Stem Cell Population Characterization. Curr. Drug Targets 2018, 20, 839–851. [Google Scholar] [CrossRef]
- Balachander, G.M.; Kotcherlakota, R.; Nayak, B.; Kedaria, D.; Rangarajan, A.; Chatterjee, K. 3D Tumor Models for Breast Cancer: Whither We Are and What We Need. ACS Biomater. Sci. Eng. 2021, 7, 3470–3486. [Google Scholar] [CrossRef]
- Murayama, T.; Gotoh, N. Patient-Derived Xenograft Models of Breast Cancer and Their Application. Cells 2019, 8, 621. [Google Scholar] [CrossRef]
- Xu, F.; Burg, K.J.L. Three-dimensional polymeric systems for cancer cell studies. Cytotechnology 2007, 54, 135–143. [Google Scholar] [CrossRef]
- Doublier, S.; Belisario, D.C.; Polimeni, M.; Annaratone, L.; Riganti, C.; Allia, E.; Ghigo, D.; Bosia, A.; Sapino, A. HIF-1 activation induces doxorubicin resistance in MCF7 3-D spheroids via P-glycoprotein expression: A potential model of the chemo-resistance of invasive micropapillary carcinoma of the breast. BMC Cancer 2012, 12, 4. [Google Scholar] [CrossRef]
- Dubois, C.; Dufour, R.; Daumar, P.; Aubel, C.; Szczepaniak, C.; Blavignac, C.; Mounetou, E.; Penault-Llorca, F.; Bamdad, M. Development and cytotoxic response of two proliferative MDA-MB-231 and non-proliferative SUM1315 three-dimensional cell culture models of triple-negative basal-like breast cancer cell lines. Oncotarget 2017, 8, 95316–95331. [Google Scholar] [CrossRef] [PubMed]
- Ham, S.L.; Thakuri, P.S.; Plaster, M.; Li, J.; Luker, K.E.; Luker, G.D.; Tavana, H. Three-dimensional tumor model mimics stromal—Breast cancer cells signaling. Oncotarget 2018, 9, 249–267. [Google Scholar] [CrossRef] [PubMed]
- Brancato, V.; Gioiella, F.; Imparato, G.; Guarnieri, D.; Urciuolo, F.; Netti, P.A. 3D breast cancer microtissue reveals the role of tumor microenvironment on the transport and efficacy of free-doxorubicin in vitro. Acta Biomater. 2018, 75, 200–212. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Jeong, E.-K.; Jeon, H.M.; Kim, C.H.; Kang, H.S. Implication of necrosis-linked p53 aggregation in acquired apoptotic resistance to 5-FU in MCF-7 multicellular tumour spheroids. Oncol. Rep. 2010, 24, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Rashidi, M.R.W.; Mehta, P.; Bregenzer, M.; Raghavan, S.; Fleck, E.M.; Horst, E.N.; Harissa, Z.; Ravikumar, V.; Brady, S.; Bild, A.; et al. Engineered 3D Model of Cancer Stem Cell Enrichment and Chemoresistance. Neoplasia 2019, 21, 822–836. [Google Scholar] [CrossRef]
- Imamura, Y.; Mukohara, T.; Shimono, Y.; Funakoshi, Y.; Chayahara, N.; Toyoda, M.; Kiyota, N.; Takao, S.; Kono, S.; Nakatsura, T.; et al. Comparison of 2D- and 3D-culture models as drug-testing platforms in breast cancer. Oncol. Rep. 2015, 33, 1837–1843. [Google Scholar] [CrossRef]
- Lovitt, C.J.; Shelper, T.B.; Avery, V.M. Evaluation of chemotherapeutics in a three-dimensional breast cancer model. J. Cancer Res. Clin. Oncol. 2015, 141, 951–959. [Google Scholar] [CrossRef]
- Lovitt, C.J.; Shelper, T.B.; Avery, V.M. Doxorubicin resistance in breast cancer cells is mediated by extracellular matrix proteins. BMC Cancer 2018, 18, 41. [Google Scholar] [CrossRef]
- Breslin, S.; O’Driscoll, L.; Breslin, S.; O’Driscoll, L. The relevance of using 3D cell cultures, in addition to 2D monolayer cultures, when evaluating breast cancer drug sensitivity and resistance. Oncotarget 2016, 7, 45745–45756. [Google Scholar] [CrossRef]
- dit Faute, M.A.; Laurent, L.; Ploton, D.; Poupon, M.-F.; Jardillier, J.-C.; Bobichon, H. Distinctive alterations of invasiveness, drug resistance and cell-cell organization in 3D-cultures of MCF-7, a human breast cancer cell line, and its multidrug resistant variant. Clin. Exp. Metastasis 2002, 19, 161–167. [Google Scholar] [CrossRef]
- Gomes, L.R.; Rocha, C.R.R.; Martins, D.J.; Fiore, A.P.Z.P.; Kinker, G.S.; Bruni-Cardoso, A.; Menck, C.F.M. ATR mediates cisplatin resistance in 3D-cultured breast cancer cells via translesion DNA synthesis modulation. Cell Death Dis. 2019, 10, 459. [Google Scholar] [CrossRef] [PubMed]
