Therapeutic Effects of microRNAs on Nonalcoholic Fatty Liver Disease (NAFLD) and Nonalcoholic Steatohepatitis (NASH): A Systematic Review and Meta-Analysis
Abstract
1. Introduction
2. Results and Discussion
2.1. Results
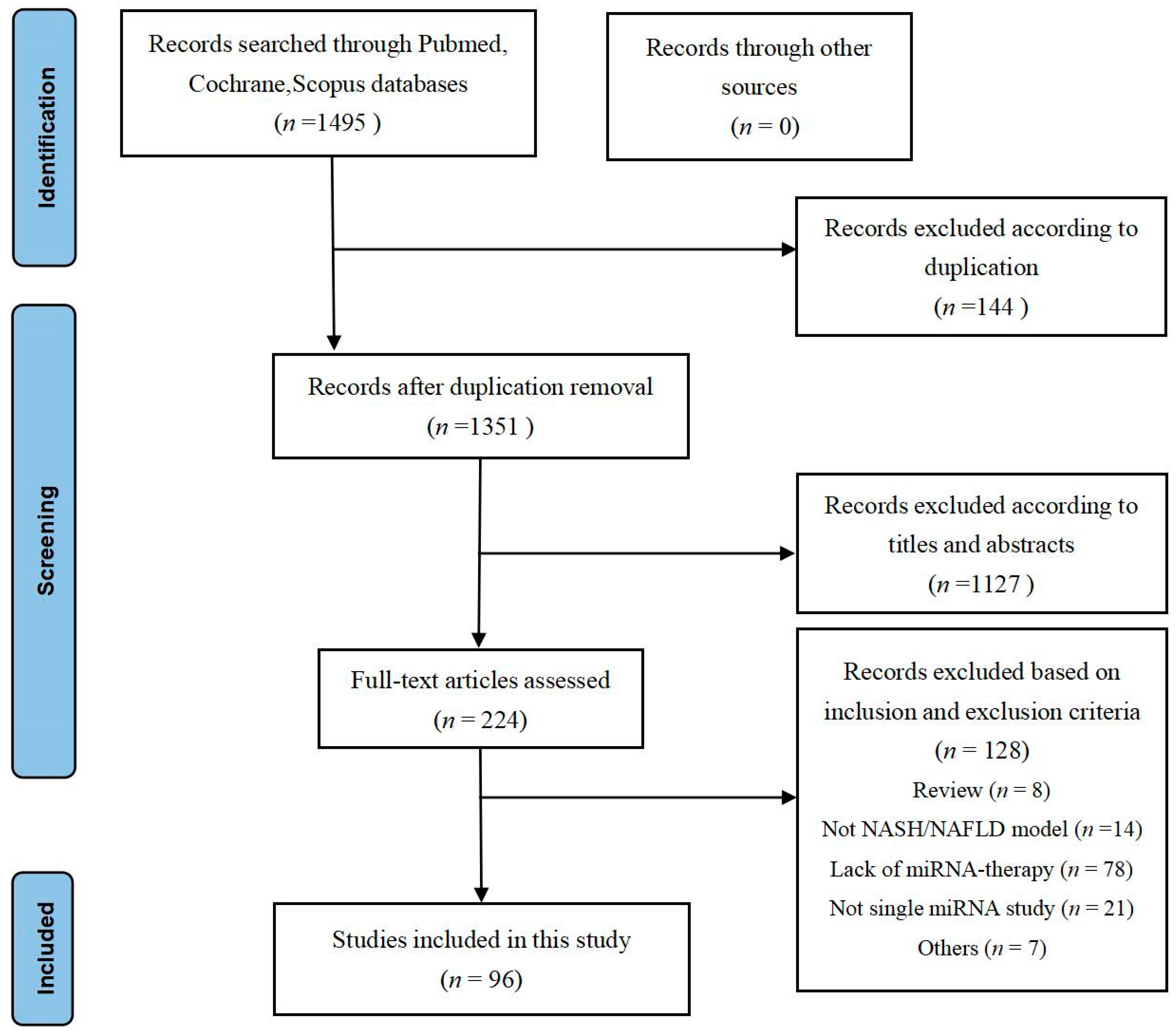
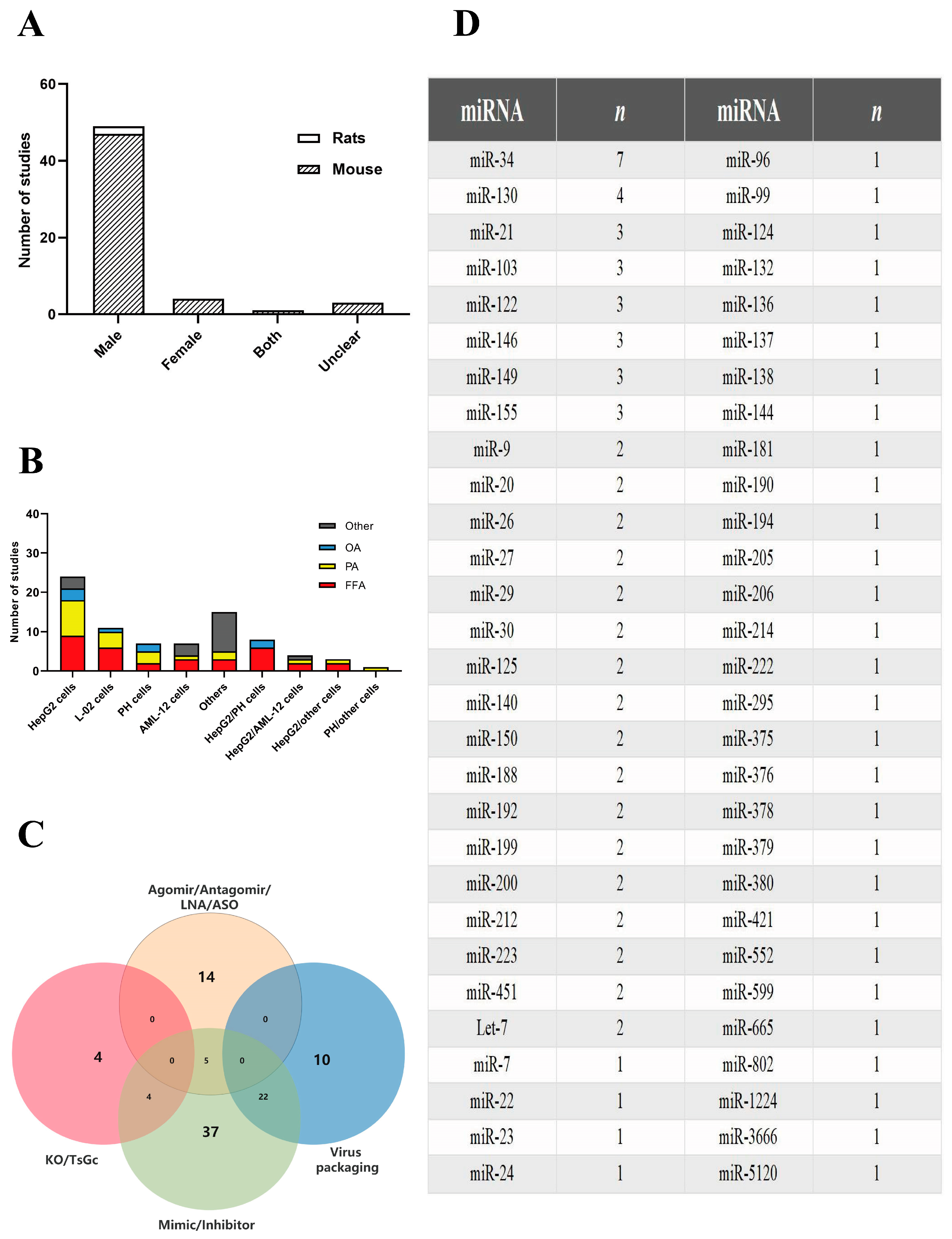
2.1.1. Study Characteristics
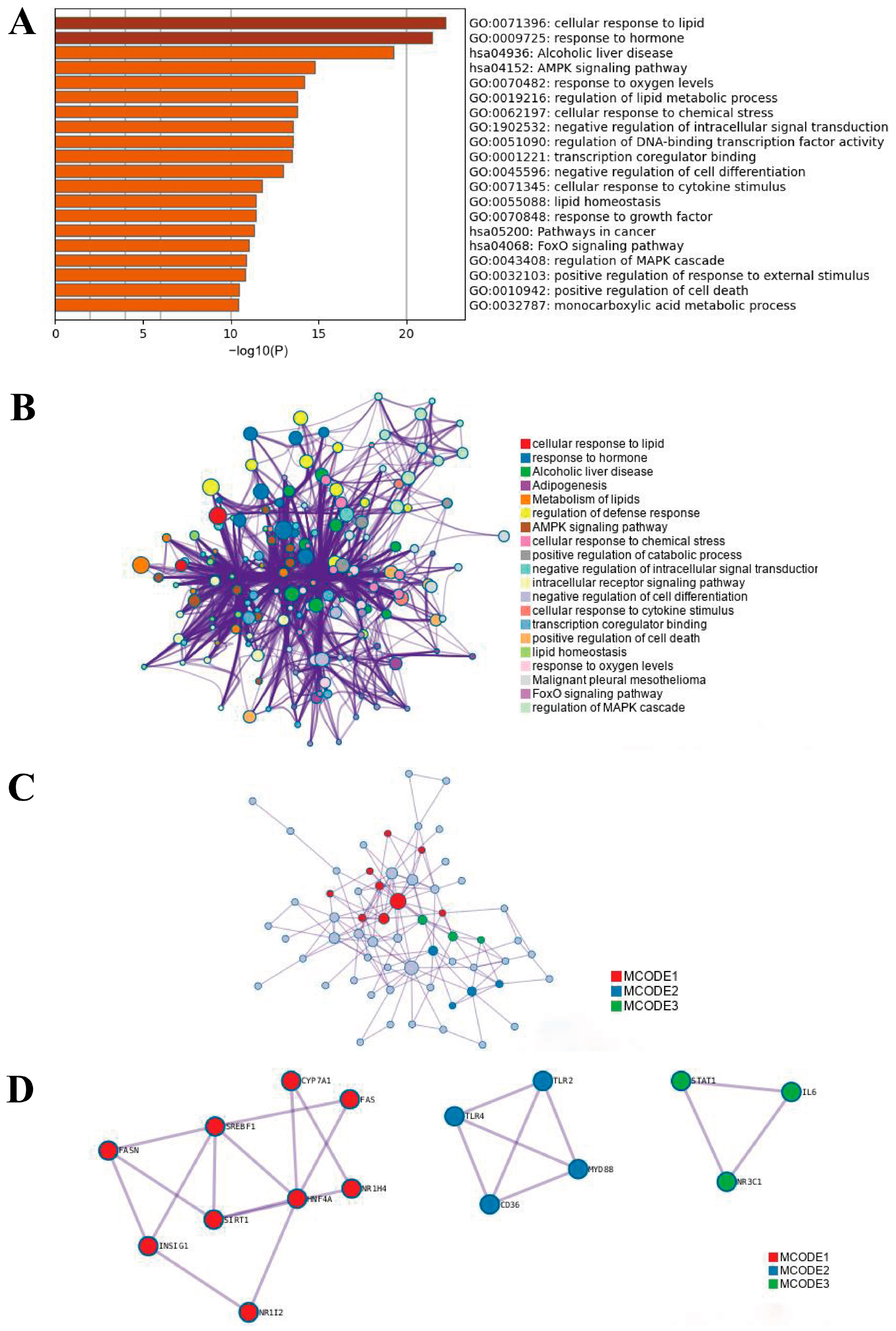
2.1.2. Functional Enrichment Analysis
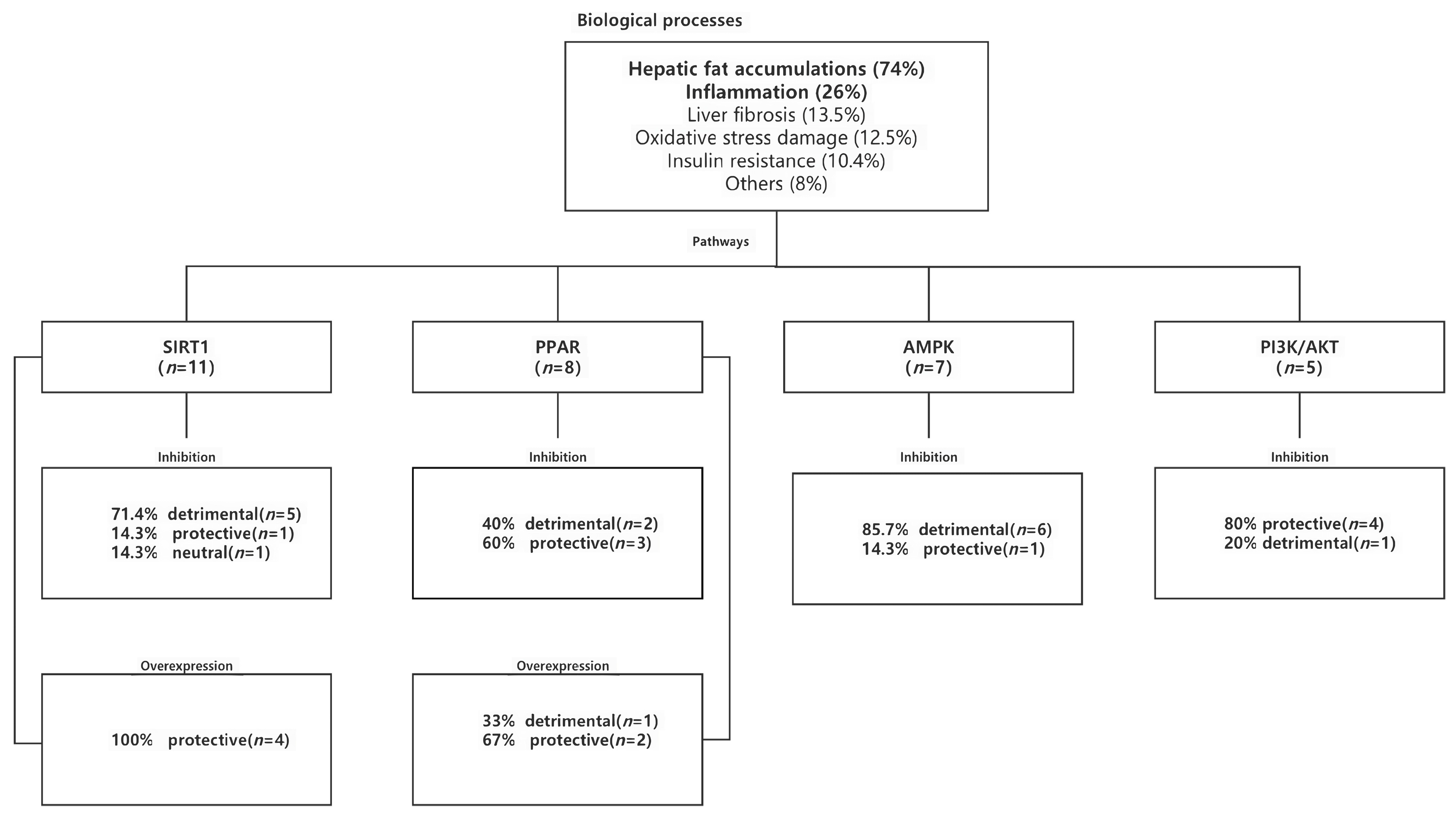
2.1.3. Biologic Processes
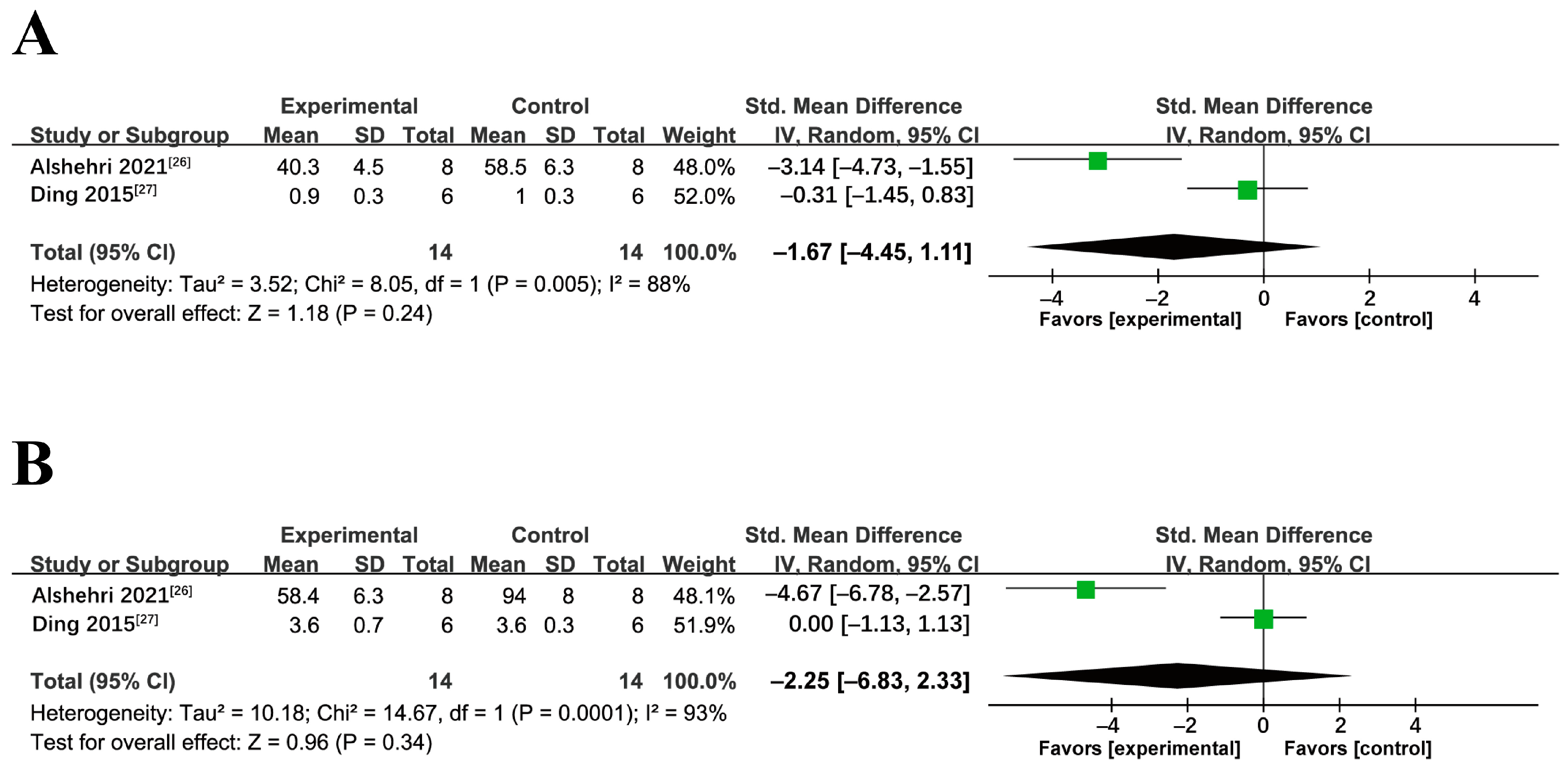
2.1.4. Meta-Analysis of miRNA Interventions on NAFLD/NASH
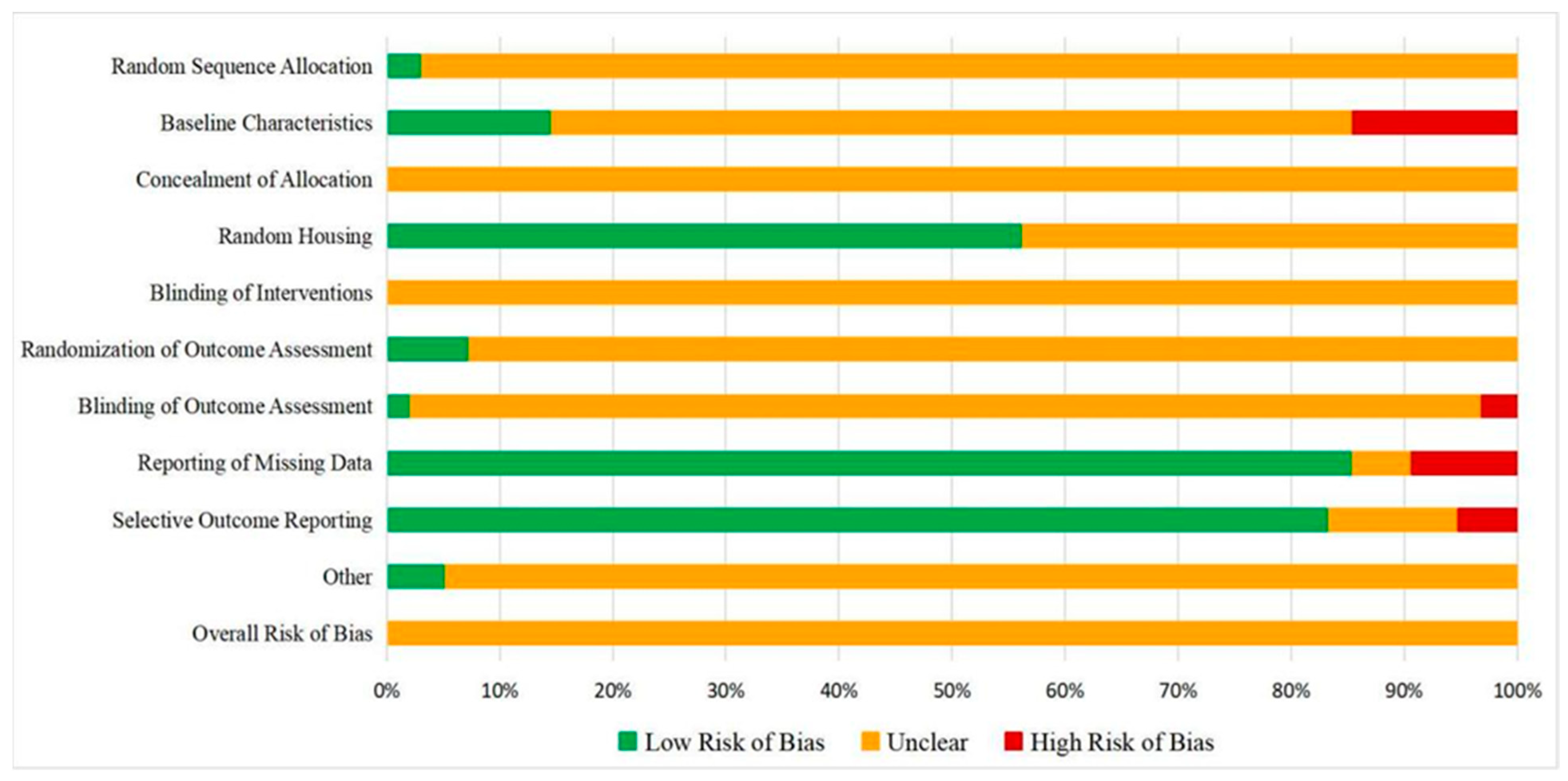
2.1.5. Quality Assessment
2.1.6. Publication Bias
2.2. Discussion
3. Materials and Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Friedman, S.L.; Neuschwander-Tetri, B.A.; Rinella, M.; Sanyal, A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 2018, 24, 908–922. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.; Anstee, Q.M.; Marietti, M.; Hardy, T.; Henry, L.; Eslam, M.; George, J.; Bugianesi, E. Global burden of NAFLD and NASH: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Eissa, A.H.A.; Nawawi, K.N.M.; Mohamad, M.S.F.; Wong, Z.; Hassan, H.H.C.; Maskon, O.; Yaacob, N.Y.; Ali, R.A.R. Association between Non Alcoholic Fatty Liver Disease and Severity of Coronary Artery Disease in Patients Aged 45 Years and Below with Acute Coronary Syndrome. J. Clin. Diagn. Res. 2021, 15, OC49–OC55. [Google Scholar] [CrossRef]
- Chin, K.Y.; Ekeuku, S.O.; Chew, D.; Trias, A. Tocotrienol in the Management of Nonalcoholic Fatty Liver Disease: A Systematic Review. Nutrients 2023, 15, 834. [Google Scholar] [CrossRef]
- Pulido, M.R.; Diaz-Ruiz, A.; Jiménez-Gómez, Y.; Garcia-Navarro, S.; Gracia-Navarro, F.; Tinahones, F.; López-Miranda, J.; Frühbeck, G.; Vázquez-Martínez, R.; Malagón, M.M. Rab18 dynamics in adipocytes in relation to lipogenesis, lipolysis and obesity. PLoS ONE 2011, 6, e22931. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Navarrete, J.M.; Escoté, X.; Ortega, F.; Serino, M.; Campbell, M.; Michalski, M.C.; Laville, M.; Xifra, G.; Luche, E.; Domingo, P.; et al. A role for adipocyte-derived lipopolysaccharide-binding protein in inflammation- and obesity-associated adipose tissue dysfunction. Diabetologia 2013, 56, 2524–2537. [Google Scholar] [CrossRef] [PubMed]
- Sabater, M.; Moreno-Navarrete, J.M.; Ortega, F.J.; Pardo, G.; Salvador, J.; Ricart, W.; Frühbeck, G.; Fernández-Real, J.M. Circulating pigment epithelium-derived factor levels are associated with insulin resistance and decrease after weight loss. J. Clin. Endocrinol. Metab. 2010, 95, 4720–4728. [Google Scholar] [CrossRef]
- Lin, S.; Huang, J.; Wang, M.; Kumar, R.; Liu, Y.; Liu, S.; Wu, Y.; Wang, X.; Zhu, Y. Comparison of MAFLD and NAFLD diagnostic criteria in real world. Liver Int. 2020, 40, 2082–2089. [Google Scholar] [CrossRef]
- Mantovani, A. MAFLD vs NAFLD: Where are we. Dig. Liver Dis. 2021, 53, 1368–1372. [Google Scholar] [CrossRef]
- Fouad, Y.; Elwakil, R.; Elsahhar, M.; Said, E.; Bazeed, S.; Ali Gomaa, A.; Hashim, A.; Kamal, E.; Mehrez, M.; Attia, D. The NAFLD-MAFLD debate: Eminence vs evidence. Liver Int. 2021, 41, 255–260. [Google Scholar] [CrossRef]
- Ayob, N.; Muhammad Nawawi, K.N.; Mohamad Nor, M.H.; Raja Ali, R.A.; Ahmad, H.F.; Oon, S.F.; Mohd Mokhtar, N. The Effects of Probiotics on Small Intestinal Microbiota Composition, Inflammatory Cytokines and Intestinal Permeability in Patients with Non-Alcoholic Fatty Liver Disease. Biomedicines 2023, 11, 640. [Google Scholar] [CrossRef] [PubMed]
- Al-Baiaty, F.; Ismail, A.; Abdul Latiff, Z.; Muhammad Nawawi, K.N.; Raja Ali, R.A.; Mokhtar, N.M. Possible Hepatoprotective Effect of Tocotrienol-Rich Fraction Vitamin E in Non-alcoholic Fatty Liver Disease in Obese Children and Adolescents. Front. Pediatr. 2021, 9, 667247. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.K.; Chin, K.Y.; Ahmad, F.; Ima-Nirwana, S. Regulation of inflammatory response and oxidative stress by tocotrienol in a rat model of non-alcoholic fatty liver disease. J. Funct. Foods 2020, 74, 104209. [Google Scholar] [CrossRef]
- Perdomo, C.M.; Cohen, R.V.; Sumithran, P.; Clément, K.; Frühbeck, G. Contemporary medical, device, and surgical therapies for obesity in adults. Lancet 2023, 401, 1116–1130. [Google Scholar] [CrossRef]
- Liu, X.L.; Cao, H.X.; Fan, J.G. MicroRNAs as biomarkers and regulators of nonalcoholic fatty liver disease. J. Dig. Dis. 2016, 17, 708–715. [Google Scholar] [CrossRef]
