Genome-Wide Identification and Functional Analysis of the AP2/ERF Transcription Factor Family in Citrus Rootstock under Waterlogging Stress
Abstract
1. Introduction
2. Results
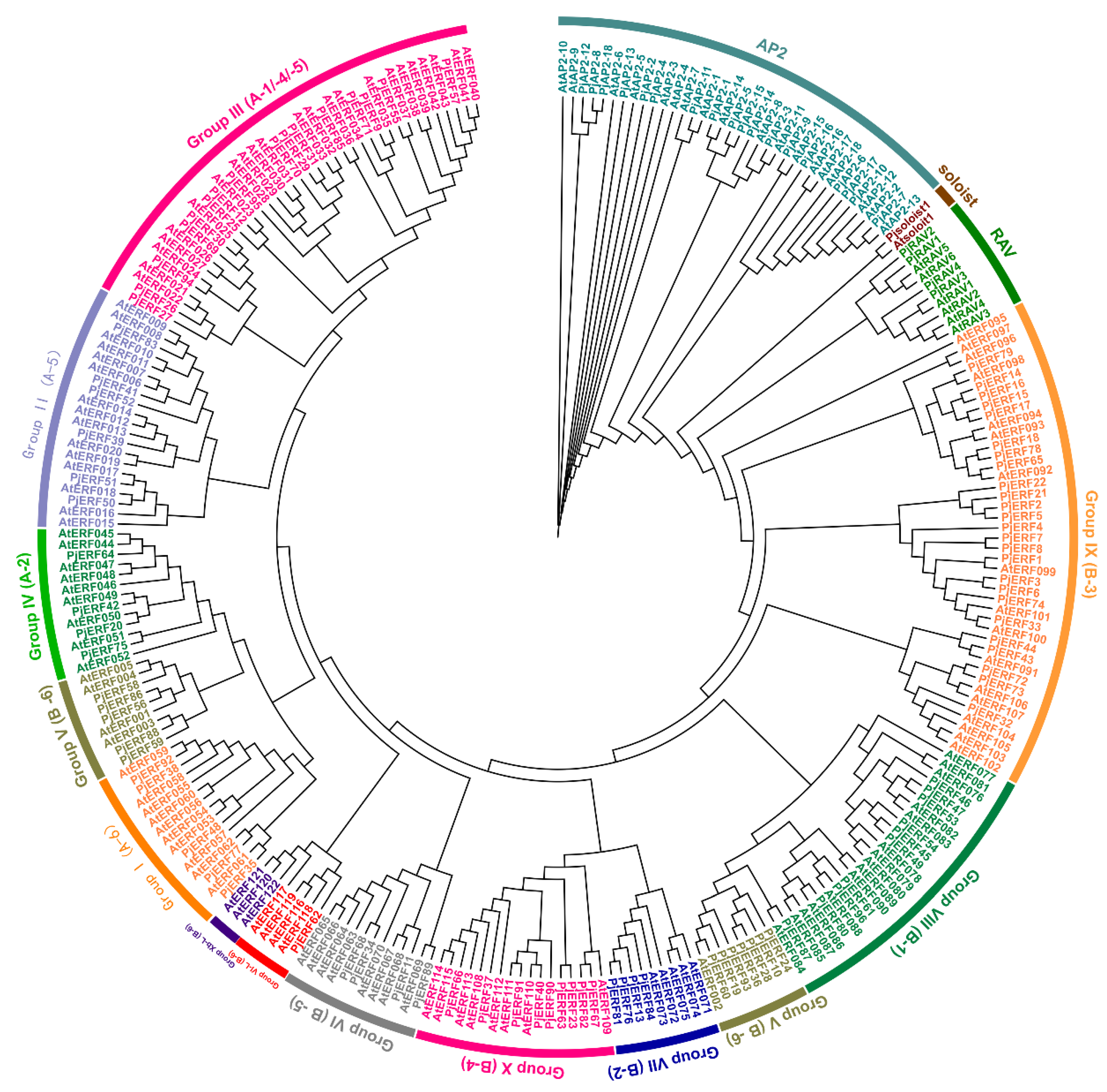
2.1. Identification and Characterization of AP2/ERFs in Citrus junos
2.2. Gene Structure and Conserved Motif Analyses of PjAP2/ERFs
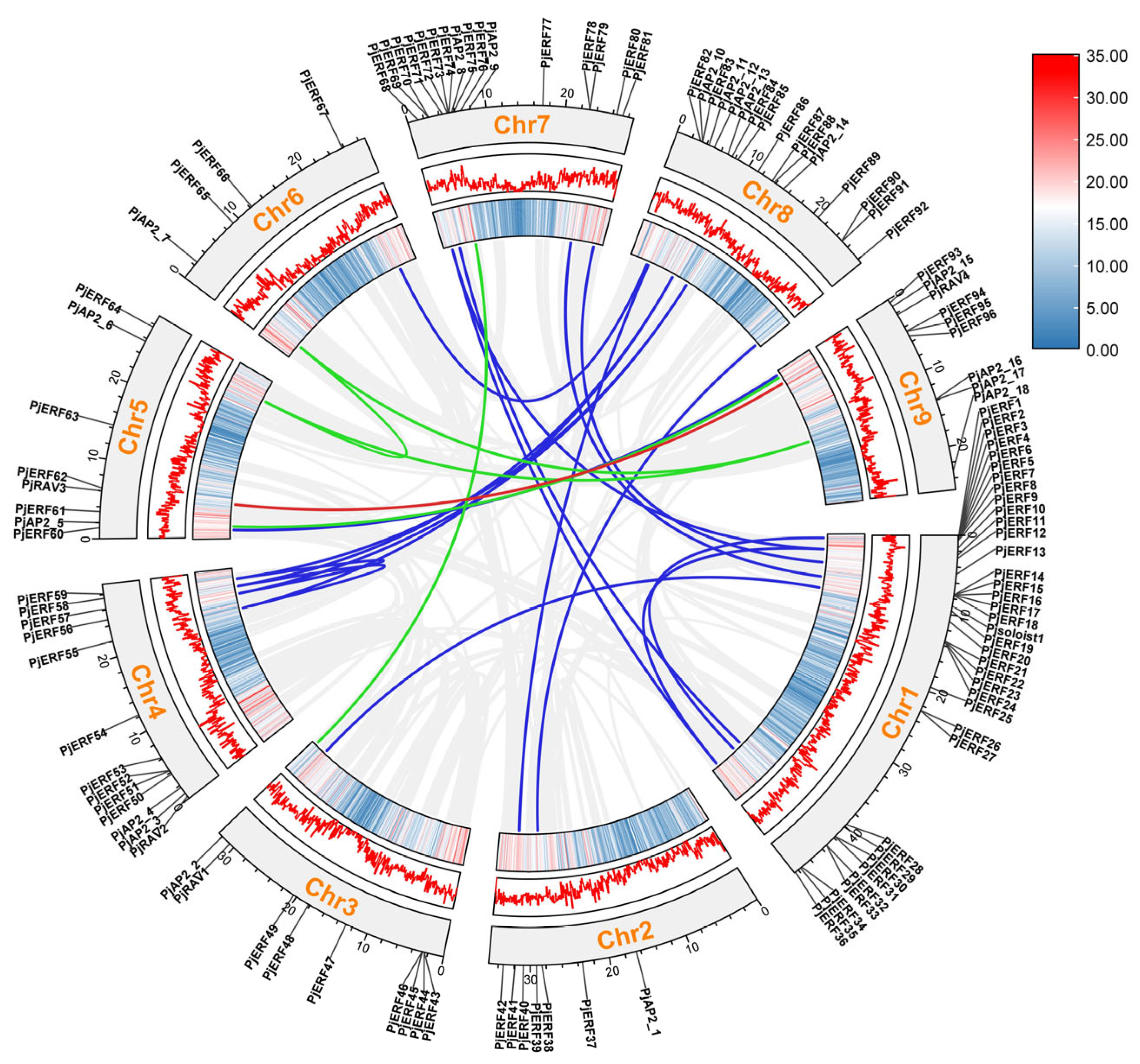
2.3. Chromosomal Location and Synteny Analysis of PjERFs
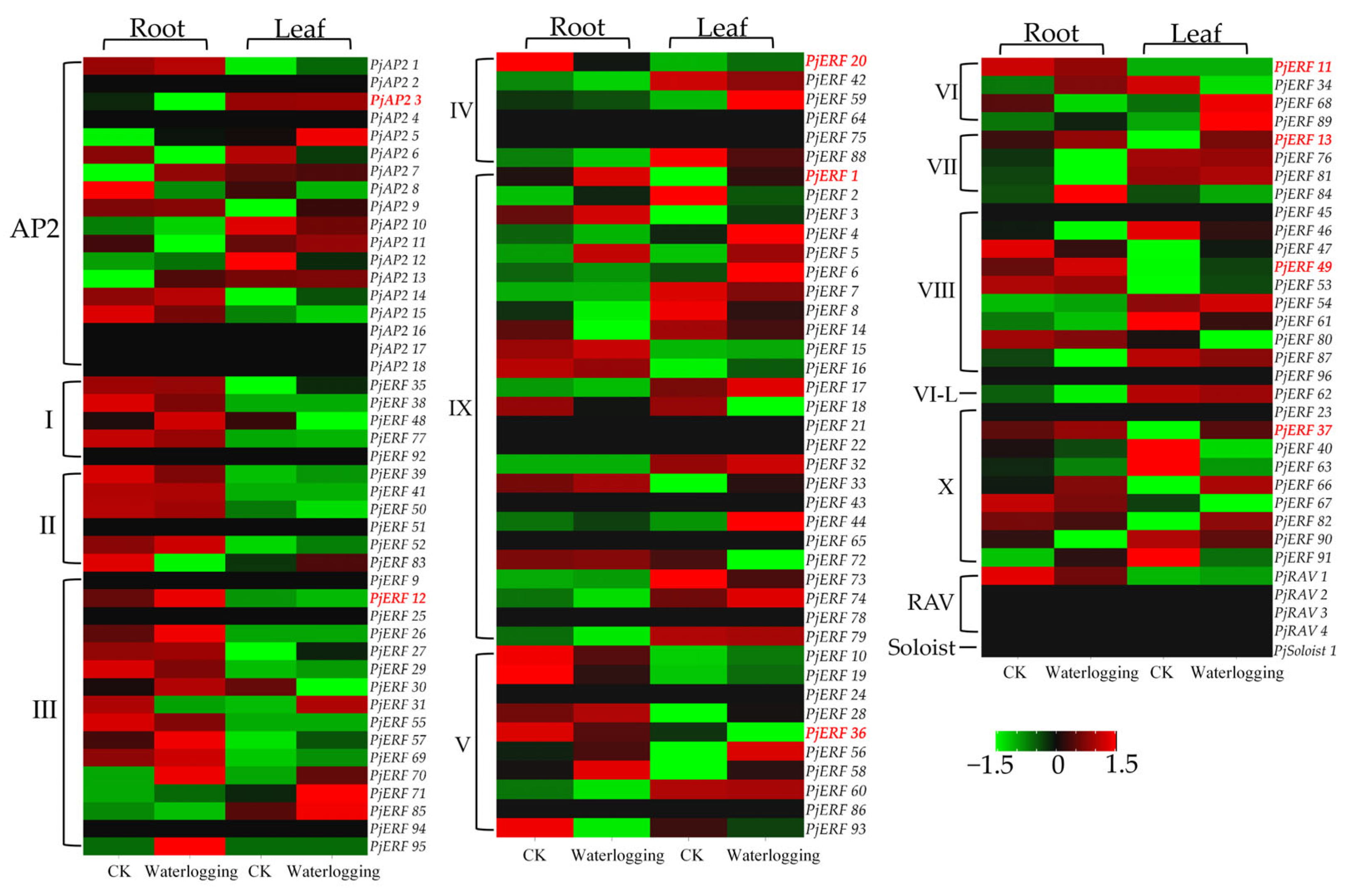
2.4. The Expression of PjAP2/ERFs in Response to Waterlogging Stress
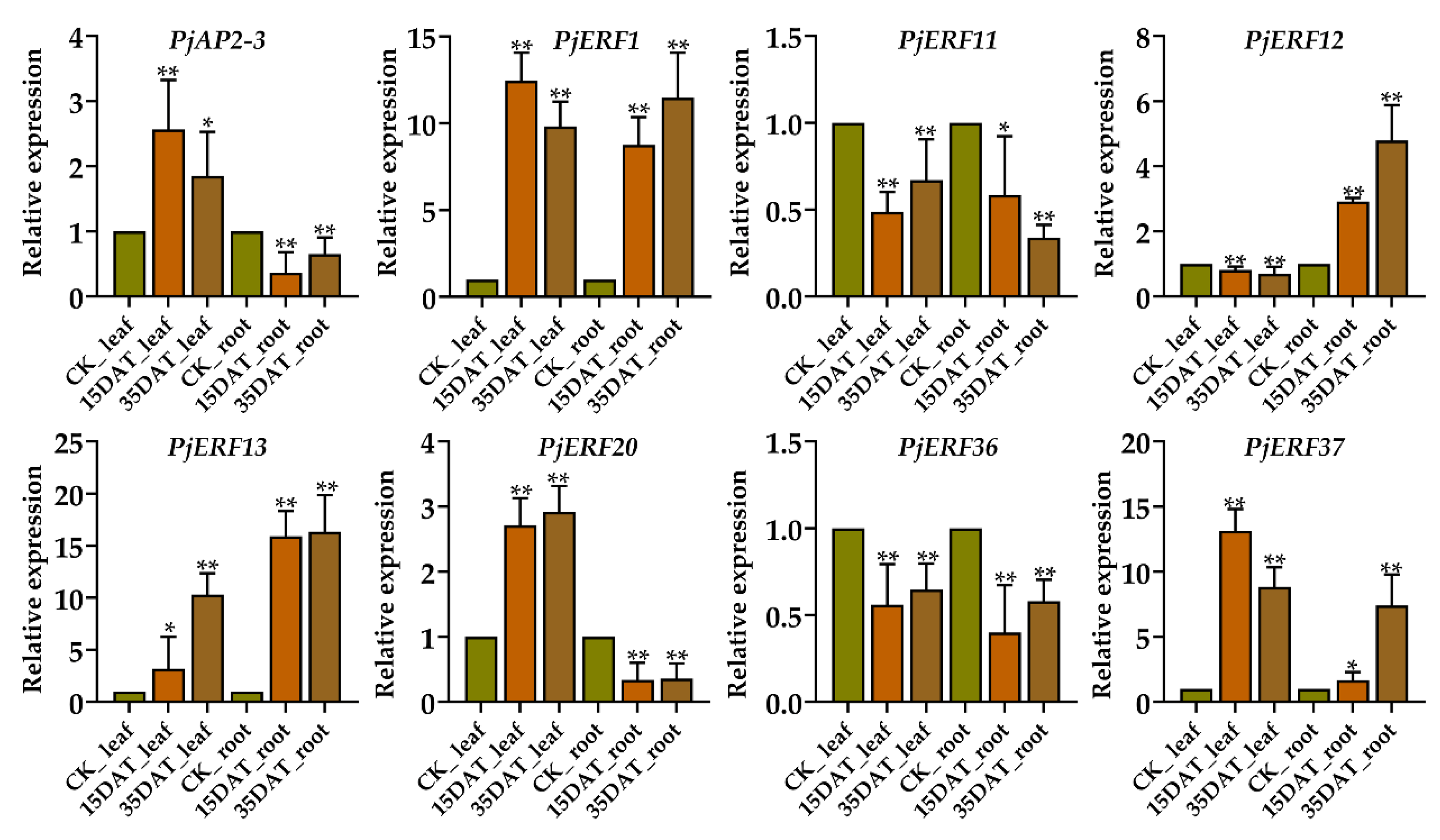
2.5. Validating the Expression of PjERFs during Waterlogging Conditions
2.6. Subcellular Localization of PjERF13
2.7. Overexpression of PjERF13 Enhanced the Tolerance of Tobacco to Waterlogging Stress
2.8. Overexpression of PjERF13 Decreased the Oxidative Damage in the Root
3. Discussion
4. Materials and Methods
4.1. Identification and Sequence Analysis of AP2/ERF Genes in Citrus junos
4.2. Phylogenetic Analysis, Chromosomal Distribution, and Conserved Motif Identification
4.3. Plant Growth and Waterlogging Stress Treatments
4.4. Analysis of PjAP2/ERFs Expression under Waterlogging Stress
4.5. Real-Time PCR Analysis
4.6. Subcellular Localization Analysis of PjERF13
4.7. Expression Pattern of PjERF13 Genes under Waterlogging Stress
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gmitter, F.G.; Chen, C.; Machado, M.A.; de Souza, A.A.; Ollitrault, P.; Froehlicher, Y.; Shimizu, T. Citrus genomics. Tree Genet. Genomes 2012, 8, 611–626. [Google Scholar] [CrossRef]