- Gangadhara, S.; Smith, C.; Barrett-Lee, P.; Hiscox, S. 3D culture of Her2+ breast cancer cells promotes AKT to MAPK switching and a loss of therapeutic response. BMC Cancer 2016, 16, 345. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Song, J.M. 3D bioprinted drug-resistant breast cancer spheroids for quantitative in situ evaluation of drug resistance. Acta Biomater. 2022, 138, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Rezakhani, L.; Rahmati, S.; Ghasemi, S.; Alizadeh, M.; Alizadeh, A. A comparative study of the effects of crab derived exosomes and doxorubicin in 2 & 3-dimensional in vivo models of breast cancer. Chem. Phys. Lipids 2022, 243, 105179. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, W.; Kuss, M.; Mirza, S.; Qi, D.; Krasnoslobodtsev, A.; Zeng, J.; Band, H.; Band, V.; Duan, B. 3D Bioprinting of Breast Cancer Models for Drug Resistance Study. ACS Biomater. Sci. Eng. 2018, 4, 4401–4411. [Google Scholar] [CrossRef]
- Horning, J.L.; Sahoo, S.K.; Vijayaraghavalu, S.; Dimitrijevic, S.; Vasir, J.K.; Jain, T.K.; Panda, A.K.; Labhasetwar, V. 3-D tumor model for in vitro evaluation of anticancer drugs. Mol. Pharm. 2008, 5, 849–862. [Google Scholar] [CrossRef]
- Gong, X.; Lin, C.; Cheng, J.; Su, J.; Zhao, H.; Liu, T.; Wen, X.; Zhao, P. Generation of Multicellular Tumor Spheroids with Microwell-Based Agarose Scaffolds for Drug Testing. PLoS ONE 2015, 10, e0130348. [Google Scholar] [CrossRef]
- Baker, A.E.G.; Tam, R.Y.; Shoichet, M.S. Independently Tuning the Biochemical and Mechanical Properties of 3D Hyaluronan-Based Hydrogels with Oxime and Diels–Alder Chemistry to Culture Breast Cancer Spheroids. Biomacromolecules 2017, 18, 4373–4384. [Google Scholar] [CrossRef]
- Arya, A.D.; Hallur, P.M.; Karkisaval, A.G.; Gudipati, A.; Rajendiran, S.; Dhavale, V.; Ramachandran, B.; Jayaprakash, A.; Gundiah, N.; Chaubey, A. Gelatin Methacrylate Hydrogels as Biomimetic Three-Dimensional Matrixes for Modeling Breast Cancer Invasion and Chemoresponse in Vitro. ACS Appl. Mater. Interfaces 2016, 8, 22005–22017. [Google Scholar] [CrossRef]
- Shin, K.; Klosterhoff, B.S.; Han, B. Characterization of Cell-Type-Specific Drug Transport and Resistance of Breast Cancers Using Tumor-Microenvironment-on-Chip. Mol. Pharm. 2016, 13, 2214–2223. [Google Scholar] [CrossRef]
- Ozcelikkale, A.; Shin, K.; Noe-Kim, V.; Elzey, B.D.; Dong, Z.; Zhang, J.T.; Kim, K.; Kwon, I.C.; Park, K.; Han, B. Differential response to doxorubicin in breast cancer subtypes simulated by a microfluidic tumor model. J. Control. Release 2017, 266, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Mao, S.; Li, M.; Li, H.F.; Lin, J.M. Near-physiological microenvironment simulation on chip to evaluate drug resistance of different loci in tumour mass. Talanta 2019, 191, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Xiao, Z.; Meng, Y.; Zhao, Y.; Han, J.; Su, G.; Chen, B.; Dai, J. The enhancement of cancer stem cell properties of MCF-7 cells in 3D collagen scaffolds for modeling of cancer and anti-cancer drugs. Biomaterials 2012, 33, 1437–1444. [Google Scholar] [CrossRef] [PubMed]
- Palomeras, S.; Rabionet, M.; Ferrer, I.; Sarrats, A.; Garcia-Romeu, M.; Puig, T.; Ciurana, J. Breast Cancer Stem Cell Culture and Enrichment Using Poly(ε-Caprolactone) Scaffolds. Molecules 2016, 21, 537. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Duan, X.; Lo, P.-K.; Liu, S.; Liu, X.; Chen, H.; Wang, Q. Expansion of breast cancer stem cells with fibrous scaffolds. Integr. Biol. 2013, 5, 768–777. [Google Scholar] [CrossRef]
- Balachander, G.M.; Balaji, S.A.; Rangarajan, A.; Chatterjee, K. Enhanced Metastatic Potential in a 3D Tissue Scaffold toward a Comprehensive in Vitro Model for Breast Cancer Metastasis. ACS Appl. Mater. Interfaces 2015, 7, 27810–27822. [Google Scholar] [CrossRef]
- Sims-Mourtada, J.; Niamat, R.A.; Samuel, S.; Eskridge, C.; Kmiec, E.B. Enrichment of breast cancer stem-like cells by growth on electrospun polycaprolactone-chitosan nanofiber scaffolds. Int. J. Nanomed. 2014, 9, 995–1003. [Google Scholar] [CrossRef]
- Malacrida, B.; Pearce, O.M.T.; Balkwill, F.R. Building in vitro 3D human multicellular models of high-grade serous ovarian cancer. STAR Protoc. 2022, 3, 101086. [Google Scholar] [CrossRef]
- Brooks, E.A.; Gencoglu, M.F.; Corbett, D.C.; Stevens, K.R.; Peyton, S.R. An omentum-inspired 3D PEG hydrogel for identifying ECM-drivers of drug resistant ovarian cancer. APL Bioeng. 2019, 3, 026106. [Google Scholar] [CrossRef]
- Velletri, T.; Villa, C.E.; Cilli, D.; Barzaghi, B.; Lo Riso, P.; Lupia, M.; Luongo, R.; López-Tobón, A.; De Simone, M.; Bonnal, R.J.P.; et al. Single cell-derived spheroids capture the self-renewing subpopulations of metastatic ovarian cancer. Cell Death Differ. 2022, 29, 614–626. [Google Scholar] [CrossRef]
- Li, S.-S.; Ip, C.K.M.; Tang, M.Y.H.; Sy, S.K.H.; Yung, S.; Chan, T.-M.; Yang, M.; Shum, H.C.; Wong, A.S.T. Modeling Ovarian Cancer Multicellular Spheroid Behavior in a Dynamic 3D Peritoneal Microdevice. J. Vis. Exp. 2017, 120, e55337. [Google Scholar] [CrossRef]
