2.3. Detailed Description of Representative Clinical Cases
Out of the 16 USC/CS patients studied with results presented in
Table 1, we selected a few representative cases to describe in more detail the patients’ disease courses in relation to treatment response, biomarkers collection (i.e., ctDNA and CA-125), and CT imaging. Visual representations of the patients’ clinical courses with CA-125 and ctDNA trends can be found in
Figure 1,
Figure 2,
Figure 3,
Figure 4 and
Figure 5.
Patient #1:
In October 2014, a 67-year-old female presented with postmenopausal bleeding and underwent robot-assisted total laparoscopic hysterectomy and bilateral salpingo-oophorectomy for a stage IA uterine serous carcinoma; notably, no lymph node or omental biopsies were taken at the time of staging. As depicted in
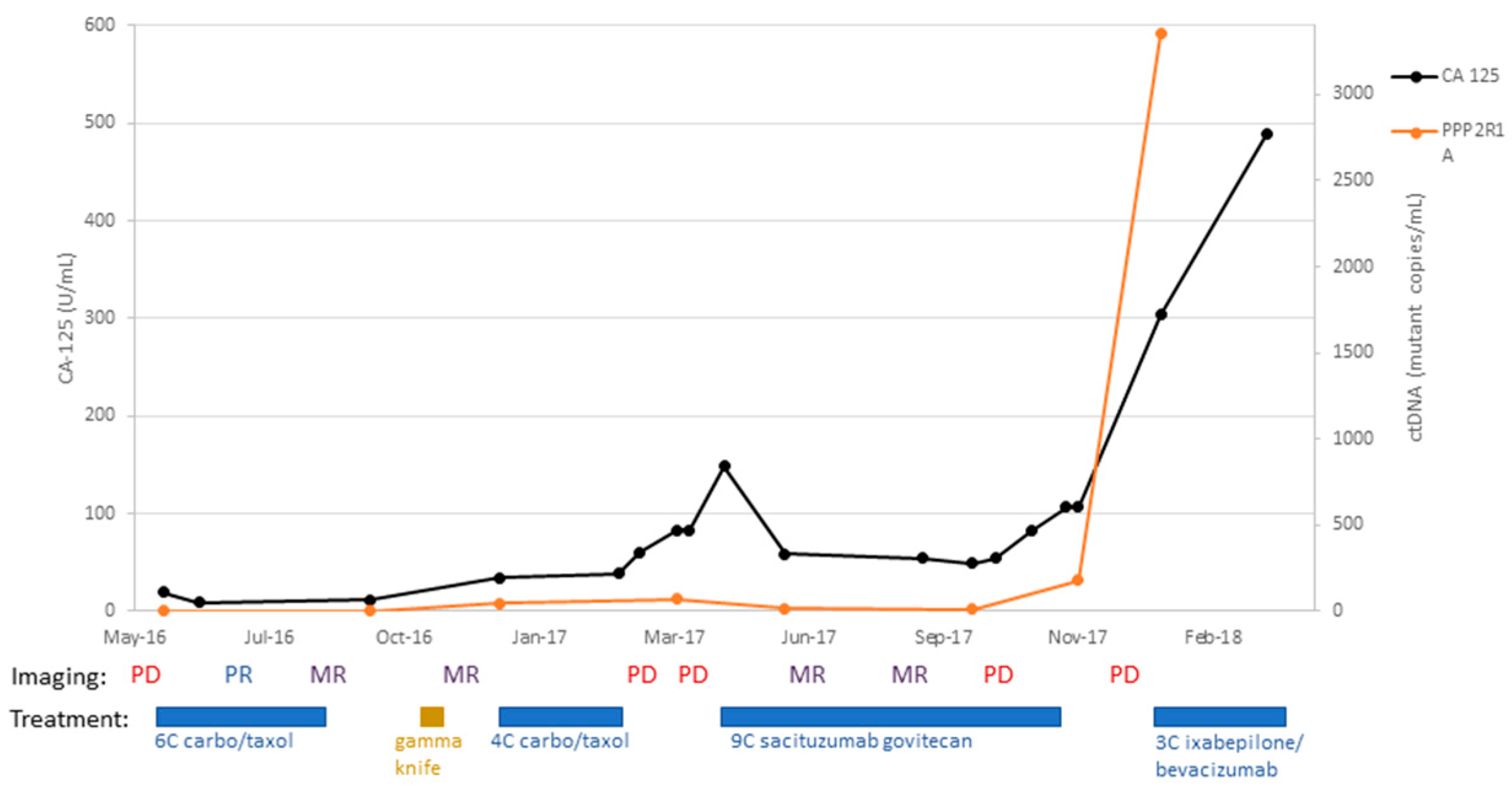
Figure 1, the patient was then treated with adjuvant paclitaxel and carboplatin for six cycles, completed in March 2015, and received vaginal apex brachytherapy (three fractions, completed 30 May 2015). A CT scan performed in June 2015 showed no evidence of disease (NED). Six months later, a surveillance CT scan (18 December 2015) showed potential metastatic disease in the pelvis and vagina, which was also detected on PET CT (30 December 2015), and recurrent USC was confirmed on lymph node biopsy. At that time, as shown in
Figure 1, CA-125 was normal at 12.7 U/mL (normal range: 0 to 35 U/mL), while ctDNA copy number was elevated to 770 mutant copy/mL. She was then enrolled in a clinical trial (NCT01367002), “Randomized Phase II Evaluation of Carboplatin/Paclitaxel with and without Trastuzumab in HER2/neu+ Patients with Advanced/Recurrent Uterine Serous Papillary Carcinoma).” She was randomized to the trial arm without trastuzumab and completed six cycles of carboplatin/paclitaxel chemotherapy. Unfortunately, a CT scan after treatment (11 October 2016) demonstrated the progression of her disease, with new pulmonary metastases, recurrent disease at vaginal apex, carcinomatosis, and worsening pelvic lymphadenopathy. As shown in