- Pan, J.; Sharif, R.; Xu, X.; Chen, X. Mechanisms of waterlogging tolerance in plants: Research progress and prospects. Front. Plant Sci. 2021, 11, 627331. [Google Scholar] [CrossRef]
- Pérez-Jiménez, M.; Pérez-Tornero, O. Short-term waterlogging in citrus rootstocks. Plants 2021, 10, 2772. [Google Scholar] [CrossRef] [PubMed]
- Habibi, F.; Liu, T.; Folta, K.; Sarkhosh, A. Physiological, biochemical, and molecular aspects of grafting in fruit trees. Hortic. Res. 2022, 9, uhac032. [Google Scholar] [CrossRef]
- Bowman, K.D.; McCollum, G.; Albrecht, U. Supersour: A new strategy for breeding superior citrus rootstocks. Front. Plant Sci. 2021, 12, 741009. [Google Scholar] [CrossRef]
- Huang, Y.; Xu, Y.; Jiang, X.; Yu, H.; Jia, H.; Tan, C.; Hu, G.; Hu, Y.; Rao, M.J.; Deng, X.; et al. Genome of a citrus rootstock and global DNA demethylation caused by heterografting. Hortic. Res. 2021, 8, 69. [Google Scholar] [CrossRef] [PubMed]
- Balfagón, D.; Terán, F.; de Oliveira, T.D.R.; Santa-Catarina, C.; Gómez-Cadenas, A. Citrus rootstocks modify scion antioxidant system under drought and heat stress combination. Plant Cell Rep. 2022, 41, 593–602. [Google Scholar] [CrossRef]
- Raiol-Junior, L.L.; de Carvalho, E.V.; Moreira, A.S.; Marques, J.P.R.; Stuchi, E.S.; Peña, L.; Girardi, E.A. Graft compatibility classification within Aurantioideae based on biometric traits and the anatomy of graft union. Agriculture 2022, 12, 76. [Google Scholar] [CrossRef]
- Shoji, T.; Yuan, L. ERF gene clusters: Working together to regulate metabolism. Trends Plant Sci. 2021, 26, 23–32. [Google Scholar] [CrossRef]
- Feng, K.; Hou, X.; Xing, G.; Liu, J.; Duan, A.; Xu, Z.; Li, M.; Zhuang, J.; Xiong, A. Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef]