- Liu, S.; Ginestier, C.; Ou, S.J.; Clouthier, S.G.; Patel, S.H.; Monville, F.; Korkaya, H.; Heath, A.; Dutcher, J.; Kleer, C.G.; et al. Breast cancer stem cells are regulated by mesenchymal stem cells through cytokine networks. Cancer Res. 2011, 71, 614–624. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cardenas, H.; Fang, F.; Condello, S.; Taverna, P.; Segar, M.; Liu, Y.; Nephew, K.P.; Matei, D. Epigenetic targeting of ovarian cancer stem cells. Cancer Res. 2014, 74, 4922–4936. [Google Scholar] [CrossRef]
- Hu, Y.; Yagüe, E.; Zhao, J.; Wang, L.; Bai, J.; Yang, Q.; Pan, T.; Zhao, H.; Liu, J.; Zhang, J. Sabutoclax, pan-active BCL-2 protein family antagonist, overcomes drug resistance and eliminates cancer stem cells in breast cancer. Cancer Lett. 2018, 423, 47–59. [Google Scholar] [CrossRef]
- DeRose, Y.S.; Wang, G.; Lin, Y.-C.; Bernard, P.S.; Buys, S.S.; Ebbert, M.T.W.; Factor, R.; Matsen, C.; Milash, B.A.; Nelson, E.; et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat. Med. 2011, 17, 1514–1520. [Google Scholar] [CrossRef] [PubMed]
- Dobrolecki, L.E.; Airhart, S.D.; Alferez, D.G.; Aparicio, S.; Behbod, F.; Bentires-Alj, M.; Brisken, C.; Bult, C.J.; Cai, S.; Clarke, R.B.; et al. Patient-derived xenograft (PDX) models in basic and translational breast cancer research. Cancer Metastasis Rev. 2016, 35, 547–573. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Patel, M.R.; Prescher, J.A.; Patsialou, A.; Qian, D.; Lin, J.; Wen, S.; Chang, Y.-F.; Bachmann, M.H.; Shimono, Y.; et al. Cancer stem cells from human breast tumors are involved in spontaneous metastases in orthotopic mouse models. Proc. Natl. Acad. Sci. USA 2010, 107, 18115–18120. [Google Scholar] [CrossRef]
- Weroha, S.J.; Becker, M.A.; Enderica-Gonzalez, S.; Harrington, S.C.; Oberg, A.L.; Maurer, M.J.; Perkins, S.E.; AlHilli, M.; Butler, K.A.; McKinstry, S.; et al. Tumorgrafts as in vivo surrogates for women with ovarian cancer. Clin. Cancer Res. 2014, 20, 1288–1297. [Google Scholar] [CrossRef]
- Flesken-Nikitin, A.; Hwang, C.-I.; Cheng, C.-Y.; Michurina, T.V.; Enikolopov, G.; Nikitin, A.Y. Ovarian surface epithelium at the junction area contains cancer-prone stem cell niche. Nature 2013, 495, 241–245. [Google Scholar] [CrossRef]
- Landberg, G.; Fitzpatrick, P.; Isakson, P.; Jonasson, E.; Karlsson, J.; Larsson, E.; Svanström, A.; Rafnsdottir, S.; Persson, E.; Gustafsson, A.; et al. Patient-derived scaffolds uncover breast cancer promoting properties of the microenvironment. Biomaterials 2020, 235, 119705. [Google Scholar] [CrossRef]
- Leiva, M.C.; Garre, E.; Gustafsson, A.; Svanström, A.; Bogestål, Y.; Håkansson, J.; Ståhlberg, A.; Landberg, G. Breast cancer patient-derived scaffolds as a tool to monitor chemotherapy responses in human tumor microenvironments. J. Cell. Physiol. 2021, 236, 4709. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, A.; Garre, E.; Leiva, M.C.; Salerno, S.; Ståhlberg, A.; Landberg, G. Patient-derived scaffolds as a drug-testing platform for endocrine therapies in breast cancer. Sci. Rep. 2021, 11, 13334. [Google Scholar] [CrossRef] [PubMed]
- Garre, E.; Gustafsson, A.; Leiva, M.C.; Håkansson, J.; Ståhlberg, A.; Kovács, A.; Landberg, G. Breast Cancer Patient-Derived Scaffolds Can Expose Unique Individual Cancer Progressing Properties of the Cancer Microenvironment Associated with Clinical Characteristics. Cancers 2022, 14, 2172. [Google Scholar] [CrossRef]
- Omole, E.B.; Aijaz, I.; Ellegate, J.; Isenhart, E.; Desouki, M.M.; Mastri, M.; Humphrey, K.; Dougherty, E.M.; Rosario, S.R.; Nastiuk, K.L.; et al. Combined BRCA2 and MAGEC3 Expression Predict Outcome in Advanced Ovarian Cancers. Cancers 2022, 14, 4724. [Google Scholar] [CrossRef] [PubMed]
- Parte, S.C.; Batra, S.K.; Kakar, S.S. Characterization of stem cell and cancer stem cell populations in ovary and ovarian tumors. J. Ovarian Res. 2018, 11, 69. [Google Scholar] [CrossRef]
- Ali, H.R.; Dawson, S.-J.; Blows, F.M.; Provenzano, E.; Pharoah, P.D.; Caldas, C. Cancer stem cell markers in breast cancer: Pathological, clinical and prognostic significance. Breast Cancer Res. 2011, 13, R118. [Google Scholar] [CrossRef]
- Abraham, B.K.; Fritz, P.; McClellan, M.; Hauptvogel, P.; Athelogou, M.; Brauch, H. Prevalence of CD44+/CD24-/low cells in breast cancer may not be associated with clinical outcome but may favor distant metastasis. Clin. Cancer Res. 2005, 11, 1154–1159. [Google Scholar] [CrossRef]