- Yamada, H.; Suzuki, K.; Ichino, N.; Ando, Y.; Sawada, A.; Osakabe, K.; Sugimoto, K.; Ohashi, K.; Teradaira, R.; Inoue, T.; et al. Associations between circulating microRNAs (miR-21, miR-34a, miR-122 and miR-451) and non-alcoholic fatty liver. Clin. Chim. Acta 2013, 424, 99–103. [Google Scholar] [CrossRef]
- Gjorgjieva, M.; Sobolewski, C.; Dolicka, D.; Correia de Sousa, M.; Foti, M. miRNAs and NAFLD: From pathophysiology to therapy. Gut 2019, 68, 2065–2079. [Google Scholar] [CrossRef]
- Dongiovanni, P.; Meroni, M.; Longo, M.; Fargion, S.; Fracanzani, A.L. miRNA Signature in NAFLD: A Turning Point for a Non-Invasive Diagnosis. Int. J. Mol. Sci. 2018, 19, 3966. [Google Scholar] [CrossRef]
- Lee, Y.H.; Jang, H.J.; Kim, S.; Choi, S.S.; Khim, K.W.; Eom, H.J.; Hyun, J.; Shin, K.J.; Chae, Y.C.; Kim, H.; et al. Hepatic MIR20B promotes nonalcoholic fatty liver disease by suppressing PPARA. eLife 2021, 10, e70472. [Google Scholar] [CrossRef]
- Sasaki, R.; Kanda, T.; Yokosuka, O.; Kato, N.; Matsuoka, S.; Moriyama, M. Exosomes and Hepatocellular Carcinoma: From Bench to Bedside. Int. J. Mol. Sci. 2019, 20, 1406. [Google Scholar] [CrossRef]
- Sodum, N.; Kumar, G.; Bojja, S.L.; Kumar, N.; Rao, C.M. Epigenetics in NAFLD/NASH: Targets and therapy. Pharmacol. Res. 2021, 167, 105484. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Du, J.; Niu, X.; Fu, N.; Wang, R.; Zhang, Y.; Zhao, S.; Sun, D.; Nan, Y. MiR-130a-3p attenuates activation and induces apoptosis of hepatic stellate cells in nonalcoholic fibrosing steatohepatitis by directly targeting TGFBR1 and TGFBR2. Cell Death Dis. 2017, 8, e2792. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chen, S.; Zhang, L. Downregulated microRNA-130b-5p prevents lipid accumulation and insulin resistance in a murine model of nonalcoholic fatty liver disease. Am. J. Physiol. Endocrinol. Metab. 2020, 319, E34–E42. [Google Scholar] [CrossRef]
- Jun, L.; Tao, T.; Guo-Dong, W.; Bo, L. LncRNA-H19 promotes hepatic lipogenesis by directly regulating miR-130a/PPARγ axis in non-alcoholic fatty liver disease. Biosci. Rep. 2019, 39, BSR20181722. [Google Scholar]
- Guo, B.; Cheng, Y.; Yao, L.; Zhang, J.; Lu, J.; Qi, H.; Chen, H. LncRNA HOTAIR regulates the lipid accumulation in non-alcoholic fatty liver disease via miR-130b-3p/ROCK1 axis. Cell. Signal. 2022, 90, 110190. [Google Scholar] [CrossRef] [PubMed]
- Alshehri, A.S.; El-Kott, A.F.; El-Kenawy, A.E.; Khalifa, H.S.; AlRamlawy, A.M. Cadmium chloride induces non-alcoholic fatty liver disease in rats by stimulating miR-34a/SIRT1/FXR/p53 axis. Sci. Total Environ. 2021, 784, 147182. [Google Scholar] [CrossRef]
- Jiexia, D.; Meng, L.; Xingyong, W.; Xi, J.; Shaohua, C.; Chaohui, Y.; Youming, L. Effect of miR-34a in regulating steatosis by targeting PPARα expression in nonalcoholic fatty liver disease. Sci. Rep. 2015, 5, 13729. [Google Scholar]
- Lee, A.T.; Yang, M.Y.; Lee, Y.J.; Yang, T.W.; Wang, C.C.; Wang, C.J. Gallic Acid Improves Diabetic Steatosis by Downregulating MicroRNA-34a-5p through Targeting NFE2L2 Expression in High-Fat Diet-Fed db/db Mice. Antioxidants 2021, 11, 92. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sun, M.; Cao, Y.; Ma, L.; Shen, Y.; Velikanova, A.A.; Li, X.; Sun, C.; Zhao, Y. miR-34a regulates lipid metabolism by targeting SIRT1 in non-alcoholic fatty liver disease with iron overload. Arch. Biochem. Biophys. 2020, 695, 108642. [Google Scholar] [CrossRef]
- Yang, X.; Munaf, Z.; Jiesi, X.; Yuanyuan, L.; Liya, Y.; Yanqiao, Z. A metabolic stress-inducible miR-34a-HNF4α pathway regulates lipid and lipoprotein metabolism. Nat. Commun. 2015, 6, 7466. [Google Scholar]
- Xu, Y.; Zhu, Y.; Hu, S.; Pan, X.; Bawa, F.C.; Wang, H.H.; Wang, D.Q.; Yin, L.; Zhang, Y. Hepatocyte miR-34a is a key regulator in the development and progression of non-alcoholic fatty liver disease. Mol. Metab. 2021, 51, 101244. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, H.; Li, D.; Sun, H.; Li, M.; Hu, H. Long noncoding RNA Mirt2 upregulates USP10 expression to suppress hepatic steatosis by sponging miR-34a-5p. Gene 2019, 700, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Simino, L.; Panzarin, C.; Fontana, M.F.; de Fante, T.; Geraldo, M.V.; Ignácio-Souza, L.M.; Milanski, M.; Torsoni, M.A.; Ross, M.G.; Desai, M.; et al. MicroRNA Let-7 targets AMPK and impairs hepatic lipid metabolism in offspring of maternal obese pregnancies. Sci. Rep. 2021, 11, 8980. [Google Scholar] [CrossRef]
- Jinfang, Z.; Lilin, H.; Wenfang, G.; Li, X.; Weijun, W.; Jing, X.; Huiqian, F.; Zhonglin, L.; Qingjing, Z.; Xiaohua, H.; et al. Hepatocyte TGF-β Signaling Inhibiting WAT Browning to Promote NAFLD and Obesity Is Associated with Let-7b-5p. Hepatol. Commun. 2022, 6, 1301–1321. [Google Scholar]
