Control of Bacterial Phenotype and Chromosomal Gene Expression by Single Plasmids of Lactococcus lactis IL594
Abstract
1. Introduction
2. Results
2.1. Presence of Plasmids Increases Ability to Utilize Carbon Compounds
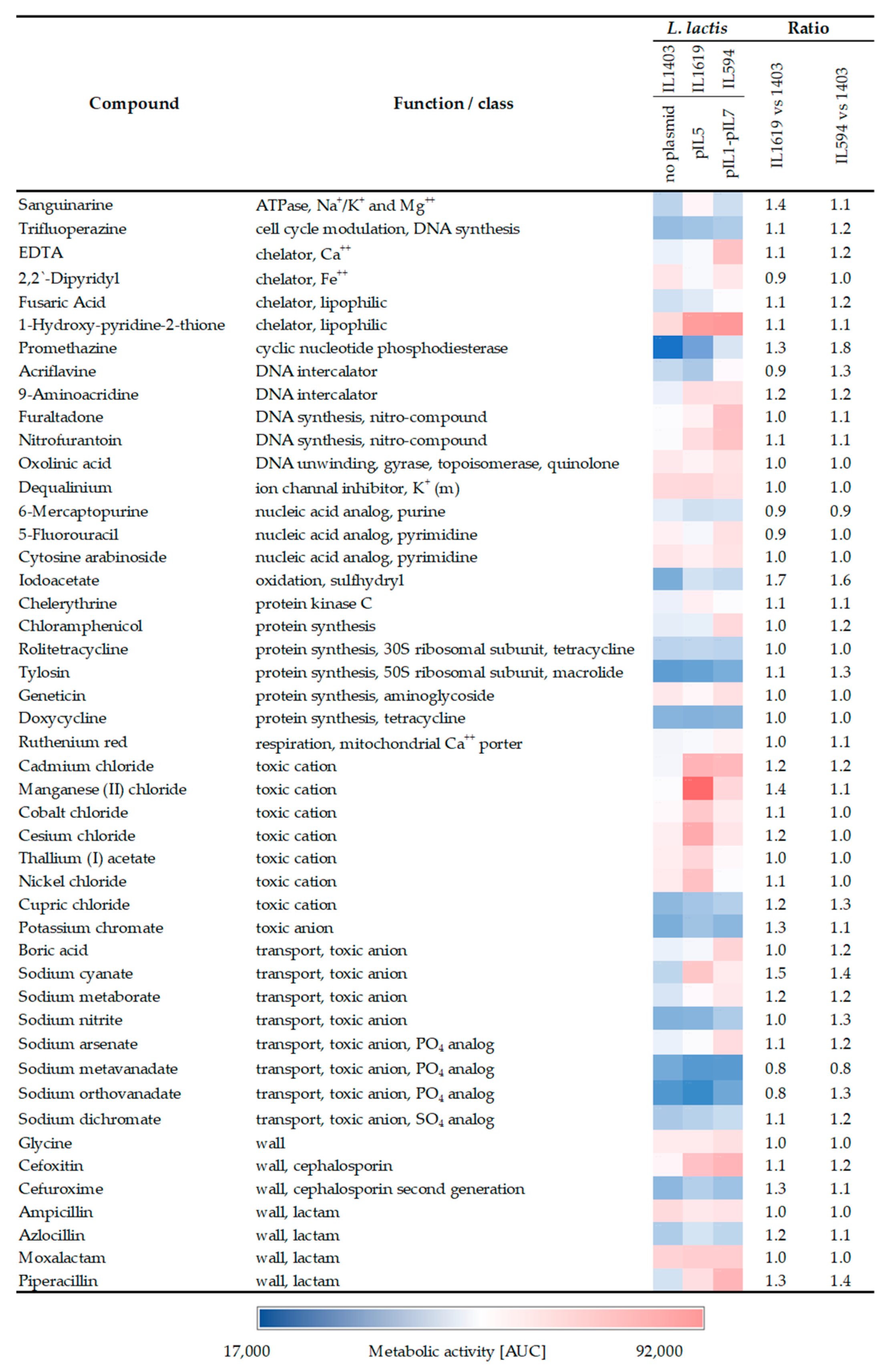
2.2. Presence of pIL5 Enhances L. lactis Tolerance to Antimicrobial Compounds
2.3. The Presence of Single Plasmids Changes the Expression of Chromosomal Genes
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Metabolic Activity and Resistance Assays
4.3. Oligonucleotide Microarrays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wels, M.; Siezen, R.; Van Hijum, S.; Kelly, W.J.; Bachmann, H. Comparative genome analysis of Lactococcus lactis indicates niche adaptation and resolves genotype/phenotype disparity. Front. Microbiol. 2019, 10, 4. [Google Scholar] [CrossRef] [PubMed]
- Tavares, L.M.; de Jesus, L.C.L.; da Silva, T.F.; Barroso, F.A.L.; Batista, V.L.; Coelho-Rocha, N.D.; Azevedo, V.; Drumond, M.M.; Mancha-Agresti, P. Novel strategies for efficient production and delivery of live biotherapeutics and biotechnological uses of Lactococcus lactis: The Lactic Acid Bacterium model. Front. Bioeng. Biotechnol. 2020, 8, 517166. [Google Scholar] [CrossRef]
- De Castro, C.P.; Drumond, M.M.; Batista, V.L.; Nunes, A.; Mancha-Agresti, P.; Azevedo, V.A.C. Vector development timeline for mucosal vaccination and treatment of disease using Lactococcus lactis and design approaches of next generation food grade plasmids. Front. Microbiol. 2018, 9, 1805. [Google Scholar] [CrossRef]
- Batista, V.L.; da Silva, T.F.; de Jesus, L.C.L.; Tapia-Costa, A.P.; Drumond, M.M.; Azevedo, V.; Mancha-Agresti, P. Lactic Acid Bacteria as delivery vehicle for therapeutics applications. Methods Mol. Biol. 2021, 2183, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Ainsworth, S.; Stockdale, S.; Bottacini, F.; Mahony, J.; van Sinderen, D. The Lactococcus lactis plasmidome: Much learnt, yet still lots to discover. FEMS Microbiol. Rev. 2014, 38, 1066–1088. [Google Scholar] [CrossRef] [PubMed]
- Van Mastrigt, O.; Di Stefano, E.; Hartono, S.; Abee, T.; Smid, E.J. Large plasmidome of dairy Lactococcus lactis subsp. lactis biovar diacetylactis FM03P encodes technological functions and appears highly unstable. BMC Genom. 2018, 19, 620. [Google Scholar] [CrossRef]
- Górecki, R.K.; Koryszewska-Bagińska, A.; Gołębiewski, M.; Żylińska, J.; Grynberg, M.; Bardowski, J.K. Adaptative potential of the Lactococcus lactis IL594 strain encoded in its 7 plasmids. PLoS ONE 2011, 6, e22238. [Google Scholar] [CrossRef]
- Kobayashi, M.; Nomura, M.; Fujita, Y.; Okamoto, T.; Ohmomo, S. Influence of lactococcal plasmid on the specific growth rate of host cells. Lett. Appl. Microbiol. 2002, 35, 403–408. [Google Scholar] [CrossRef]