- Cheng, L.; Ramesh, A.V.; Flesken-Nikitin, A.; Choi, J.; Nikitin, A.Y. Mouse models for cancer stem cell research. Toxicol. Pathol. 2010, 38, 62–71. [Google Scholar] [CrossRef]
- Abdullah, L.N.; Chow, E.K.-H. Mechanisms of chemoresistance in cancer stem cells. Clin. Transl. Med. 2013, 2, 3. [Google Scholar] [CrossRef]
- Zhang, S.; Cui, B.; Lai, H.; Liu, G.; Ghia, E.M.; Widhopf, G.F.; Zhang, Z.; Wu, C.C.N.; Chen, L.; Wu, R.; et al. Ovarian cancer stem cells express ROR1, which can be targeted for anti-cancer-stem-cell therapy. Proc. Natl. Acad. Sci. USA 2014, 111, 17266–17271. [Google Scholar] [CrossRef]
- Konrad, C.V.; Murali, R.; Varghese, B.A.; Nair, R. The role of cancer stem cells in tumor heterogeneity and resistance to therapy. Can. J. Physiol. Pharmacol. 2017, 95, 1–15. [Google Scholar] [CrossRef]
- Mao, X.-D.; Wei, X.; Xu, T.; Li, T.-P.; Liu, K.-S. Research progress in breast cancer stem cells: Characterization and future perspectives. Am. J. Cancer Res. 2022, 12, 3208–3222. [Google Scholar] [PubMed]
- Kim, W.-T.; Ryu, C.J. Cancer stem cell surface markers on normal stem cells. BMB Rep. 2017, 50, 285–298. [Google Scholar] [CrossRef]
- Yang, L.; Shi, P.; Zhao, G.; Xu, J.; Peng, W.; Zhang, J.; Zhang, G.; Wang, X.; Dong, Z.; Chen, F.; et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Target. Ther. 2020, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Palomeras, S.; Ruiz-Martínez, S.; Puig, T. Targeting Breast Cancer Stem Cells to Overcome Treatment Resistance. Molecules 2018, 23, 2193. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Yin, X.; Zhou, X.; Niu, X.; Wang, Y.; Su, M. Salinomycin-Loaded High-Density Lipoprotein Exerts Promising Anti-Ovarian Cancer Effects by Inhibiting Epithelial–Mesenchymal Transition. Int. J. Nanomed. 2022, 17, 4059–4071. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, H.A.; Iliopoulos, D.; Tsichlis, P.N.; Struhl, K. Metformin Selectively Targets Cancer Stem Cells, and Acts Together with Chemotherapy to Block Tumor Growth and Prolong Remission. Cancer Res. 2009, 69, 7507–7511. [Google Scholar] [CrossRef]
- Liu, Q.; Hodge, J.; Wang, J.; Wang, Y.; Wang, L.; Singh, U.P.; Li, Y.; Yao, Y.; Wang, D.; Ai, W.; et al. Emodin reduces Breast Cancer Lung Metastasis by suppressing Macrophage-induced Breast Cancer Cell Epithelial-mesenchymal transition and Cancer Stem Cell formation. Theranostics 2020, 10, 8365–8381. [Google Scholar] [CrossRef]
- Long, H.; Chen, H.; Yan, J.; Cheng, H. Emodin exerts antitumor effects in ovarian cancer cell lines by preventing the development of cancer stem cells via epithelial mesenchymal transition. Oncol. Lett. 2022, 23, 95. [Google Scholar] [CrossRef]
- Shan, N.L.; Wahler, J.; Lee, H.J.; Bak, M.J.; Gupta, S.D.; Maehr, H.; Suh, N. Vitamin D compounds inhibit cancer stem-like cells and induce differentiation in triple negative breast cancer. J. Steroid Biochem. Mol. Biol. 2017, 173, 122–129. [Google Scholar] [CrossRef]
- Ji, M.; Liu, L.; Hou, Y.; Li, B. 1α,25-Dihydroxyvitamin D3 restrains stem cell-like properties of ovarian cancer cells by enhancing vitamin D receptor and suppressing CD44. Oncol. Rep. 2019, 41, 3393–3403. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.-J.; Tsai, H.-Y.; Tsai, C.-C.; Chen, T.-Y.; Hsieh, T.-H.; Chen, C.-L.; Mbuyisa, L.; Huang, Y.-B.; Lin, M.-W. Luteolin Inhibits Breast Cancer Stemness and Enhances Chemosensitivity through the Nrf2-Mediated Pathway. Molecules 2021, 26, 6452. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Zhu, G.-Y.; Lu, Y.; Yang, A.; Chen, D.; Huang, H.-J.; Peng, S.-X.; Chen, L.-W.; Li, Y.-W. Luteolin suppresses epithelial-mesenchymal transition and migration of triple-negative breast cancer cells by inhibiting YAP/TAZ activity. Biomed. Pharmacother. 2020, 129, 110462. [Google Scholar] [CrossRef]
- Li, Y.; Hu, Y.; Yang, L.; Liu, J.; Cui, C.; Yang, M.; Zou, D.; Zhou, L.; Zhou, Q.; Ge, W.; et al. Luteolin directly binds to KDM4C and attenuates ovarian cancer stemness via epigenetic suppression of PPP2CA/YAP axis. Biomed. Pharmacother. 2023, 160, 114350. [Google Scholar] [CrossRef] [PubMed]
- McClements, L.; Annett, S.; Yakkundi, A.; O’Rourke, M.; Valentine, A.; Moustafa, N.; Alqudah, A.; Simões, B.M.; Furlong, F.; Short, A.; et al. FKBPL and its peptide derivatives inhibit endocrine therapy resistant cancer stem cells and breast cancer metastasis by downregulating DLL4 and Notch4. BMC Cancer 2019, 19, 351. [Google Scholar] [CrossRef] [PubMed]