- Zhuang, J.; Peng, R.; Cheng, Z.M.; Zhang, J.; Cai, B.; Zhang, Z.; Gao, F.; Zhu, B.; Fu, X.; Jin, X.; et al. Genome-wide analysis of the putative AP2/ERF family genes in Vitis vinifera. Sci. Hortic. 2009, 123, 73–81. [Google Scholar] [CrossRef]
- Bai, D.; Li, Z.; Hu, C.; Zhang, Y.; Muhammad, A.; Zhong, Y.; Fang, J. Transcriptome-wide identification and expression analysis of ERF family genes in Actinidia valvata during waterlogging stress. Sci. Hortic. 2021, 281, 109994. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, C.; Deng, M.; Li, S.; Chen, Y.; Gu, X.; Tang, G.; Lin, Y.; Wang, Y.; He, W.; et al. Genome-wide analysis of the ERF family and identification of potential genes involved in fruit ripening in octoploid strawberry. Int. J. Mol. Sci. 2022, 23, 10550. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.H.; Shangguan, L.F.; Ma, R.J.; Sun, X.; Tao, R.; Guo, L.; Korir, N.K.; Yu, M.L. Genome-wide analysis of the AP2/ERF superfamily in peach (Prunus persica). Genet. Mol. Res. 2012, 11, 4789. [Google Scholar] [CrossRef]
- Licausi, F.; Ohme Takagi, M.; Perata, P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef]
- Van Veen, H.; Akman, M.; Jamar, D.C.L.; Vreugdenhil, D.; Kooiker, M.; van Tienderen, P.; Voesenek, L.A.C.J.; Schranz, M.E.; Sasidharan, R. Group VII Ethylene Response Factor diversification and regulation in four species from flood-prone environments. Plant Cell Environ. 2014, 37, 2421–2432. [Google Scholar] [CrossRef]
- Yu, F.; Liang, K.; Fang, T.; Zhao, H.; Han, X.; Cai, M.; Qiu, F. A group VII ethylene response factor gene, ZmEREB180, coordinates waterlogging tolerance in maize seedlings. Plant Biotechnol. J. 2019, 17, 2286–2298. [Google Scholar] [CrossRef]