Figure 1, at the time of the CT scan, the CA-125 tumor marker was 14.8 U/mL (normal range:0 to 35 U/mL), while the ctDNA copy number was 109 mutant copy/mL. She then subsequently started another clinical trial of afatinib for patients with recurrent USC overexpressing HER2. She received afatinib therapy from 13 October to 20 December 2016; unfortunately, a CT scan imaging from 6 January 2017 demonstrated progression of her disease, with increased chest adenopathy, increased size of pulmonary nodules, and vaginal cuff recurrence. At this time, the patient also described increased vaginal bleeding. A vaginal apex biopsy 12 January 2017 was consistent with vaginal recurrence with HER2 3+ expression. As shown in
Figure 1, at the time of the vaginal biopsy confirming recurrent disease, CA-125 was negative at 17.2 U/mL (normal range: 0 to 35 U/mL), while ctDNA copy number was elevated to 457 mutant copy/mL. She then went on to receive trastuzumab and carboplatin for five cycles (12 January to 2 May 2017), as well as vaginal apex radiation to treat her vaginal recurrence (10 fractions, completed 11 April 2017). Unfortunately, a CT scan after her fifth cycle (19 May 2017) demonstrated interval growth of pulmonary metastases (an index lesion measured 4.4 cm, increased from 2.3 cm). With this worsening of the disease, carboplatin/trastuzumab was stopped, and she initiated treatment in an open-label clinical trial of sacituzumab govitecan for patients with recurrent uterine serous carcinoma, IMMU-132. She received two cycles of this experimental therapy but unfortunately experienced further progression of her disease (CT 10 August 2017). She was removed from the clinical trial secondary to progression, with a plan to start weekly abraxane. Unfortunately, in the 2 weeks after stopping IMMU-132, the patient developed difficulty walking and was found to have a large posterior fossa metastasis in the brain. The patient was discharged to hospice care on 31 August 2017. Notably, as described in
Figure 1, the patient’s ctDNA levels were found to strongly correlate to the clinical evidence of recurrent disease as evaluated by both imaging and confirmed with biopsies, while CA-125 levels remained within normal limits and without any significant increase throughout the course of her disease.
Figure 1.
Timeline of patient #1 disease course with treatment: C/T, carboplatin/paclitaxel; PD, progression of disease; MR, mixed response; SD, stable disease; PR, partial response; NED, no evidence of disease.