- Annett, S.; Moore, G.; Short, A.; Marshall, A.; McCrudden, C.; Yakkundi, A.; Das, S.; McCluggage, W.G.; Nelson, L.; Harley, I.; et al. FKBPL-based peptide, ALM201, targets angiogenesis and cancer stem cells in ovarian cancer. Br. J. Cancer 2020, 122, 361–371. [Google Scholar] [CrossRef]
- Lacerda, L.; Reddy, J.P.; Liu, D.; Larson, R.; Li, L.; Masuda, H.; Brewer, T.; Debeb, B.G.; Xu, W.; Hortobágyi, G.N.; et al. Simvastatin Radiosensitizes Differentiated and Stem-Like Breast Cancer Cell Lines and Is Associated with Improved Local Control in Inflammatory Breast Cancer Patients Treated with Postmastectomy Radiation. Stem Cells Transl. Med. 2014, 3, 849–856. [Google Scholar] [CrossRef]
- Ehmsen, S.; Pedersen, M.H.; Wang, G.; Terp, M.G.; Arslanagic, A.; Hood, B.L.; Conrads, T.P.; Leth-Larsen, R.; Ditzel, H.J. Increased Cholesterol Biosynthesis Is a Key Characteristic of Breast Cancer Stem Cells Influencing Patient Outcome. Cell Rep. 2019, 27, 3927–3938.e6. [Google Scholar] [CrossRef]
- Kato, S.; Liberona, M.F.; Cerda-Infante, J.; Sánchez, M.; Henríquez, J.; Bizama, C.; Bravo, M.L.; Gonzalez, P.; Gejman, R.; Brañes, J.; et al. Simvastatin interferes with cancer ‘stem-cell’ plasticity reducing metastasis in ovarian cancer. Endocr. Relat. Cancer 2018, 25, 821–836. [Google Scholar] [CrossRef]
- Yan, Y.; Li, Z.; Xu, X.; Chen, C.; Wei, W.; Fan, M.; Chen, X.; Li, J.J.; Wang, Y.; Huang, J. All-trans retinoic acids induce differentiation and sensitize a radioresistant breast cancer cells to chemotherapy. BMC Complement. Altern. Med. 2016, 16, 113. [Google Scholar] [CrossRef]
- Schech, A.; Kazi, A.; Yu, S.; Shah, P.; Sabnis, G. Histone Deacetylase Inhibitor Entinostat Inhibits Tumor-Initiating Cells in Triple-Negative Breast Cancer Cells. Mol. Cancer Ther. 2015, 14, 1848–1857. [Google Scholar] [CrossRef]
- Wang, D.; Li, W.; Zhao, R.; Chen, L.; Liu, N.; Tian, Y.; Zhao, H.; Xie, M.; Lu, F.; Fang, Q.; et al. Stabilized Peptide HDAC Inhibitors Derived from HDAC1 Substrate H3K56 for the Treatment of Cancer Stem–Like Cells In Vivo. Cancer Res. 2019, 79, 1769–1783. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, P.; Barua, A.; Roy, A.; Pattanayak, R.; Bhattacharyya, M.; Saha, P. Eugenol restricts Cancer Stem Cell population by degradation of β-catenin via N-terminal Ser37 phosphorylation-an in vivo and in vitro experimental evaluation. Chem. Biol. Interact. 2020, 316, 108938. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.S.; Aboussekhra, A. Sequential combination of cisplatin with eugenol targets ovarian cancer stem cells through the Notch-Hes1 signalling pathway. J. Exp. Clin. Cancer Res. 2019, 38, 382. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.H.; Lin, C.; Liu, M.M.; Zhang, J.; Tao, Z.H.; Hu, X.C. Src inhibition can synergize with gemcitabine and reverse resistance in triple negative breast cancer cells via the AKT/c-jun pathway. PLoS ONE 2016, 11, e0169230. [Google Scholar] [CrossRef] [PubMed]
- Simpkins, F.; Jang, K.; Yoon, H.; Hew, K.E.; Kim, M.; Azzam, D.J.; Sun, J.; Zhao, D.; Ince, T.A.; Liu, W.; et al. Dual Src and MEK Inhibition Decreases Ovarian Cancer Growth and Targets Tumor Initiating Stem-Like Cells. Clin. Cancer Res. 2018, 24, 4874–4886. [Google Scholar] [CrossRef]
- Chen, L.; Yang, G.; Dong, H. Everolimus Reverses Palbociclib Resistance in ER+ Human Breast Cancer Cells by Inhibiting Phosphatidylinositol 3-Kinase(PI3K)/Akt/Mammalian Target of Rapamycin (mTOR) Pathway. Med. Sci. Monit. 2019, 25, 77–86. [Google Scholar] [CrossRef]
- Phan, N.L.-C.; Van Trinh, N.; Pham, P. Van Low concentrations of 5-aza-2’-deoxycytidine induce breast cancer stem cell differentiation by triggering tumor suppressor gene expression. Onco. Targets. Ther. 2016, 9, 49–59. [Google Scholar] [CrossRef]
- Ho, C.-M.; Lee, F.-K.; Yen, T.-L.; Huang, S.-H.; Cheng, W.-F. Everolimus combined with 5-aza-2-deoxycytidine generated potent anti-tumor effects on ovarian clear cell cancer stem-like/spheroid cells by inhibiting the COL6A3-AKT-mTOR pathway. Am. J. Cancer Res. 2022, 12, 1686–1706. [Google Scholar]
- Liu, P.; Kumar, I.S.; Brown, S.; Kannappan, V.; Tawari, P.E.; Tang, J.Z.; Jiang, W.; Armesilla, A.L.; Darling, J.L.; Wang, W. Disulfiram targets cancer stem-like cells and reverses resistance and cross-resistance in acquired paclitaxel-resistant triple-negative breast cancer cells. Br. J. Cancer 2013, 109, 1876–1885. [Google Scholar] [CrossRef]