- Wei, X.; Xu, H.; Rong, W.; Ye, X.; Zhang, Z. Constitutive expression of a stabilized transcription factor group VII ethylene response factor enhances waterlogging tolerance in wheat without penalizing grain yield. Plant Cell Environ. 2019, 42, 1471–1485. [Google Scholar] [CrossRef]
- Liu, J.; Chen, Y.; Wang, W.; Liu, J.; Zhu, C.; Zhong, Y.; Zhang, H.; Liu, X.; Yin, X. Transcription factors AcERF74/75 respond to waterlogging stress and trigger alcoholic fermentation-related genes in kiwifruit. Plant Sci. 2022, 314, 111115. [Google Scholar] [CrossRef]
- Zhu, K.; Sun, Q.; Chen, H.; Mei, X.; Lu, S.; Ye, J.; Chai, L.; Xu, Q.; Deng, X. Ethylene activation of carotenoid biosynthesis by a novel transcription factor CsERF061. J. Exp. Bot. 2021, 72, 3137–3154. [Google Scholar] [CrossRef]
- Li, S.; Xie, X.; Liu, S.; Chen, K.; Yin, X. Auto- and mutual-regulation between two CitERFs contribute to ethylene-induced citrus fruit degreening. Food Chem. 2019, 299, 125163. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Hu, J.; Dahro, B.; Ming, R.; Zhang, Y.; Wang, Y.; Alhag, A.; Li, C.; Liu, J.H. ERF108 from Poncirus trifoliata (L.) Raf. functions in cold tolerance by modulating raffinose synthesis through transcriptional regulation of PtrRafS. Plant J. 2021, 108, 705–724. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ming, R.; Khan, M.; Wang, Y.; Dahro, B.; Xiao, W.; Li, C.; Liu, J.H. ERF9 of Poncirus trifoliata (L.) Raf. undergoes feedback regulation by ethylene and modulates cold tolerance via regulating aglutathione S-transferase U17 gene. Plant Biotechnol. J. 2022, 20, 183–200. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Dai, W.; Du, J.; Ming, R.; Dahro, B.; Liu, J.H. ERF109 of trifoliate orange (Poncirus trifoliata (L.) Raf.) contributes to cold tolerance by directly regulating expression of Prx1 involved in antioxidative process. Plant Biotechnol. J. 2019, 17, 1316–1332. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.; He, W.; Chai, J.; Luo, L.; Wang, Y.; Chen, Q.; Tang, H.; Wang, X. A study of scion phenotypes in pummelo grafted onto a new citrus rootstock Citrus junos ‘Pujiang Xiangcheng’. Horticulturae 2022, 8, 1039. [Google Scholar] [CrossRef]
- Xie, R.; Zheng, L.; Jiao, Y.; Huang, X. Understanding physiological and molecular mechanisms of citrus rootstock seedlings in response to root zone hypoxia by RNA-Seq. Environ. Exp. Bot. 2021, 192, 104647. [Google Scholar] [CrossRef]
- Xie, X.; Shen, S.; Yin, X.; Xu, Q.; Sun, C.; Grierson, D.; Ferguson, I.; Chen, K. Isolation, classification and transcription profiles of the AP2/ERF transcription factor superfamily in citrus. Mol. Biol. Rep. 2014, 41, 4261–4271. [Google Scholar] [CrossRef]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.Z.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Li, J.; Li, H.; Quan, X.; Shan, Q.; Wang, W.; Yin, N.; Wang, S.; Wang, Z.; He, W. Comprehensive analysis of cucumber C-repeat/dehydration-responsive element binding factor family genes and their potential roles in cold tolerance of cucumber. BMC Plant Biol. 2022, 22, 270. [Google Scholar] [CrossRef]