Figure 1.
Timeline of patient #1 disease course with treatment: C/T, carboplatin/paclitaxel; PD, progression of disease; MR, mixed response; SD, stable disease; PR, partial response; NED, no evidence of disease.
Patient #3:
A 68-year-old female with stage IVB uterine serous carcinoma underwent surgical staging with total abdominal hysterectomy, bilateral salpingo-oophorectomy, subtotal omentectomy, and pelvic and periaortic lymph node biopsy on 31 March 2014. Her tumor was found to overexpress HER2, and thus she received adjuvant carboplatin, paclitaxel, and trastuzumab for six cycles. A follow-up CT scan (8 October 2015) after adjuvant therapy demonstrated no evidence of disease. She completed adjuvant brachytherapy to vaginal apex in March 2015 and completed 12 months of trastuzumab maintenance with arimidex (i.e., an aromatase inhibitor). Unfortunately, a CT scan performed on 8 October 2015 demonstrated an enlarged periaortic lymph node suspicious for recurrent disease. To treat this recurrence, she completed radiotherapy (4320cGy) on 10 February 2016; her CA-125 decreased to 23.5 U/mL (2 March 2016) from 104 U/mL (31 December 2015). Unfortunately, a subsequent CT scan (13 April 2016) demonstrated several new areas of metastatic disease in the liver and lung. She was then started on dose-dense paclitaxel with carboplatin; after three cycles, a CT scan (17 June 2016) demonstrated a partial response, with interval improvement in pulmonary metastases and hepatic implant. Imaging at the conclusion of six cycles (22 August 2016) demonstrated an overall mixed response, with decreased size of pulmonary and hepatic metastases but increased size of a mediastinal lymph node. A further mixed response was demonstrated on a subsequent CT scan (7 November 2016). Unfortunately, at this time, two brain metastases were discovered in CT and MRI imaging studies. She went on to complete gamma knife radiation to her brain lesions (22 November 2016), which remained stable-to-improved on imaging 3 months later (6 March 2017). At that time, as depicted in
Figure 2, CA-125 was still within the normal limit at 34.4 U/mL (normal range: 0 to 35 U/mL), while ctDNA copy number was elevated to 46 mutant copy/mL. She completed four subsequent cycles of dose-dense paclitaxel/carboplatin. Unfortunately, a CT scan after these four cycles (6 March 2017) showed disease progression with enlarging lung metastases and increased retroperitoneal lymphadenopathy. At this point, the patient’s CA-125 was elevated to 83 U/mL while her ctDNA copy number was further increased to 69 mutant copy/mL. She then enrolled in the IMMU-132 (sacituzumab govitecan) open-label clinical trial and completed nine cycles of treatment. Imaging studies initially demonstrated a mixed response (22 June and 21 August 2017) but went on to demonstrate progressive disease (15 December 2018). She was removed from the IMMU-132 trial and was started on ixabepilone with bevacizumab. The patient required multiple delays in treatment because of leukopenia, symptomatic anemia, and dehydration. She completed three cycles of treatment (27 February 2018) and then chose to discontinue treatment due to her symptoms. The patient was discharged to hospice care (31 March 2018). As demonstrated in
Figure 2, her CA-125 levels as well as ctDNA levels were found to be concordant with disease response and progression.
Figure 2.
Timeline of patient #3 disease course with treatment: C/T, carboplatin/paclitaxel; PD, progression of disease; MR, mixed response; SD, stable disease; PR, partial response; NED, no evidence of disease.
Figure 2.
Timeline of patient #3 disease course with treatment: C/T, carboplatin/paclitaxel; PD, progression of disease; MR, mixed response; SD, stable disease; PR, partial response; NED, no evidence of disease.