- Guo, F.; Yang, Z.; Sehouli, J.; Kaufmann, A.M. Blockade of ALDH in Cisplatin-Resistant Ovarian Cancer Stem Cells In Vitro Synergistically Enhances Chemotherapy-Induced Cell Death. Curr. Oncol. 2022, 29, 2808–2822. [Google Scholar] [CrossRef] [PubMed]
- Spizzo, G.; Fong, D.; Wurm, M.; Ensinger, C.; Obrist, P.; Hofer, C.; Mazzoleni, G.; Gastl, G.; Went, P. EpCAM expression in primary tumour tissues and metastases: An immunohistochemical analysis. J. Clin. Pathol. 2011, 64, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Heiss, M.M.; Murawa, P.; Koralewski, P.; Kutarska, E.; Kolesnik, O.O.; Ivanchenko, V.V.; Dudnichenko, A.S.; Aleknaviciene, B.; Razbadauskas, A.; Gore, M.; et al. The trifunctional antibody catumaxomab for the treatment of malignant ascites due to epithelial cancer: Results of a prospective randomized phase II/III trial. Int. J. Cancer 2010, 127, 2209–2221. [Google Scholar] [CrossRef]
- Kubo, M.; Umebayashi, M.; Kurata, K.; Mori, H.; Kai, M.; Onishi, H.; Katano, M.; Nakamura, M.; Morisaki, T. Catumaxomab with Activated T-cells Efficiently Lyses Chemoresistant EpCAM-positive Triple-negative Breast Cancer Cell Lines. Anticancer Res. 2018, 38, 4273–4279. [Google Scholar] [CrossRef]
- Skubitz, A.P.N.; Taras, E.P.; Boylan, K.L.M.; Waldron, N.N.; Oh, S.; Panoskaltsis-Mortari, A.; Vallera, D.A. Targeting CD133 in an in vivo ovarian cancer model reduces ovarian cancer progression. Gynecol. Oncol. 2013, 130, 579–587. [Google Scholar] [CrossRef] [PubMed]
- Tume, L.; Paco, K.; Ubidia-Incio, R.; Moya, J. CD133 in breast cancer cells and in breast cancer stem cells as another target for immunotherapy. Gac. Mex. Oncol. 2016, 15, 22–30. [Google Scholar] [CrossRef]
- Vora, P.; Venugopal, C.; Salim, S.K.; Tatari, N.; Bakhshinyan, D.; Singh, M.; Seyfrid, M.; Upreti, D.; Rentas, S.; Wong, N.; et al. The Rational Development of CD133-Targeting Immunotherapies for Glioblastoma. Cell Stem Cell 2020, 26, 832–844.e6. [Google Scholar] [CrossRef]
- Ahmed, N.; Kadife, E.; Raza, A.; Short, M.; Jubinsky, P.T.; Kannourakis, G. Ovarian Cancer, Cancer Stem Cells and Current Treatment Strategies: A Potential Role of Magmas in the Current Treatment Methods. Cells 2020, 9, 719. [Google Scholar] [CrossRef]
- Bellio, C.; DiGloria, C.; Foster, R.; James, K.; Konstantinopoulos, P.A.; Growdon, W.B.; Rueda, B.R. PARP Inhibition Induces Enrichment of DNA Repair–Proficient CD133 and CD117 Positive Ovarian Cancer Stem Cells. Mol. Cancer Res. 2019, 17, 431–445. [Google Scholar] [CrossRef]
- Zeniou, M.; Nguekeu-Zebaze, L.; Dantzer, F. Therapeutic considerations of PARP in stem cell biology: Relevance in cancer and beyond. Biochem. Pharmacol. 2019, 167, 107–115. [Google Scholar] [CrossRef]
- Liu, Y.; Burness, M.L.; Martin-Trevino, R.; Guy, J.; Bai, S.; Harouaka, R.; Brooks, M.D.; Shang, L.; Fox, A.; Luther, T.K.; et al. RAD51 Mediates Resistance of Cancer Stem Cells to PARP Inhibition in Triple-Negative Breast Cancer. Clin. Cancer Res. 2017, 23, 514–522. [Google Scholar] [CrossRef] [PubMed]
- McClements, L.; Yakkundi, A.; Papaspyropoulos, A.; Harrison, H.; Ablett, M.P.; Jithesh, P.V.; McKeen, H.D.; Bennett, R.; Donley, C.; Kissenpfennig, A.; et al. Targeting Treatment-Resistant Breast Cancer Stem Cells with FKBPL and Its Peptide Derivative, AD-01, via the CD44 Pathway. Clin. Cancer Res. 2013, 19, 3881–3893. [Google Scholar] [CrossRef] [PubMed]
- Chesnokov, M.S.; Khan, I.; Park, Y.; Ezell, J.; Mehta, G.; Yousif, A.; Hong, L.J.; Buckanovich, R.J.; Takahashi, A.; Chefetz, I. The MEK1/2 Pathway as a Therapeutic Target in High-Grade Serous Ovarian Carcinoma. Cancers 2021, 13, 1369. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarty, A.; Bhola, N.E.; Sutton, C.; Ghosh, R.; Kuba, M.G.; Dave, B.; Chang, J.C.; Arteaga, C.L. Trastuzumab-resistant cells rely on a HER2-PI3K-FoxO-survivin axis and are sensitive to PI3K inhibitors. Cancer Res. 2013, 73, 1190–1200. [Google Scholar] [CrossRef] [PubMed]
- Shank, J.J.; Yang, K.; Ghannam, J.; Cabrera, L.; Johnston, C.J.; Reynolds, R.K.; Buckanovich, R.J. Metformin targets ovarian cancer stem cells in vitro and in vivo. Gynecol. Oncol. 2012, 127, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Hampsch, R.A.; Wells, J.D.; Traphagen, N.A.; McCleery, C.F.; Fields, J.L.; Shee, K.; Dillon, L.M.; Pooler, D.B.; Lewis, L.D.; Demidenko, E.; et al. AMPK Activation by Metformin Promotes Survival of Dormant ERþ Breast Cancer Cells. Clin. Cancer Res. 2020, 26, 3707–3719. [Google Scholar] [CrossRef]