- Ao, R.; Wang, Y.; Tong, J.; Wang, B.F. Altered microRNA-9 Expression Level is Directly Correlated with Pathogenesis of Nonalcoholic Fatty Liver Disease by Targeting Onecut2 and SIRT1. Med. Sci. Monit. 2016, 22, 3804–3819. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Niu, Q.; Wang, T.; Dong, H.; Bian, C. Lipotoxic hepatocytes promote nonalcoholic fatty liver disease progression by delivering microRNA-9-5p and activating macrophages. Int. J. Biol. Sci. 2021, 17, 3745–3759. [Google Scholar] [CrossRef]
- Wang, X.; Ma, Y.; Yang, L.Y.; Zhao, D. MicroRNA-20a-5p Ameliorates Non-alcoholic Fatty Liver Disease via Inhibiting the Expression of CD36. Front. Cell Dev. Biol. 2020, 8, 596329. [Google Scholar] [CrossRef]
- He, Q.; Li, F.; Li, J.; Li, R.; Zhan, G.; Li, G.; Du, W.; Tan, H. MicroRNA-26a-interleukin (IL)-6-IL-17 axis regulates the development of non-alcoholic fatty liver disease in a murine model. Clin. Exp. Immunol. 2017, 187, 174–184. [Google Scholar] [CrossRef]
- Xu, H.; Tian, Y.; Tang, D.; Zou, S.; Liu, G.; Song, J.; Zhang, G.; Du, X.; Huang, W.; He, B.; et al. An Endoplasmic Reticulum Stress-MicroRNA-26a Feedback Circuit in NAFLD. Hepatology 2021, 73, 1327–1345. [Google Scholar] [CrossRef]
- Murata, Y.; Yamashiro, T.; Kessoku, T.; Jahan, I.; Usuda, H.; Tanaka, T.; Okamoto, T.; Nakajima, A.; Wada, K. Up-Regulated MicroRNA-27b Promotes Adipocyte Differentiation via Induction of Acyl-CoA Thioesterase 2 Expression. Biomed. Res. Int. 2019, 2019, 2916243. [Google Scholar] [CrossRef]
- Law, D.G. A History of the Modern Chinese Navy, 1840–2020; Routledge Studies in the Modern History of Asia; The Mariner’s Mirror; Routledge: Oxfordshire, UK, 2022; p. 108. [Google Scholar]
- Lin, H.Y.; Wang, F.S.; Yang, Y.L.; Huang, Y.H. MicroRNA-29a Suppresses CD36 to Ameliorate High Fat Diet-Induced Steatohepatitis and Liver Fibrosis in Mice. Cells 2019, 8, 1298. [Google Scholar] [CrossRef]
- Dai, L.L.; Li, S.D.; Ma, Y.C.; Tang, J.R.; Lv, J.Y.; Zhang, Y.Q.; Miao, Y.L.; Ma, Y.Q.; Li, C.M.; Chu, Y.Y.; et al. MicroRNA-30b regulates insulin sensitivity by targeting SERCA2b in non-alcoholic fatty liver disease. Liver Int. 2019, 39, 1504–1513. [Google Scholar] [CrossRef]
- Fan, J.; Li, H.; Nie, X.; Yin, Z.; Zhao, Y.; Chen, C.; Wen Wang, D. MiR-30c-5p ameliorates hepatic steatosis in leptin receptor-deficient (db/db) mice via down-regulating FASN. Oncotarget 2017, 8, 13450–13463. [Google Scholar] [CrossRef]
- Zhang, Q.; Yu, K.; Cao, Y.; Luo, Y.; Liu, Y.; Zhao, C. miR-125b promotes the NF-κB-mediated inflammatory response in NAFLD via directly targeting TNFAIP3. Life Sci. 2021, 270, 119071. [Google Scholar] [CrossRef]
- Sun, Y.; Song, Y.; Liu, C.; Geng, J. LncRNA NEAT1-MicroRNA-140 axis exacerbates nonalcoholic fatty liver through interrupting AMPK/SREBP-1 signaling. Biochem. Biophys. Res. Commun. 2019, 516, 584–590. [Google Scholar] [CrossRef]
- Wolfson, B.; Lo, P.K.; Yao, Y.; Li, L.; Wang, H.; Zhou, Q. Impact of miR-140 Deficiency on Non-Alcoholic Fatty Liver Disease. Mol. Nutr. Food Res. 2018, 62, e1800189. [Google Scholar] [CrossRef]
- Zhang, N.; Qu, Y.; Qin, B. Sodium butyrate ameliorates non-alcoholic fatty liver disease by upregulating miR-150 to suppress CXCR4 expression. Clin. Exp. Pharmacol. Physiol. 2021, 48, 1125–1136. [Google Scholar] [CrossRef]
- Zhuge, B.; Li, G. MiR-150 deficiency ameliorated hepatosteatosis and insulin resistance in nonalcoholic fatty liver disease via targeting CASP8 and FADD-like apoptosis regulator. Biochem. Biophys. Res. Commun. 2017, 494, 687–692. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, X.; Xiao, Y.; Li, C.; Huang, Y.; Guo, Q.; Su, T.; Fu, L.; Luo, L. miR-188 promotes liver steatosis and insulin resistance via the autophagy pathway. J. Endocrinol. 2020, 245, 411–423. [Google Scholar] [CrossRef]
- Riaz, F.; Chen, Q.; Lu, K.; Osoro, E.K.; Wu, L.; Feng, L.; Zhao, R.; Yang, L.; Zhou, Y.; He, Y.; et al. Inhibition of miR-188-5p alleviates hepatic fibrosis by significantly reducing the activation and proliferation of HSCs through PTEN/PI3K/AKT pathway. J. Cell. Mol. Med. 2021, 25, 4073–4087. [Google Scholar] [CrossRef]
- Lin, Y.; Ding, D.; Huang, Q.; Liu, Q.; Lu, H.; Lu, Y.; Chi, Y.; Sun, X.; Ye, G.; Zhu, H.; et al. Downregulation of miR-192 causes hepatic steatosis and lipid accumulation by inducing SREBF1: Novel mechanism for bisphenol A-triggered non-alcoholic fatty liver disease. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 869–882. [Google Scholar] [CrossRef]