Patient #9:
A 69-year-old patient underwent staging surgery for stage IVB uterine serous carcinoma (with a 20% clear cell component) on 8 August 2012. For adjuvant therapy, she received six cycles of carboplatin and paclitaxel. She completed adjuvant therapy and remained disease-free for almost 4 years. Starting in June of 2016, the patient’s CA-125 was noted to rise above its normal value (54.6 U/mL 7 June 2016). However, a CT scan acquired on 10 June 2016 did not demonstrate any visible recurrent disease. Importantly, as described in
Figure 3, evaluation of ctDNA at this time point demonstrated a copy number of 114 for PIK3CA vs. a copy number of 2 for TP53 mutant copy/mL. These findings triggered a PET CT within a few weeks that demonstrated two enlarging hypermetabolic mesenteric lymph nodes, consistent with recurrent metastatic disease. She then underwent rectosigmoid resection (1 September 2016), and the final pathology demonstrated recurrent USC, HER2 2+, metastatic to the bowel. She then had several scans demonstrating no evidence of disease until an 28 November 2017 PET CT demonstrated increased avidity in a 1 cm soft tissue nodule adjacent to the ascending colon. She then started trastuzumab therapy on 19 December 2017. Notably, her CA-125 did not rise as expected with the new metastatic implant; from that point forward in her course, CA-125 was no more a reliable marker of disease status, and accordingly, ctDNA and serial imaging studies were used to guide treatment decisions. Circulating tumor DNA with a TP53 probe was present for the first two collection time points (1 mutation/mL on 8 March 2016 and 2 mutation/mL on 7 June 2016) but then was undetectable for the remaining time points, even when PIK3CA mutations were consistently increasing in patient’s plasma (
Figure 3). Her subsequent imaging demonstrated stable oligometastatic disease with the 1 cm implant. Starting on 5 April 2019, her PIK3CA ctDNA values began to rise; 14 mut/mL on 5 April 2019 to 88 mut/mL on 28 June 2019. Despite this increase, there was no evidence of disease on a 17 June 2019 CT scan. She completed 31 cycles of trastzumab on 20 September 2019. Unfortunately, CT imaging on 20 September 2019 demonstrated increased size of the prior 1 cm implant, now 3.6 cm. On 28 October 2019, she underwent laparoscopic right hemicolectomy, and pathology confirmed metastatic disease; notably, HER2 was not expressed. PIK3CA ctDNA copies decreased from 1599 mut/mL before the surgery to 22 mut/mL after the surgery. She then went on to receive pembrolizumab (17 December 2019) with lenvatinib added for cycle two (12 January 2020). Notably, during the COVID-19 pandemic, lab collections were kept to a minimum and so while the patient continued treatment, we ceased ctDNA collection after 7 January 2020. Lenvatinib was discontinued in June 2020 due to side effects, and the patient continued pembrolizumab for 12 total cycles (completed 4 August 2020). A PET CT from 24 September 2020 showed new recurrent disease with an intensely avid anterior abdominal wall nodule. She was then started on the IMMU-132 trial (sacituzumab govitecan) (16 October 2020). Her subsequent CT scans demonstrated a decreased size of the lesion (11 December 2020) and stable disease (5 February 2021). On 2 April 2021, her abdominal wall metastasis was noted to have increased in size significantly, and she was removed from the IMMU-132 trial after eight cycles (31 March 2021). As the patient again had oligometastatic disease, she underwent tumor debulking of her port site metastasis on 23 April 2021. As her postop PET CT was concerning for ongoing FDG avidity along her abdominal wall, she was started on weekly temsirolimus (25 mg) on 29 June 2021 because of her PIK3CA 1007 K hotspot mutation. Her subsequent scans have all shown no evidence of disease. She remains free of disease on temsirolimus 20 mg/weekly and was last seen in our clinic on 21 February 2023.
Figure 3.
Timeline of patient #9 disease course with treatment: C/T, carboplatin/paclitaxel; PD, progression of disease; MR, mixed response; SD, stable disease; PR, partial response; NED, no evidence of disease.
Figure 3.
Timeline of patient #9 disease course with treatment: C/T, carboplatin/paclitaxel; PD, progression of disease; MR, mixed response; SD, stable disease; PR, partial response; NED, no evidence of disease.