- Licausi, F.; Van Dongen, J.T.; Giuntoli, B.; Novi, G.; Santaniello, A.; Geigenberger, P.; Perata, P. HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J. 2010, 62, 302–315. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.; Chou, S.; Shih, M. Ethylene plays an essential role in the recovery of Arabidopsis during post-anaerobiosis reoxygenation. Plant Cell Environ. 2014, 37, 2391–2405. [Google Scholar] [CrossRef]
- Phukan, U.J.; Jeena, G.S.; Tripathi, V.; Shukla, R.K. MaRAP2-4, a waterlogging-responsive ERF from Mentha, regulates bidirectional sugar transporter AtSWEET10 to modulate stress response in Arabidopsis. Plant Biotechnol. J. 2018, 16, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Alpuerto, J.B.; Hussain, R.M.F.; Fukao, T. The key regulator of submergence tolerance, SUB1A, promotes photosynthetic and metabolic recovery from submergence damage in rice leaves. Plant Cell Environ. 2016, 39, 672–684. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Wang, G.; Wang, T.; Jia, Z.; Guo, Z.; Zhang, J. AdRAP2.3, a novel ethylene response factor VII from Actinidia deliciosa, enhances waterlogging resistance in transgenic tobacco through improving expression levels of PDC and ADH genes. Int. J. Mol. Sci. 2019, 20, 1189. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Fukao, T.; Gibbs, D.J.; Holdsworth, M.J.; Lee, S.C.; Licausi, F.; Perata, P.; Voesenek, L.A.C.J.; van Dongen, J.T. Making sense of low oxygen sensing. Trends Plant Sci. 2012, 17, 129–138. [Google Scholar] [CrossRef]
- Seok, H.; Tran, H.T.; Lee, S.; Moon, Y. AtERF71/HRE2, an Arabidopsis AP2/ERF transcription factor gene, contains both positive and negative cis-regulatory elements in its promoter region involved in hypoxia and salt stress responses. Int. J. Mol. Sci. 2022, 23, 5310. [Google Scholar] [CrossRef]