- Wang, Z.; Miu, K.K.; Zhang, X.; Wan, A.T.; Lu, G.; Cheung, H.H.; Lee, H.M.; Kong, A.P.; Chan, J.C.; Chan, W.Y. Hepatic miR-192-3p reactivation alleviates steatosis by targeting glucocorticoid receptor. JHEP Rep. 2020, 2, 100179. [Google Scholar] [CrossRef]
- Cheng, Y.; Huang, L.; Ping, J.; Chen, T.; Chen, J. MicroRNA-199a-3p attenuates hepatic lipogenesis by targeting Sp1. Am. J. Transl. Res. 2017, 9, 1905–1913. [Google Scholar]
- Li, B.; Zhang, Z.; Zhang, H.; Quan, K.; Lu, Y.; Cai, D.; Ning, G. Aberrant miR199a-5p/caveolin1/PPARα axis in hepatic steatosis. J. Mol. Endocrinol. 2014, 53, 393–403. [Google Scholar] [CrossRef]
- Li, Y.; Cen, C.Q.; Liu, B.; Zhou, L.; Huang, X.M.; Liu, G.Y. Overexpression of circ PTK2 suppresses the progression of nonalcoholic fatty liver disease via the miR-200c/SIK2/PI3K/Akt axis. Kaohsiung J. Med. Sci. 2022, 38, 869–878. [Google Scholar] [CrossRef]
- Wang, Y.; Zeng, Z.; Guan, L.; Ao, R. GRHL2 induces liver fibrosis and intestinal mucosal barrier dysfunction in non-alcoholic fatty liver disease via microRNA-200 and the MAPK pathway. J. Cell. Mol. Med. 2020, 24, 6107–6119. [Google Scholar] [CrossRef]
- Guo, Y.; Yu, J.; Wang, C.; Li, K.; Liu, B.; Du, Y.; Xiao, F.; Chen, S.; Guo, F. miR-212-5p suppresses lipid accumulation by targeting FAS and SCD1. J. Mol. Endocrinol. 2017, 59, 205–217. [Google Scholar] [CrossRef]
- Xiao, J.; Bei, Y.; Liu, J.; Dimitrova-Shumkovska, J.; Kuang, D.; Zhou, Q.; Li, J.; Yang, Y.; Xiang, Y.; Wang, F.; et al. miR-212 downregulation contributes to the protective effect of exercise against non-alcoholic fatty liver via targeting FGF-21. J. Cell. Mol. Med. 2016, 20, 204–216. [Google Scholar] [CrossRef]
- He, Y.; Hwang, S.; Cai, Y.; Kim, S.J.; Xu, M.; Yang, D.; Guillot, A.; Feng, D.; Seo, W.; Hou, X.; et al. MicroRNA-223 Ameliorates Nonalcoholic Steatohepatitis and Cancer by Targeting Multiple Inflammatory and Oncogenic Genes in Hepatocytes. Hepatology 2019, 70, 1150–1167. [Google Scholar] [CrossRef]
- Niu, Q.; Wang, T.; Wang, Z.; Wang, F.; Huang, D.; Sun, H.; Liu, H. Adipose-derived mesenchymal stem cell-secreted extracellular vesicles alleviate non-alcoholic fatty liver disease via delivering miR-223-3p. Adipocyte 2022, 11, 572–587. [Google Scholar] [CrossRef]
- Hur, W.; Lee, J.H.; Kim, S.W.; Kim, J.H.; Bae, S.H.; Kim, M.; Hwang, D.; Kim, Y.S.; Park, T.; Um, S.J.; et al. Downregulation of microRNA-451 in non-alcoholic steatohepatitis inhibits fatty acid-induced proinflammatory cytokine production through the AMPK/AKT pathway. Int. J. Biochem. Cell Biol. 2015, 64, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Tan, X.; Zhu, L.; Qin, K.; Gao, H.; Bai, H. Swimming prevents nonalcoholic fatty liver disease by reducing migration inhibitory factor through Akt suppression and autophagy activation. Am. J. Transl. Res. 2019, 11, 4315–4325. [Google Scholar] [PubMed]
- Rodrigues, P.M.; Afonso, M.B.; Simão, A.L.; Carvalho, C.C.; Trindade, A.; Duarte, A.; Borralho, P.M.; Machado, M.V.; Cortez-Pinto, H.; Rodrigues, C.M.; et al. miR-21 ablation and obeticholic acid ameliorate nonalcoholic steatohepatitis in mice. Cell Death Dis. 2017, 8, e2748. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.M.; Wang, X.Y.; Huang, Y.M.; Chen, X.; Lü, M.H.; Shi, L.; Li, C.P. Role and mechanisms of action of microRNA-21 as regards the regulation of the WNT/β-catenin signaling pathway in the pathogenesis of non-alcoholic fatty liver disease. Int. J. Mol. Med. 2019, 44, 2201–2212. [Google Scholar] [CrossRef]
- Wu, H.; Ng, R.; Chen, X.; Steer, C.J.; Song, G. MicroRNA-21 is a potential link between non-alcoholic fatty liver disease and hepatocellular carcinoma via modulation of the HBP1-p53-Srebp1c pathway. Gut 2016, 65, 1850–1860. [Google Scholar] [CrossRef]
- Chu, K.; Gu, J. microRNA-103a-3p promotes inflammation and fibrosis in nonalcoholic fatty liver disease by targeting HBP1. Immunopharmacol. Immunotoxicol. 2022, 44, 993–1003. [Google Scholar] [CrossRef]
- Ding, J.; Xia, C.; Cen, P.; Li, S.; Yu, L.; Zhu, J.; Jin, J. MiR-103-3p promotes hepatic steatosis to aggravate nonalcoholic fatty liver disease by targeting of ACOX1. Mol. Biol. Rep. 2022, 49, 7297–7305. [Google Scholar] [CrossRef]
- Zhang, M.; Tang, Y.; Tang, E.; Lu, W. MicroRNA-103 represses hepatic de novo lipogenesis and alleviates NAFLD via targeting FASN and SCD1. Biochem. Biophys. Res. Commun. 2020, 524, 716–722. [Google Scholar] [CrossRef]
- Chai, C.; Cox, B.; Yaish, D.; Gross, D.; Rosenberg, N.; Amblard, F.; Shemuelian, Z.; Gefen, M.; Korach, A.; Tirosh, O.; et al. Agonist of RORA Attenuates Nonalcoholic Fatty Liver Progression in Mice via Up-regulation of MicroRNA 122. Gastroenterology 2020, 159, 999–1014.e9. [Google Scholar] [CrossRef]