Patient #13:
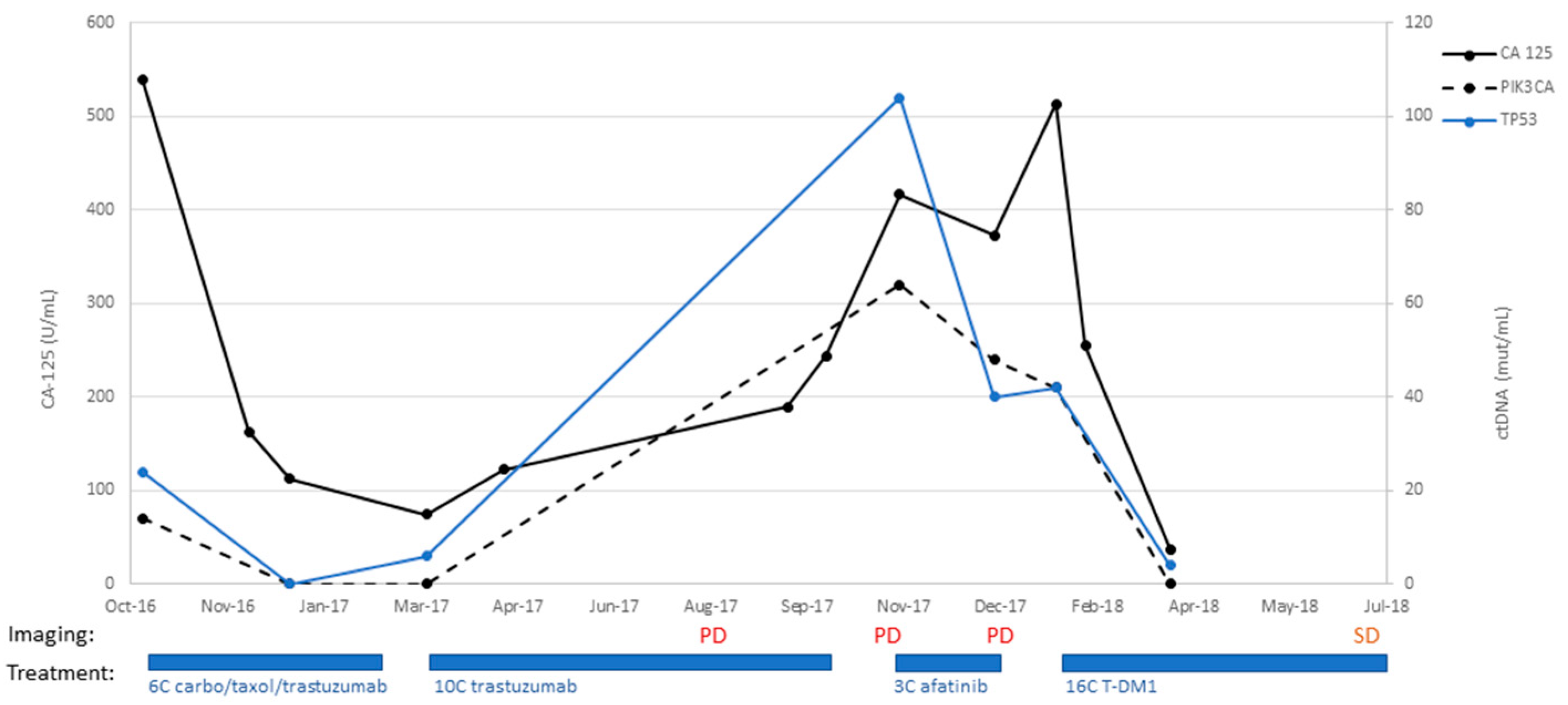
An 82-year-old patient underwent staging surgery on 26 September 2016 with a total laparoscopic hysterectomy, bilateral salpingo-oophorectomy, pelvic and periaortic lymph node biopsies, and omentectomy for a stage IVB uterine serous carcinoma, with HER2 overexpression. Postoperatively, she received six cycles of adjuvant carboplatin, paclitaxel, and trastuzumab. She then transitioned to maintenance trastuzumab and completed eight cycles. On 22 August 2017, she underwent a surveillance CT scan, which unfortunately demonstrated the progression of the disease. This was preceded by an increase in CA-125 to 122 (18 April 2017) from 74.7 (9 March 2017), as shown in
Figure 4. She completed an additional two cycles of trastuzumab (completed 5 October 2017). On 2 November 2017, a CT scan demonstrated interval growth of lung nodules, as well as increased size of a bladder mass. She then went on to begin treatment with afatinib on 11 November 2017. She received three cycles but, unfortunately, had increasing CA-125 values and progressive disease on imaging. On 18 January 2018, she had vaginal cuff biopsies consistent with serous carcinoma. She then began treatment with the antibody-drug-conjugate trastuzumab emtansine (T-DM1) on 29 January 2018. She received 16 cycles and experienced decreased CA-125 (513 U/mL on 29 January 2018 to 37.2 U/mL on 29 March 2018). Her ctDNA was followed with two separate probes for PIK3CA and TP53, both of which decreased dramatically during her treatment with T-DM1 as well and correlated to her CA-125 trends throughout her disease course (
Figure 4). She had one CT scan after initiating T-DM1 treatment on 6 July 2018, which demonstrated stable disease in the omentum and lower abdomen, with a slight increase in the size of a right upper-lobe metastasis. The patient, unfortunately, succumbed to injuries sustained in a motor vehicle accident on 21 December 2018.