- Xu, K.; Xu, X.; Fukao, T.; Canlas, P.; Maghirang-Rodriguez, R.; Heuer, S.; Ismail, A.M.; Bailey-Serres, J.; Ronald, P.C.; Mackill, D.J. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 2006, 442, 705–708. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- He, W.; Wang, Y.; Chen, Q.; Sun, B.; Tang, H.; Pan, D.; Wang, X. Dissection of the mechanism for compatible and incompatible graft combinations of Citrus grandis (L.) Osbeck (‘Hongmian Miyou’). Int. J. Mol. Sci. 2018, 19, 505. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. Genome Biol. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Xie, R.; Li, H.; Wang, Y.; Chen, Q.; Lin, Y.; Zhang, Y.; Luo, Y.; Zhang, Y.; Tang, H.; et al. Evaluation of suitable qRT-PCR normalization genes for various citrus rootstocks. Plant Biotechnol. Rep. 2021, 16, 101–111. [Google Scholar] [CrossRef]
- Ye, Y.; Liu, Y.; Li, X.; Wang, G.; Zhou, Q.; Chen, Q.; Li, J.; Wang, X.; Tang, H. An evolutionary analysis of B-Box transcription factors in strawberry reveals the role of FaBBx28c1 in the regulation of flowering time. Int. J. Mol. Sci. 2021, 22, 11766. [Google Scholar] [CrossRef]
- Shahid, M.A.; Balal, R.M.; Khan, N.; Simón-Grao, S.; Alfosea-Simón, M.; Cámara-Zapata, J.M.; Mattson, N.S.; Garcia-Sanchez, F. Rootstocks influence the salt tolerance of Kinnow mandarin trees by altering the antioxidant defense system, osmolyte concentration, and toxic ion accumulation. Sci. Hortic. 2019, 250, 1–11. [Google Scholar] [CrossRef]
- Zaher-Ara, T.; Boroomand, N.; Sadat-Hosseini, M. Physiological and morphological response to drought stress in seedlings of ten citrus. Trees 2016, 30, 985–993. [Google Scholar] [CrossRef]