- Hu, Y.; Peng, X.; Du, G.; Zhang, Z.; Zhai, Y.; Xiong, X.; Luo, X. MicroRNA-122-5p Inhibition Improves Inflammation and Oxidative Stress Damage in Dietary-Induced Non-alcoholic Fatty Liver Disease Through Targeting FOXO3. Front. Physiol. 2022, 13, 803445. [Google Scholar] [CrossRef]
- Long, J.K.; Dai, W.; Zheng, Y.W.; Zhao, S.P. miR-122 promotes hepatic lipogenesis via inhibiting the LKB1/AMPK pathway by targeting Sirt1 in non-alcoholic fatty liver disease. Mol. Med. 2019, 25, 26. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Tan, X.R.; Li, S.J.; Zhang, X.X. LncRNA NEAT1 promotes hepatic lipid accumulation via regulating miR-146a-5p/ROCK1 in nonalcoholic fatty liver disease. Life Sci. 2019, 235, 116829. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Guo, W.; Deng, F.; Chen, K.; Jiang, Y.; Dong, M.; Peng, L.; Chen, X. Targeted delivery of microRNA 146b mimic to hepatocytes by lactosylated PDMAEMA nanoparticles for the treatment of NAFLD. Artif. Cells Nanomed. Biotechnol. 2018, 46, 217–228. [Google Scholar] [CrossRef]
- Li, K.; Zhao, B.; Wei, D.; Wang, W.; Cui, Y.; Qian, L.; Liu, G. miR-146a improves hepatic lipid and glucose metabolism by targeting MED1. Int. J. Mol. Med. 2020, 45, 543–555. [Google Scholar] [CrossRef]
- Chen, S.; Chen, D.; Yang, H.; Wang, X.; Wang, J.; Xu, C. Uric acid induced hepatocytes lipid accumulation through regulation of miR-149-5p/FGF21 axis. BMC Gastroenterol. 2020, 20, 39. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Liu, Y.; Yang, L.; Liu, P.; Zhang, Y.; Wang, X. MiR-149 attenuates endoplasmic reticulum stress-induced inflammation and apoptosis in nonalcoholic fatty liver disease by negatively targeting ATF6 pathway. Immunol. Lett. 2020, 222, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Lv, D.; Zhao, Y.; Chen, X.; Song, M.; Liu, J.; Bei, Y.; Wang, F.; Yang, W.; Yang, C. miR-149 controls non-alcoholic fatty liver by targeting FGF-21. J. Cell. Mol. Med. 2016, 20, 1603–1608. [Google Scholar] [CrossRef]
- Bala, S.; Ganz, M.; Babuta, M.; Zhuang, Y.; Csak, T.; Calenda, C.D.; Szabo, G. Steatosis, inflammasome upregulation, and fibrosis are attenuated in miR-155 deficient mice in a high fat-cholesterol-sugar diet-induced model of NASH. Lab. Investig. 2021, 101, 1540–1549. [Google Scholar] [CrossRef]
- Csak, T.; Bala, S.; Lippai, D.; Kodys, K.; Catalano, D.; Iracheta-Vellve, A.; Szabo, G. MicroRNA-155 Deficiency Attenuates Liver Steatosis and Fibrosis without Reducing Inflammation in a Mouse Model of Steatohepatitis. PLoS ONE 2015, 10, e0129251. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, N.; Wang, Z.; Ai, D.M.; Cao, Z.Y.; Pan, H.P. Decreased MiR-155 Level in the Peripheral Blood of Non-Alcoholic Fatty Liver Disease Patients may Serve as a Biomarker and may Influence LXR Activity. Cell. Physiol. Biochem. 2016, 39, 2239–2248. [Google Scholar] [CrossRef]
- Chen, T.; Yan, D.; Cheng, X.; Ji, X.; Bian, J.; Yin, W. miR-1224-5p Enhances Hepatic Lipogenesis by Targeting Adenosine Monophosphate-Activated Protein Kinase α1 in Male Mice. Endocrinology 2018, 159, 2008–2021. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ma, H.; Gao, Y.; Jin, Y.; Ning, W.; Hou, Y.; Su, J. Long non-coding RNA AC012668 suppresses non-alcoholic fatty liver disease by competing for microRNA miR-380-5p with lipoprotein-related protein LRP2. Bioengineered 2021, 12, 6738–6747. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Mai, J.; Hou, T.; Ping, J. MicroRNA-421 induces hepatic mitochondrial dysfunction in non-alcoholic fatty liver disease mice by inhibiting sirtuin 3. Biochem. Biophys. Res. Commun. 2016, 474, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Yu, C.; Ma, N.; Xu, X.; Wu, Q.; Lu, H.; Gong, L.; Chen, J.; Ren, J. MicroRNA-379-5p regulates free cholesterol accumulation and relieves diet induced-liver damage in db/db mice via STAT1/HMGCS1 axis. Mol. Biomed. 2022, 3, 25. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Lai, R.; Ma, N.; Dong, Y.; Li, Y.; Wu, Q.; Qiao, J.; Lu, H.; Gong, L.; Tao, Z.; et al. miR-552-3p modulates transcriptional activities of FXR and LXR to ameliorate hepatic glycolipid metabolism disorder. J. Hepatol. 2021, 74, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Hanin, G.; Yayon, N.; Tzur, Y.; Haviv, R.; Bennett, E.R.; Udi, S.; Krishnamoorthy, Y.R.; Kotsiliti, E.; Zangen, R.; Efron, B.; et al. miRNA-132 induces hepatic steatosis and hyperlipidaemia by synergistic multitarget suppression. Gut 2018, 67, 1124–1134. [Google Scholar] [CrossRef]