Figure 4.
Timeline of patient #13 disease course with treatment: C/T, carboplatin/paclitaxel; PD, progression of disease; MR, mixed response; SD, stable disease; PR, partial response; NED, no evidence of disease.
Figure 4.
Timeline of patient #13 disease course with treatment: C/T, carboplatin/paclitaxel; PD, progression of disease; MR, mixed response; SD, stable disease; PR, partial response; NED, no evidence of disease.
Patient #14:
A 64-year-old patient underwent a staging laparoscopic hysterectomy, bilateral salpingo-oophorectomy, sentinel lymph node biopsy, and partial omentectomy on 3 January 2018. Pathology yielded stage IA carcinosarcoma of the uterus, with endometrial adenocarcinoma and homologous sarcomatous components and positive pelvic washings. She completed six cycles of adjuvant carboplatin and paclitaxel and brachytherapy to the vaginal cuff. She underwent surveillance CT scans every 3 months, as CA-125 was proven to not be a marker of disease for her. Next-generation sequencing yielded a mutation in PTEN (R130G) in her tumor. For 6 months after the completion of her adjuvant therapy, she had persistently negative results for ctDNA, CA-125, and CT imaging (
Figure 5). Starting in February 2019, a 1.5 cm cystic lesion adjacent to the third portion of the duodenum was identified on a CT scan. As the patient’s ctDNA results remained negative throughout this time, and thus clinical suspicion for recurrence was low, the mass was followed with serial CT scans. The mass was unchanged on imaging until 29 April 2020 when the patient presented with partial small bowel obstruction (SBO) symptoms. On CT imaging at that time, the mass was identified to have increased in size from 1.5 cm to 1.7 cm. The patient underwent diagnostic laparoscopy and lysis of adhesions. What had appeared as a cystic mass on CT imaging, concerning for a potential recurrence, was found to be a fibrotic band causing an SBO. Multiple biopsies were sent for pathologic evaluation, all of which were negative for recurrent carcinosarcoma. This patient remains disease-free after 4 years from the initial diagnosis.
Figure 5.
Timeline of patient #14 disease course with treatment: C/T, carboplatin/paclitaxel; PD, progression of disease; MR, mixed response; SD, stable disease; PR, partial response; NED, no evidence of disease; SBO, small bowel obstruction.
Figure 5.
Timeline of patient #14 disease course with treatment: C/T, carboplatin/paclitaxel; PD, progression of disease; MR, mixed response; SD, stable disease; PR, partial response; NED, no evidence of disease; SBO, small bowel obstruction.
Additional Patients:
Two additional patients (i.e., patients #2 and #6) were both diagnosed with advanced CS and USC (
Table 1) and were similarly treated with surgical staging/debulking followed by gold standard carboplatin and paclitaxel chemotherapy. These patients also underwent ctDNA, CA-125, and CT imaging evaluation every 6 months after completion of their adjuvant therapy and had persistently negative results for ctDNA, CA-125, and CT imaging. These two patients, for whom ctDNA was undetectable at 6 months after the completion of treatment, remain disease-free after 5 years (patient #2) and 9 years (patient #6) from the initial diagnosis.