- Lee, H.G.; Shin, S.-J.J.; Chung, H.-W.W.; Kwon, S.-H.H.; Cha, S.-D.D.; Lee, J.-E.E.; Cho, C.-H.H. Salinomycin reduces stemness and induces apoptosis on human ovarian cancer stem cell. J. Gynecol. Oncol. 2017, 28, e14. [Google Scholar] [CrossRef]
- Mi, Y.; Huang, Y.; Deng, J. The enhanced delivery of salinomycin to CD133+ ovarian cancer stem cells through CD133 antibody conjugation with poly(lactic-co-glycolic acid)-poly(ethylene glycol) nanoparticles. Oncol. Lett. 2018, 15, 6611–6621. [Google Scholar] [CrossRef]
- Lee, H.; Kim, J.W.; Kim, D.K.; Choi, D.K.; Lee, S.; Yu, J.H.; Kwon, O.-B.; Lee, J.; Lee, D.-S.; Kim, J.H.; et al. Calcium Channels as Novel Therapeutic Targets for Ovarian Cancer Stem Cells. Int. J. Mol. Sci. 2020, 21, 2327. [Google Scholar] [CrossRef]
- Alqudah, M.; Al-Samman, R.; Azaizeh, M.; Alzoubi, K. Amlodipine inhibits proliferation, invasion, and colony formation of breast cancer cells. Biomed. Rep. 2022, 16, 50. [Google Scholar] [CrossRef]
- Chang, Y.-H.; Lin, Y.-J.; Huang, C.-Y.; Harnod, T.; Ding, D.-C. Shikonin impedes type 2 ovarian cancer progression via FasL/caspase-8 and mir-874-3p/XIAP axis and prohibits the properties of stemness. Am. J. Cancer Res. 2022, 12, 4584–4601. [Google Scholar] [PubMed]
- Thakur, R.; Trivedi, R.; Rastogi, N.; Singh, M.; Mishra, D.P. Inhibition of STAT3, FAK and Src mediated signaling reduces cancer stem cell load, tumorigenic potential and metastasis in breast cancer. Sci. Rep. 2015, 5, 10194. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zhao, F.; Wu, D.; Zhang, Y.; Bao, X.; Shi, F.; Cai, Y.; Dou, J. Eliciting effective tumor immunity against ovarian cancer by cancer stem cell vaccination. Biomed. Pharmacother. 2023, 161, 114547. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Yu, X.; Wang, J.; Hui, X.; Zhang, Y.; Cai, Y.; Ren, M.; Guo, M.; Zhao, F.; Dou, J. Ovarian Cancer Stem Cells with High ROR1 Expression Serve as a New Prophylactic Vaccine for Ovarian Cancer. J. Immunol. Res. 2019, 2019, 9394615. [Google Scholar] [CrossRef]
- Wu, D.; Wang, J.; Cai, Y.; Ren, M.; Zhang, Y.; Shi, F.; Zhao, F.; He, X.; Pan, M.; Yan, C.; et al. Effect of targeted ovarian cancer immunotherapy using ovarian cancer stem cell vaccine. J. Ovarian Res. 2015, 8, 68. [Google Scholar] [CrossRef]
- Ruiu, R.; Di Lorenzo, A.; Cavallo, F.; Conti, L. Are Cancer Stem Cells a Suitable Target for Breast Cancer Immunotherapy? Front. Oncol. 2022, 12, 877384. [Google Scholar] [CrossRef]
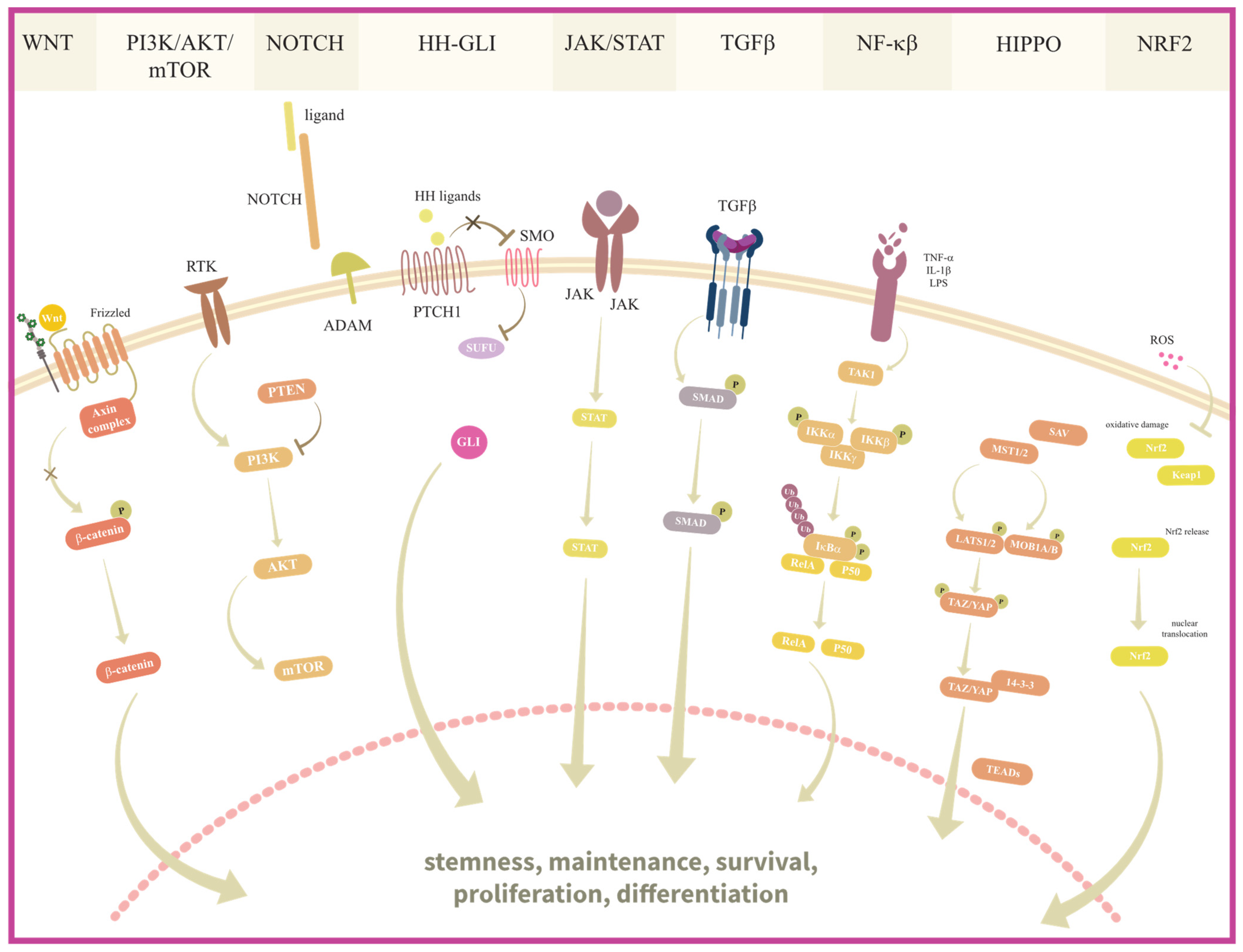
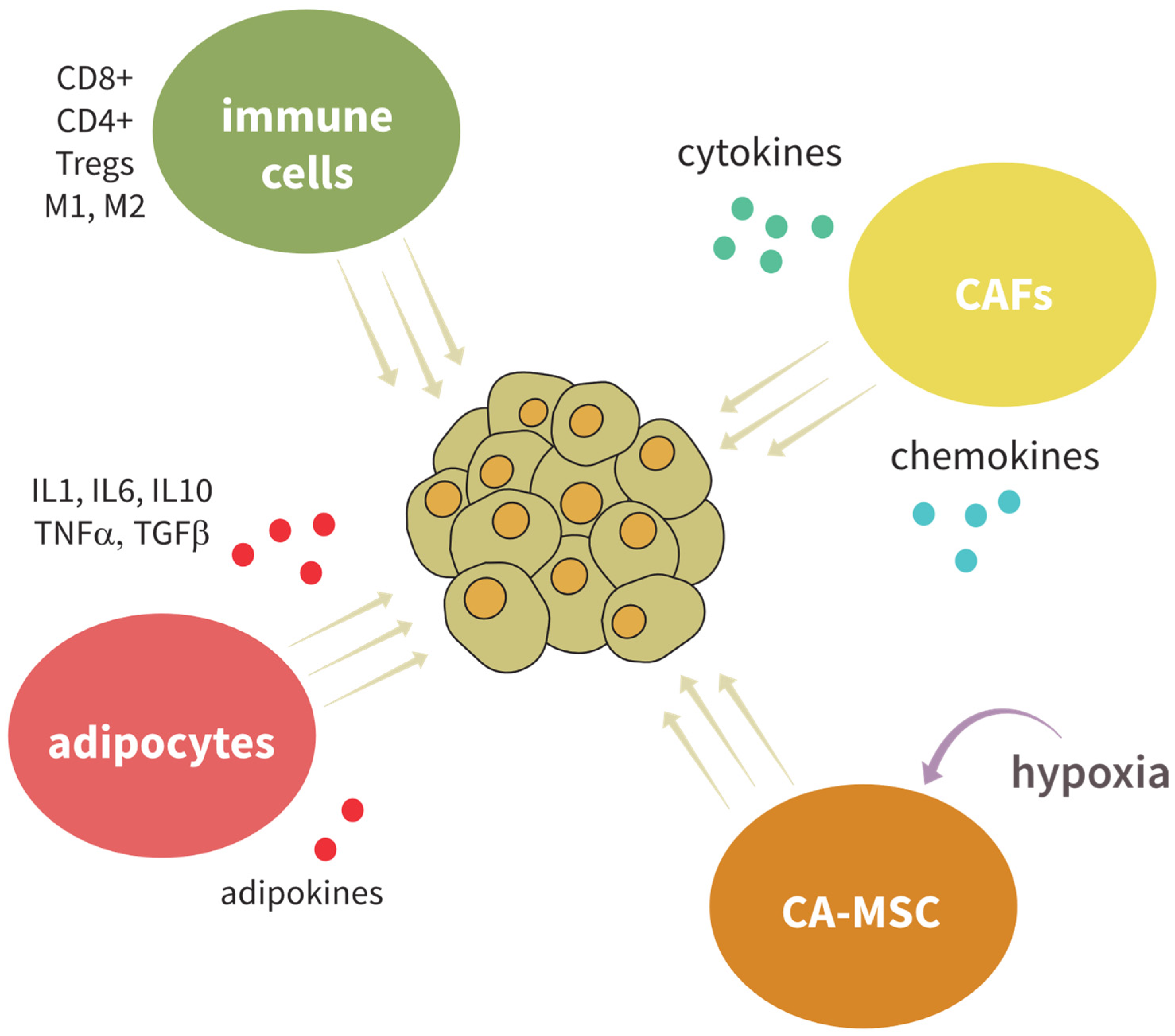
| Compound | Mode of Action | Experimental Model | Effect |
|---|---|---|---|
| Salinomycin | Membrane ionophore antibiotic | BC cell lines | Reduces BCSC proportion, and improves survival [416]. |
| Salinomycin-HDL | OC cell line (CD133+ population), normal ovarian epithelial line, xenografts | Downregulation of stemness markers (MYCN, NANOG, OCT4, SOX2) in vitro and in vivo, induction of apoptosis [417]. | |
| Metformin | Antidiabetic agent | BC cell lines (spheroids) | Decreases CD44+/CD24−/population and mammospheres formation [418]. |
| OC cell lines (spheroids), xenografts | Inhibits the CD44+ CD117+ population, no effect on CD44+ ALDH+ population, inhibits EMT [163]. | ||
| Emodin | Anti-malarial and anti-allergic agent | BC cell lines and mouse models | Suppresses TGF-β1 production, reduces macrophage-induced EMT and BCSC formation, and reduces breast cancer lung metastasis [419]. |
| OC cell lines, xenografts | Inhibits the growth of OC cells in vitro and in vivo, reduces the number of CD44+/CD24− OCSC [420]. | ||
| 1α,25-dihydroxyvitamin D3 | Vitamin | BC cell lines | Inhibition of spheroid formation, downregulation of stemness markers (OCT4, CD44, NOTCH1, NOTCH2, NOTCH3) [421]. |
| OC cell lines, xenografts | Inhibition of spheroid formation, downregulation of stemness markers (OCT4, CD44, NANOG, SOX2, KLF4, ABCG2), delays onset of tumor formation [422]. | ||
| Luteolin | Flavonoid, antioxidant | BC cell lines, xenografts | Inhibition of stemness and chemoresistance via the NRF2-mediated pathway [423] and suppresses EMT and migration of TNBC cells by inhibition of Hippo/YAP pathway [424]. |
| OC cell line, primary OC (CD133+ ALDH+ populations), xenografts | Inhibition of stemness, inhibition of Hippo/YAP pathway, sensitization to paclitaxel and carboplatin [425]. | ||
| ALM201 | Anti-angiogenic therapeutic peptide | BC cell lines (spheroids), xenografts | Inhibition of spheroid formation, decrease in in vivo lung metastasis, and sensitization to tamoxifen by downregulating NOTCH4 and DLL4 [426]. |