- Hu, Y.; Ye, H.; Shi, L.X. MicroRNA-205 ameliorates lipid accumulation in non-alcoholic fatty liver disease through targeting NEU1. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 10072–10082. [Google Scholar] [CrossRef]
- Lingbo, K.; Xinyu, A.; Lingxi, H.; Siyu, Z.; Lingdi, L.; Suxian, Z.; Rongqi, W.; Yuemin, N. Resveratrol ameliorates nutritional steatohepatitis through the mmu-miR-599/PXR pathway. Int. J. Mol. Med. 2022, 49, 5102. [Google Scholar] [CrossRef]
- Hye, L.D.; Hyun, P.S.; Jiyun, A.; Pyo, H.S.; Eunyoung, L.; Jin, J.Y.; Youl, H.T.; Hoon, H.Y.; Yeon, H.S.; Il, J.T.; et al. Mir214-3p and Hnf4a/Hnf4α reciprocally regulate Ulk1 expression and autophagy in nonalcoholic hepatic steatosis. Autophagy 2020, 17, 2415–2431. [Google Scholar]
- Lei, L.; Zhou, C.; Yang, X.; Li, L. Down-regulation of microRNA-375 regulates adipokines and inhibits inflammatory cytokines by targeting AdipoR2 in non-alcoholic fatty liver disease. Clin. Exp. Pharmacol. Physiol. 2018, 45, 819–831. [Google Scholar] [CrossRef]
- Li, D.; Wang, X.; Lan, X.; Li, Y.; Liu, L.; Yi, J.; Li, J.; Sun, Q.; Wang, Y.; Li, H.; et al. Down-regulation of miR-144 elicits proinflammatory cytokine production by targeting toll-like receptor 2 in nonalcoholic steatohepatitis of high-fat-diet-induced metabolic syndrome E3 rats. Mol. Cell. Endocrinol. 2015, 402, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, X.; Yu, S.; Hu, Y.; Xu, L.; Wang, T.; Yang, X.; Sun, X.; Zhao, B. miR-23b Ameliorates nonalcoholic steatohepatitis by targeting Acyl-CoA thioesterases 4. Exp. Cell Res. 2021, 407, 112787. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Fan, C.; Cai, Y.; Fang, S.; Zeng, Y.; Zhang, Y.; Lin, X.; Zhang, H.; Xue, Y.; Guan, M. Transplantation of brown adipose tissue up-regulates miR-99a to ameliorate liver metabolic disorders in diabetic mice by targeting NOX4. Adipocyte 2020, 9, 57–67. [Google Scholar] [CrossRef]
- Li, R.; Ye, Z.; She, D.; Fang, P.; Zong, G.; Hu, K.; Kong, D.; Xu, W.; Li, L.; Zhou, Y.; et al. Semaglutide May Alleviate Hepatic Steatosis in T2DM Combined with NFALD Mice via miR-5120/ABHD6. Drug. Des. Dev. Ther. 2022, 16, 3557–3572. [Google Scholar] [CrossRef] [PubMed]
- Mittal, S.; Inamdar, S.; Acharya, J.; Pekhale, K.; Kalamkar, S.; Boppana, R.; Ghaskadbi, S. miR-3666 inhibits development of hepatic steatosis by negatively regulating PPARγ. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2020, 1865, 158777. [Google Scholar] [CrossRef]
- Ng, R.; Wu, H.; Xiao, H.; Chen, X.; Willenbring, H.; Steer, C.J.; Song, G. Inhibition of microRNA-24 expression in liver prevents hepatic lipid accumulation and hyperlipidemia. Hepatology 2014, 60, 554–564. [Google Scholar] [CrossRef]
- Nie, H.; Song, C.; Wang, D.; Cui, S.; Ren, T.; Cao, Z.; Liu, Q.; Chen, Z.; Chen, X.; Zhou, Y. MicroRNA-194 inhibition improves dietary-induced non-alcoholic fatty liver disease in mice through targeting on FXR. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 3087–3094. [Google Scholar] [CrossRef]
- Sun, H.; Seok, S.; Jung, H.; Kemper, B.; Kemper, J.K. Obesity-induced miR-802 directly targets AMPK and promotes nonalcoholic steatohepatitis in mice. Mol. Metab. 2022, 66, 101603. [Google Scholar] [CrossRef]
- Wang, G.; Zou, H.; Lai, C.; Huang, X.; Yao, Y.; Xiang, G. Repression of MicroRNA-124-3p Alleviates High-Fat Diet-Induced Hepatosteatosis by Targeting Pref-1. Front. Endocrinol. 2020, 11, 589994. [Google Scholar] [CrossRef]
- Wang, J.J.; Zhang, Y.T.; Tseng, Y.J.; Zhang, J. miR-222 targets ACOX1, promotes triglyceride accumulation in hepatocytes. Hepatobil. Pancreat. Dis. Int. 2019, 18, 360–365. [Google Scholar] [CrossRef]
- Lu, W.; Xiaohui, W.; Lina, K.; Yingying, L.; Kai, H.; Jingjing, W.; Changyuan, W.; Huijun, S.; Pengyuan, S.; Jiangning, G.; et al. Activation of PGC-1α via isoliquiritigenin-induced downregulation of miR-138-5p alleviates nonalcoholic fatty liver disease. Phytother. Res. 2022, 36, 899–913. [Google Scholar]
- Wang, X.; Wang, J. High-content hydrogen water-induced downregulation of miR-136 alleviates non-alcoholic fatty liver disease by regulating Nrf2 via targeting MEG3. Biol. Chem. 2018, 399, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Lu, L.J.; Li, Y.M.; Xu, C.F. MicroRNA-376b-3p ameliorates nonalcoholic fatty liver disease by targeting FGFR1 and regulating lipid oxidation in hepatocytes. Life Sci. 2022, 308, 120925. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhu, K.; Yu, W.; Wang, H.; Liu, L.; Wu, Q.; Li, S.; Guo, J. MiR-181b regulates steatosis in nonalcoholic fatty liver disease via targeting SIRT1. Biochem. Biophys. Res. Commun. 2017, 493, 227–232. [Google Scholar] [CrossRef]