- Muñoz, N.; González, C.; Molina, A.; Zirulnik, F.; Luna, C.M. Cadmium-induced early changes in O2−, H2O2 and antioxidative enzymes in soybean (Glycine max L.) leaves. Plant Growth Regul. 2008, 56, 159–166. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, W.; Luo, L.; Xie, R.; Chai, J.; Wang, H.; Wang, Y.; Chen, Q.; Wu, Z.; Yang, S.; Li, M.; et al. Genome-Wide Identification and Functional Analysis of the AP2/ERF Transcription Factor Family in Citrus Rootstock under Waterlogging Stress. Int. J. Mol. Sci. 2023, 24, 8989. https://doi.org/10.3390/ijms24108989
He W, Luo L, Xie R, Chai J, Wang H, Wang Y, Chen Q, Wu Z, Yang S, Li M, et al. Genome-Wide Identification and Functional Analysis of the AP2/ERF Transcription Factor Family in Citrus Rootstock under Waterlogging Stress. International Journal of Molecular Sciences. 2023; 24(10):8989. https://doi.org/10.3390/ijms24108989
Chicago/Turabian StyleHe, Wen, Liang Luo, Rui Xie, Jiufeng Chai, Hao Wang, Yan Wang, Qing Chen, Zhiwei Wu, Shaofeng Yang, Mengyao Li, and et al. 2023. "Genome-Wide Identification and Functional Analysis of the AP2/ERF Transcription Factor Family in Citrus Rootstock under Waterlogging Stress" International Journal of Molecular Sciences 24, no. 10: 8989. https://doi.org/10.3390/ijms24108989
APA StyleHe, W., Luo, L., Xie, R., Chai, J., Wang, H., Wang, Y., Chen, Q., Wu, Z., Yang, S., Li, M., Lin, Y., Zhang, Y., Luo, Y., Zhang, Y., Tang, H., & Wang, X. (2023). Genome-Wide Identification and Functional Analysis of the AP2/ERF Transcription Factor Family in Citrus Rootstock under Waterlogging Stress. International Journal of Molecular Sciences, 24(10), 8989. https://doi.org/10.3390/ijms24108989