- Bolotin, A.; Wincker, P.; Mauger, S.; Jaillon, O.; Malarme, K.; Weissenbach, J.; Ehrlich, S.D.; Sorokin, A. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 2001, 11, 731–753. [Google Scholar] [CrossRef]
- Johansen, E. Challenges when transferring technology from Lactococcus laboratory strains to industrial strains. Genet. Mol. Res. 2003, 2, 112–116. [Google Scholar]
- Kosiorek, K.; Koryszewska-Bagińska, A.; Skoneczny, M.; Stasiak-Różańska, L.; Aleksandrzak-Piekarczyk, T. The presence of plasmids in Lactococcus lactis IL594 determines changes in the host phenotype and expression of chromosomal genes. Int. J. Mol. Sci. 2023, 24, 793. [Google Scholar] [CrossRef] [PubMed]
- Chopin, A.; Chopin, M.C.; Moillo-Batt, A.; Langella, P. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid 1984, 11, 260–263. [Google Scholar] [CrossRef]
- Barrière, C.; Veiga-da-Cunha, M.; Pons, N.; Guédon, E.; van Hijum, S.A.F.T.; Kok, J.; Kuipers, O.P.; Ehrlich, D.S.; Renault, P. Fructose Utilization in Lactococcus lactis as a model for low-GC Gram-positive bacteria: Its regulator, signal, and DNA-binding Site. J. Bacteriol. 2005, 187, 3752–3761. [Google Scholar] [CrossRef] [PubMed]
- Tymoszewska, A.; Diep, D.B.; Wirtek, P.; Aleksandrzak-Piekarczyk, T. The non-lantibiotic bacteriocin Garvicin Q targets Man-PTS in a broad spectrum of sensitive bacterial genera. Sci. Rep. 2017, 7, 8359. [Google Scholar] [CrossRef] [PubMed]
- Castro, R.; Neves, A.R.; Fonseca, L.L.; Pool, W.A.; Kok, J.; Kuipers, O.P.; Santos, H. Characterization of the individual glucose uptake systems of Lactococcus lactis: Mannose-PTS, cellobiose-PTS and the novel GlcU permease. Mol. Microbiol. 2009, 71, 795–806. [Google Scholar] [CrossRef]
- Aleksandrzak-Piekarczyk, T.; Szatraj, K.; Kosiorek, K. GlaR (YugA)-a novel RpiR-family transcription activator of the Leloir pathway of galactose utilization in Lactococcus lactis IL1403. Microbiologyopen 2019, 8, e00714. [Google Scholar] [CrossRef]
- Siezen, R.J.; Starrenburg, M.J.C.; Boekhorst, J.; Renckens, B.; Molenaar, D.; van Hylckama Vlieg, J.E.T. Genome-scale genotype-phenotype matching of two Lactococcus lactis isolates from plants identifies mechanisms of adaptation to the plant niche. Appl. Environ. Microbiol. 2008, 74, 424–436. [Google Scholar] [CrossRef]
- Christen, S.; Srinivas, A.; Bähler, P.; Zeller, A.; Pridmore, R.; Bieniossek, C.; Baumann, U.; Erni, B. Regulation of Dha operon of Lactococcus lactis A deviation from the rule followed by the TetR family of transcription regulators. J. Biol. Chem. 2006, 281, 23129–23137. [Google Scholar] [CrossRef]
- Stasiak-Różańska, L.; Kupiec, M. Industrial applications of wild and genetically-modified strains of acetic acid bacteria. Postępy Mikrobiol.-Adv. Microbiol. 2018, 57, 398–402. [Google Scholar] [CrossRef]
- Crupper, S.S.; Worrell, V.; Stewart, G.C.; Iandolo, J.J. Cloning and expression of cadD, a new cadmium resistance gene of Staphylococcus aureus. J. Bacteriol. 1999, 181, 4071–4075. [Google Scholar] [CrossRef]