- Wu, H.; Zhang, T.; Pan, F.; Steer, C.J.; Li, Z.; Chen, X.; Song, G. MicroRNA-206 prevents hepatosteatosis and hyperglycemia by facilitating insulin signaling and impairing lipogenesis. J. Hepatol. 2017, 66, 816–824. [Google Scholar] [CrossRef]
- Xu, M.; Zheng, X.M.; Jiang, F.; Qiu, W.Q. MicroRNA-190b regulates lipid metabolism and insulin sensitivity by targeting IGF-1 and ADAMTS9 in non-alcoholic fatty liver disease. J. Cell. Biochem. 2018, 119, 5864–5874. [Google Scholar] [CrossRef]
- Yang, Z.; Qin, W.; Huo, J.; Zhuo, Q.; Wang, J.; Wang, L. MiR-22 modulates the expression of lipogenesis-related genes and promotes hepatic steatosis in vitro. FEBS Open. Bio 2021, 11, 322–332. [Google Scholar] [CrossRef]
- Yuanjie, Y.; Chunping, H.; Shiyun, T.; Mengjun, H.; Yitian, G.; Ming, L.; Qian, Z. MicroRNA-137-3p Improves Nonalcoholic Fatty Liver Disease through Activating AMPKα. Anal. Cell. Pathol. 2021, 2021, 4853355. [Google Scholar]
- Yu, Y.; Tian, T.; Tan, S.; Wu, P.; Guo, Y.; Li, M.; Huang, M. MicroRNA-665-3p exacerbates nonalcoholic fatty liver disease in mice. Bioengineered 2022, 13, 2927–2942. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, J.; Li, H.; Zhang, Y.; Chu, X.; Yang, J.; Li, Y. LncRNA Gm12664-001 ameliorates nonalcoholic fatty liver through modulating miR-295-5p and CAV1 expression. Nutr. Metab. 2020, 17, 13. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, X.; Steer, C.J.; Yan, G.; Song, G. A negative feedback loop between microRNA-378 and Nrf1 promotes the development of hepatosteatosis in mice treated with a high fat diet. Metab. Clin. Exp. 2018, 85, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, C.; Lu, J.; Jin, Y.; Xu, C.; Meng, Q.; Liu, Q.; Dong, D.; Ma, X.; Liu, K.; et al. Targeting of miR-96-5p by catalpol ameliorates oxidative stress and hepatic steatosis in LDLr-/- mice via p66shc/cytochrome C cascade. Aging 2020, 12, 2049–2069. [Google Scholar] [CrossRef] [PubMed]
- Zuo, R.; Ye, L.F.; Huang, Y.; Song, Z.Q.; Wang, L.; Zhi, H.; Zhang, M.Y.; Li, J.Y.; Zhu, L.; Xiao, W.J.; et al. Hepatic small extracellular vesicles promote microvascular endothelial hyperpermeability during NAFLD via novel-miRNA-7. J. Nanobiotechnol. 2021, 19, 396. [Google Scholar] [CrossRef]
- Meng, Z.; Zhou, D.; Gao, Y.; Zeng, M.; Wang, W. miRNA delivery for skin wound healing. Adv. Drug Deliv. Rev. 2018, 129, 308–318. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.L.; Zheng, M.H.; Shi, K.Q.; Yang, T.; Chen, Y.P. A new strategy for treatment of liver fibrosis: Letting MicroRNAs do the job. BioDrugs 2013, 27, 25–34. [Google Scholar] [CrossRef]
- Wang, R.; Ma, J.; Wu, Q.; Xia, J.; Miele, L.; Sarkar, F.H.; Wang, Z. Functional role of miR-34 family in human cancer. Curr. Drug Targets 2013, 14, 1185–1191. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Yalcin, A.; Meyer, J.; Lendeckel, W.; Tuschl, T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002, 12, 735–739. [Google Scholar] [CrossRef] [PubMed]
- Wen, F.; An, C.; Wu, X.; Yang, Y.; Xu, J.; Liu, Y.; Wang, C.; Nie, L.; Fang, H.; Yang, Z. MiR-34a regulates mitochondrial content and fat ectopic deposition induced by resistin through the AMPK/PPARα pathway in HepG2 cells. Int. J. Biochem. Cell Biol. 2018, 94, 133–145. [Google Scholar] [CrossRef]
- Thounaojam, M.C.; Jadeja, R.N.; Warren, M.; Powell, F.L.; Raju, R.; Gutsaeva, D.; Khurana, S.; Martin, P.M.; Bartoli, M. MicroRNA-34a (miR-34a) Mediates Retinal Endothelial Cell Premature Senescence through Mitochondrial Dysfunction and Loss of Antioxidant Activities. Antioxidants 2019, 8, 328. [Google Scholar] [CrossRef]
- Torres, L.F.; Cogliati, B.; Otton, R. Green Tea Prevents NAFLD by Modulation of miR-34a and miR-194 Expression in a High-Fat Diet Mouse Model. Oxid. Med. Cell. Longev. 2019, 2019, 4168380. [Google Scholar] [CrossRef]
- Jia, N.; Lin, X.; Ma, S.; Ge, S.; Mu, S.; Yang, C.; Shi, S.; Gao, L.; Xu, J.; Bo, T.; et al. Amelioration of hepatic steatosis is associated with modulation of gut microbiota and suppression of hepatic miR-34a in Gynostemma pentaphylla (Thunb.) Makino treated mice. Nutr. Metab. 2018, 15, 86. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.Y.; Chen, J.N.; Sun, F.; Wang, Y.Q.; Pan, Q.; Fan, J.G. circRNA_0046367 Prevents Hepatoxicity of Lipid Peroxidation: An Inhibitory Role against Hepatic Steatosis. Oxid. Med. Cell. Longev. 2017, 2017, 3960197. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.Y.; Ding, X.Q.; Gu, T.T.; Guo, W.J.; Jiao, R.Q.; Song, L.; Sun, Y.; Pan, Y.; Kong, L.D. Pterostilbene Improves Hepatic Lipid Accumulation via the MiR-34a/Sirt1/SREBP-1 Pathway in Fructose-Fed Rats. J. Agric. Food Chem. 2020, 68, 1436–1446. [Google Scholar] [CrossRef] [PubMed]