| OC cell lines, xenografts | Inhibition of spheroid formation, inhibition of CD44+ CD117+ population, induction of differentiation [427]. | ||
| Simvastatin | Inhibition of cholesterol synthesis | BC cell lines (spheroids), PDX | Radiosensitizes mammospheres of inflammatory BC (IBC) and TNBC, yet radioprotects HR+ and HER2+ [428]; inhibition of spheroid formation in PDX-derived TNBC [429]. |
| OC cell lines, primary cultures, xenografts | Inhibition of spheroid formation, downregulation of CD44 and ALDH1A1, reduction of tumorigenesis and metastasis in vivo [430]. | ||
| All-trans retinoic acid (ATRA) | Vitamin A metabolite | BC cell lines, xenografts | Induces differentiation, inhibits invasiveness and migration of BCSC, and sensitizes to chemotherapy [431]. |
| OC cell lines (ALDH-high and ALDH-low), xenografts | Inhibition of stemness properties, inhibition of p62/NRF2 axis, decreases in vivo tumor growth only in ALDH-high OCSC [248]. | ||
| Entinostat | HDAC inhibitors | BC cell lines, mouse model | Reduces CD44+/CD24− cell population, ALDH-1 activity, BMI-1, NANOG, and OCT4. Reduces tumor formation at the primary site and lung metastasis [432]. |
| 16cyc-HxA, 16lin-HxA, and 16KA | OC cell lines, xenografts | Induction of apoptosis in OCSC, reduction of tumor size in vivo [433]. | |
| Eugenol | Inhibition of NOTCH pathway | BC cell lines, mouse model | Downregulation of stemness of secondary mammosphere, lower CD44+/CD24−/low population, stemness suppression [434]. |
| OC cell lines, xenografts | Reduction of chemotherapy-induced spheroid formation, inhibition of CD44+ and ALDH+ cells, increases tumor-free survival [435]. | ||
| Saracatinib + gemcitabine | Inhibition of SRC + chemotherapeutic | BC cell lines, mouse model | Synergistic effect, reverse drug resistance, inhibition tumor stemness, metastasis, and growth of BCSC (CD44+ OCT4+) [436]. |
| Saracatinib + selumetinib | SRC inhibitor + MEK inhibitor | OC cell lines, primary cell lines, xenografts | Synergistic effect on cell cycle arrest, induction of apoptosis and autophagy, decrease in ALDH1+ population in vitro and in vivo [437]. |
| Everolimus | Inhibitor of PI3K/Akt/mTOR signaling pathway | BC cell lines | Reverses Palbociclib resistance, decreases the expression of ALDH1 and NANOG, decreases cell migration, self-renewal, and EMT [438]. |
| 5-aza-2′-deoxycytidine | Inhibitor of DNA methylation | BC cell lines | Decreases the expression of CD44+/CD24−, activation of P53 expression, increases sensitivity to chemotherapy [439] |
| Everolimus + 5-aza-2-deoxycytidine | mTOR inhibitor + inhibitor of DNA methylation | OC cell lines, xenografts | Synergistic effect on OC tumorspheres (ALDH1+ CD44+) and tumor formation in vivo, induction of apoptosis [440]. |
| Disulfiram + cisplatin + paclitaxel | ALDH inhibitor + chemotherapy | BC cell lines | Decreases ALDH activity, increases expression of SOX2 and NANOG. Disulfiram sensitizes BCSCs to chemotherapeutics [441]. |
| OC cell lines | Synergistic effect on OC cells, re-sensitization of cisplatin-resistant lines [442]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lučić, I.; Kurtović, M.; Mlinarić, M.; Piteša, N.; Čipak Gašparović, A.; Sabol, M.; Milković, L. Deciphering Common Traits of Breast and Ovarian Cancer Stem Cells and Possible Therapeutic Approaches. Int. J. Mol. Sci. 2023, 24, 10683. https://doi.org/10.3390/ijms241310683
Lučić I, Kurtović M, Mlinarić M, Piteša N, Čipak Gašparović A, Sabol M, Milković L. Deciphering Common Traits of Breast and Ovarian Cancer Stem Cells and Possible Therapeutic Approaches. International Journal of Molecular Sciences. 2023; 24(13):10683. https://doi.org/10.3390/ijms241310683
Chicago/Turabian StyleLučić, Ivan, Matea Kurtović, Monika Mlinarić, Nikolina Piteša, Ana Čipak Gašparović, Maja Sabol, and Lidija Milković. 2023. "Deciphering Common Traits of Breast and Ovarian Cancer Stem Cells and Possible Therapeutic Approaches" International Journal of Molecular Sciences 24, no. 13: 10683. https://doi.org/10.3390/ijms241310683
APA StyleLučić, I., Kurtović, M., Mlinarić, M., Piteša, N., Čipak Gašparović, A., Sabol, M., & Milković, L. (2023). Deciphering Common Traits of Breast and Ovarian Cancer Stem Cells and Possible Therapeutic Approaches. International Journal of Molecular Sciences, 24(13), 10683. https://doi.org/10.3390/ijms241310683