- Monachese, M.; Burton, J.P.; Reid, G. Bioremediation and tolerance of humans to heavy metals through microbial processes: A potential role for probiotics? Appl. Environ. Microbiol. 2012, 78, 6397–6404. [Google Scholar] [CrossRef] [PubMed]
- Pajarillo, E.A.B.; Lee, E.; Kang, D.-K. Trace metals and animal health: Interplay of the gut microbiota with iron, manganese, zinc, and copper. Anim. Nutr. 2021, 7, 750–761. [Google Scholar] [CrossRef] [PubMed]
- Nelson, N. Metal ion transporters and homeostasis. EMBO J. 1999, 18, 4361–4371. [Google Scholar] [CrossRef]
- Nawaz, M.; Wang, J.; Zhou, A.; Ma, C.; Wu, X.; Moore, J.E.; Millar, B.C.; Xu, J. Characterization and transfer of antibiotic resistance in lactic acid bacteria from fermented food products. Curr. Microbiol. 2011, 62, 1081–1089. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, Y.; Ponce-Alquicira, E. Antibiotic resistance in Lactic Acid Bacteria. In Antimicrobial Resistance-a Global Threat; IntechOpen: London, UK, 2018. [Google Scholar]
- Gad, G.F.M.; Abdel-Hamid, A.M.; Farag, Z.S.H. Antibiotic resistance in lactic acid bacteria isolated from some pharmaceutical and dairy products. Braz. J. Microbiol. 2014, 45, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Lubelski, J.; Mazurkiewicz, P.; van Merkerk, R.; Konings, W.N.; Driessen, A.J.M. ydaG and ydbA of Lactococcus lactis encode a heterodimeric ATP-binding cassette-type multidrug transporter. J. Biol. Chem. 2004, 279, 34449–34455. [Google Scholar] [CrossRef]
- Poelarends, G.J.; Mazurkiewicz, P.; Konings, W.N. Multidrug transporters and antibiotic resistance in Lactococcus lactis. Biochim. Biophys. Acta 2002, 1555, 1–7. [Google Scholar] [CrossRef]
- Kramer, N.E.; van Hijum, S.A.F.T.; Knol, J.; Kok, J.; Kuipers, O.P. Transcriptome analysis reveals mechanisms by which Lactococcus lactis acquires nisin resistance. Antimicrob. Agents Chemother. 2006, 50, 1753–1761. [Google Scholar] [CrossRef]
- Giaouris, E.; Briandet, R.; Meyrand, M.; Courtin, P.; Chapot-Chartier, M.-P. Variations in the degree of alanylation of teichoic acids in Lactococcus lactis alter resistance to cationic antimicrobials but have no effect on bacterial surface hydrophobicity and charge. Appl. Environ. Microbiol. 2008, 74, 4764–4767. [Google Scholar] [CrossRef]
- McBride, S.M.; Sonenshein, A.L. The dlt operon confers resistance to cationic antimicrobial peptides in Clostridium difficile. Microbiology 2011, 157, 1457–1465. [Google Scholar] [CrossRef]
- Campelo, A.B.; López-González, M.J.; Escobedo, S.; Janzen, T.; Neves, A.R.; Rodríguez, A.; Martínez, B. Mutations selected after exposure to bacteriocin Lcn972 activate a bce-like bacitracin resistance module in Lactococcus lactis. Front. Microbiol. 2020, 11, 1805. [Google Scholar] [CrossRef] [PubMed]