- Tao, W.; Yu-hua, L.; Yu-cai, F.; Xing-mu, L.; Xiao-hui, Z. Direct evidence of sirtuin downregulation in the liver of non-alcoholic fatty liver disease patients. Ann. Clin. Lab. Sci. 2014, 44, 410–418. [Google Scholar]
- Min, H.K.; Kapoor, A.; Fuchs, M.; Mirshahi, F.; Zhou, H.; Maher, J.; Kellum, J.; Warnick, R.; Contos, M.J.; Sanyal, A.J. Increased hepatic synthesis and dysregulation of cholesterol metabolism is associated with the severity of nonalcoholic fatty liver disease. Cell Metab. 2012, 15, 665–674. [Google Scholar] [CrossRef]
- Han, H.S.; Choi, B.H.; Kim, J.S.; Kang, G.; Koo, S.H. Hepatic Crtc2 controls whole body energy metabolism via a miR-34a-Fgf21 axis. Nat. Commun. 2017, 8, 1878. [Google Scholar] [CrossRef]
- Guo, L.; Yu, J.; Yu, H.; Zhao, Y.; Chen, S.; Xu, C.; Chen, F. Evolutionary and expression analysis of miR-#-5p and miR-#-3p at the miRNAs/isomiRs levels. Biomed. Res. Int. 2015, 2015, 168358. [Google Scholar] [CrossRef]
- Tazawa, H.; Kagawa, S.; Fujiwara, T. MicroRNAs as potential target gene in cancer gene therapy of gastrointestinal tumors. Expert. Opin. Biol. Ther. 2011, 11, 145–155. [Google Scholar] [CrossRef]
- Becker, P.P.; Rau, M.; Schmitt, J.; Malsch, C.; Hammer, C.; Bantel, H.; Müllhaupt, B.; Geier, A. Performance of Serum microRNAs -122, -192 and -21 as Biomarkers in Patients with Non-Alcoholic Steatohepatitis. PLoS ONE 2015, 10, e0142661. [Google Scholar] [CrossRef]
- Marques-Rocha, J.L.; Samblas, M.; Milagro, F.I.; Bressan, J.; Martínez, J.A.; Marti, A. Noncoding RNAs, cytokines, and inflammation-related diseases. FASEB J. 2015, 29, 3595–3611. [Google Scholar] [CrossRef]
- Zhang, W.; Jia, J.; Liu, Z.; Si, D.; Ma, L.; Zhang, G. Circulating microRNAs as biomarkers for Sepsis secondary to pneumonia diagnosed via Sepsis 3.0. BMC Pulm. Med. 2019, 19, 93. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Gao, J.; Zheng, R.; Yu, M.; Ren, Y.; Yan, T.; Huang, Y.; Li, Y. Antagonizing circRNA_002581-miR-122-CPEB1 axis alleviates NASH through restoring PTEN-AMPK-mTOR pathway regulated autophagy. Cell Death Dis. 2020, 11, 123. [Google Scholar] [CrossRef]
- Nakamura, M.; Kanda, T.; Sasaki, R.; Haga, Y.; Jiang, X.; Wu, S.; Nakamoto, S.; Yokosuka, O. MicroRNA-122 Inhibits the Production of Inflammatory Cytokines by Targeting the PKR Activator PACT in Human Hepatic Stellate Cells. PLoS ONE 2015, 10, e0144295. [Google Scholar] [CrossRef]
- van der Ree, M.H.; van der Meer, A.J.; van Nuenen, A.C.; de Bruijne, J.; Ottosen, S.; Janssen, H.L.; Kootstra, N.A.; Reesink, H.W. Miravirsen dosing in chronic hepatitis C patients results in decreased microRNA-122 levels without affecting other microRNAs in plasma. Aliment. Pharmacol. Ther. 2016, 43, 102–113. [Google Scholar] [CrossRef] [PubMed]
- van der Ree, M.H.; van der Meer, A.J.; de Bruijne, J.; Maan, R.; van Vliet, A.; Welzel, T.M.; Zeuzem, S.; Lawitz, E.J.; Rodriguez-Torres, M.; Kupcova, V.; et al. Long-term safety and efficacy of microRNA-targeted therapy in chronic hepatitis C patients. Antivir. Res. 2014, 111, 53–59. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Tan, J.K.; Wong, S.K.; Goon, J.A. Therapeutic Effects of microRNAs on Nonalcoholic Fatty Liver Disease (NAFLD) and Nonalcoholic Steatohepatitis (NASH): A Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2023, 24, 9168. https://doi.org/10.3390/ijms24119168
Zhu Y, Tan JK, Wong SK, Goon JA. Therapeutic Effects of microRNAs on Nonalcoholic Fatty Liver Disease (NAFLD) and Nonalcoholic Steatohepatitis (NASH): A Systematic Review and Meta-Analysis. International Journal of Molecular Sciences. 2023; 24(11):9168. https://doi.org/10.3390/ijms24119168
Chicago/Turabian StyleZhu, Yuezhi, Jen Kit Tan, Sok Kuan Wong, and Jo Aan Goon. 2023. "Therapeutic Effects of microRNAs on Nonalcoholic Fatty Liver Disease (NAFLD) and Nonalcoholic Steatohepatitis (NASH): A Systematic Review and Meta-Analysis" International Journal of Molecular Sciences 24, no. 11: 9168. https://doi.org/10.3390/ijms24119168
APA StyleZhu, Y., Tan, J. K., Wong, S. K., & Goon, J. A. (2023). Therapeutic Effects of microRNAs on Nonalcoholic Fatty Liver Disease (NAFLD) and Nonalcoholic Steatohepatitis (NASH): A Systematic Review and Meta-Analysis. International Journal of Molecular Sciences, 24(11), 9168. https://doi.org/10.3390/ijms24119168