- Tymoszewska, A.; Ovchinnikov, K.V.; Diep, D.B.; Słodownik, M.; Maron, E.; Martínez, B.; Aleksandrzak-Piekarczyk, T. Lactococcus lactis resistance to Aureocin A53- and enterocin L50-like bacteriocins and membrane-targeting peptide antibiotics relies on the YsaCB-KinG-LlrG four-component system. Antimicrob. Agents Chemother. 2021, 65, e00921-21. [Google Scholar] [CrossRef] [PubMed]
- Van Domselaar, G.H.; Stothard, P.; Shrivastava, S.; Cruz, J.A.; Guo, A.; Dong, X.; Lu, P.; Szafron, D.; Greiner, R.; Wishart, D.S. BASys: A web server for automated bacterial genome annotation. Nucleic Acids Res. 2005, 33, W455–W459. [Google Scholar] [CrossRef] [PubMed]


| L. lactis Strain | IL1618 | IL1421 | IL2661 | IL1619 | IL1420 | IL1530 | IL1392 | IL594 |
|---|---|---|---|---|---|---|---|---|
| Plasmid (s) | pIL1 | pIL3 | pIL4 | pIL5 | pIL6 | pIL7 | pIL1, pIL2, pIL3, pIL5 | pIL1–pIL7 |
| Upregulated genes | 90 | 98 | 89 | 88 | 37 | 25 | 148 | 21 |
| Downregulated genes | 30 | 33 | 30 | 28 | 31 | 17 | 41 | 23 |
| Total genes affected | 120 | 131 | 119 | 116 | 68 | 42 | 189 | 44 |
| % of chromosomal genes | 5% | 6% | 5% | 5% | 3% | 2% | 9% | 2% |
| L. lactis Strain | Genotypic Characteristics | Source |
|---|---|---|
| IL594 | wild-type strain carrying seven plasmids: pIL1–pIL7 | INRA |
| IL1618 | carrying pIL1 plasmid | INRA |
| IL1392 | carrying four plasmids: pIL1, pIL2, pIL3, pIL5 | INRA |
| IL1421 | carrying pIL3 plasmid | INRA |
| IL2661 | carrying pIL4 plasmid | INRA |
| IL1619 | carrying pIL5 plasmid | INRA |
| IL1420 | carrying pIL6 plasmid | INRA |
| IL1530 | carrying pIL7 plasmid | INRA |
| IL1403 | plasmid-free strain, derivative of IL594 | INRA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kosiorek, K.; Koryszewska-Bagińska, A.; Skoneczny, M.; Aleksandrzak-Piekarczyk, T. Control of Bacterial Phenotype and Chromosomal Gene Expression by Single Plasmids of Lactococcus lactis IL594. Int. J. Mol. Sci. 2023, 24, 9877. https://doi.org/10.3390/ijms24129877
Kosiorek K, Koryszewska-Bagińska A, Skoneczny M, Aleksandrzak-Piekarczyk T. Control of Bacterial Phenotype and Chromosomal Gene Expression by Single Plasmids of Lactococcus lactis IL594. International Journal of Molecular Sciences. 2023; 24(12):9877. https://doi.org/10.3390/ijms24129877
Chicago/Turabian StyleKosiorek, Katarzyna, Anna Koryszewska-Bagińska, Marek Skoneczny, and Tamara Aleksandrzak-Piekarczyk. 2023. "Control of Bacterial Phenotype and Chromosomal Gene Expression by Single Plasmids of Lactococcus lactis IL594" International Journal of Molecular Sciences 24, no. 12: 9877. https://doi.org/10.3390/ijms24129877
APA StyleKosiorek, K., Koryszewska-Bagińska, A., Skoneczny, M., & Aleksandrzak-Piekarczyk, T. (2023). Control of Bacterial Phenotype and Chromosomal Gene Expression by Single Plasmids of Lactococcus lactis IL594. International Journal of Molecular Sciences, 24(12), 9877. https://doi.org/10.3390/ijms24129877






