Identification of Endometrial Cancer-Specific microRNA Biomarkers in Endometrial Fluid
Abstract
1. Introduction
2. Results
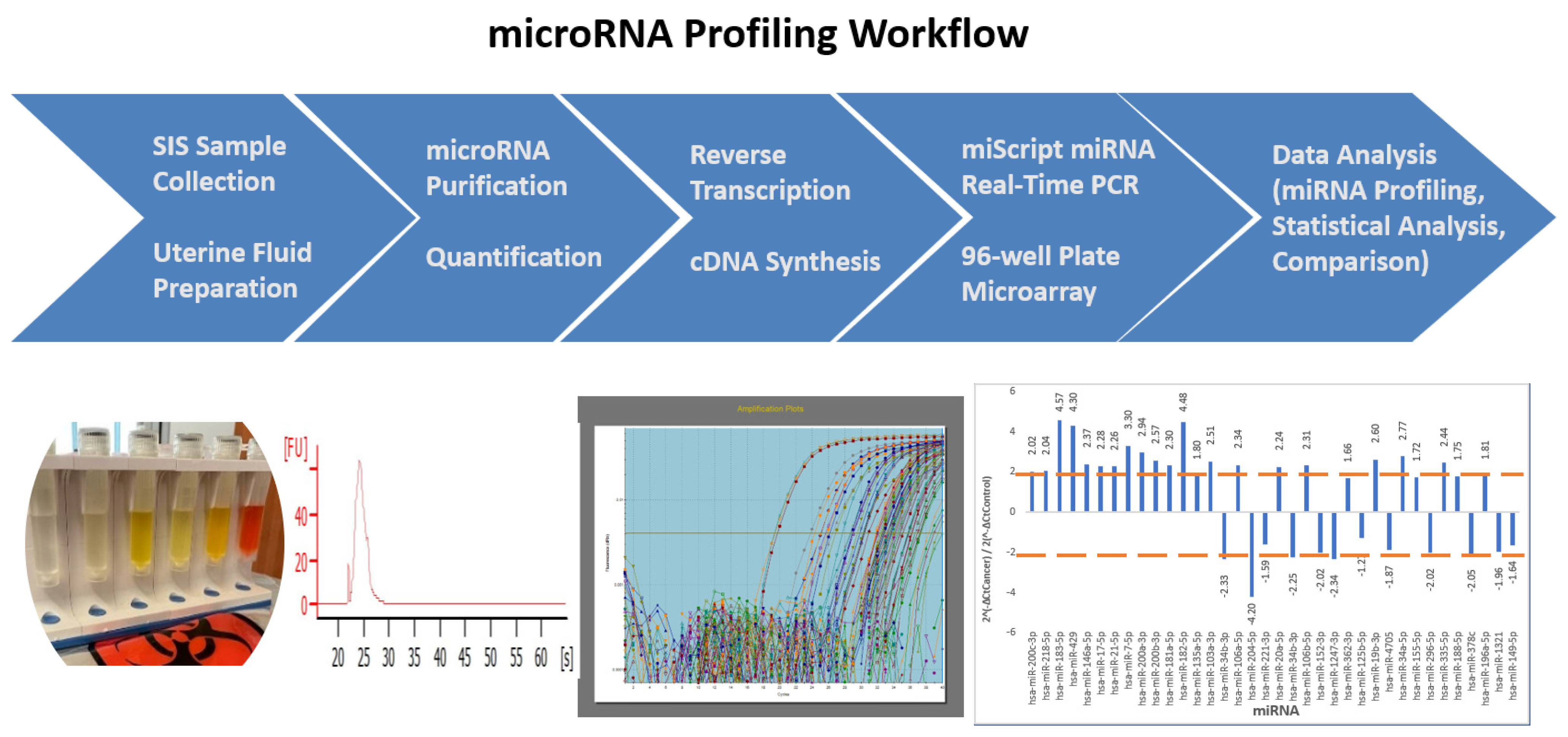
2.1. Study Design and Workflow
2.2. miRNA Extraction and Selection
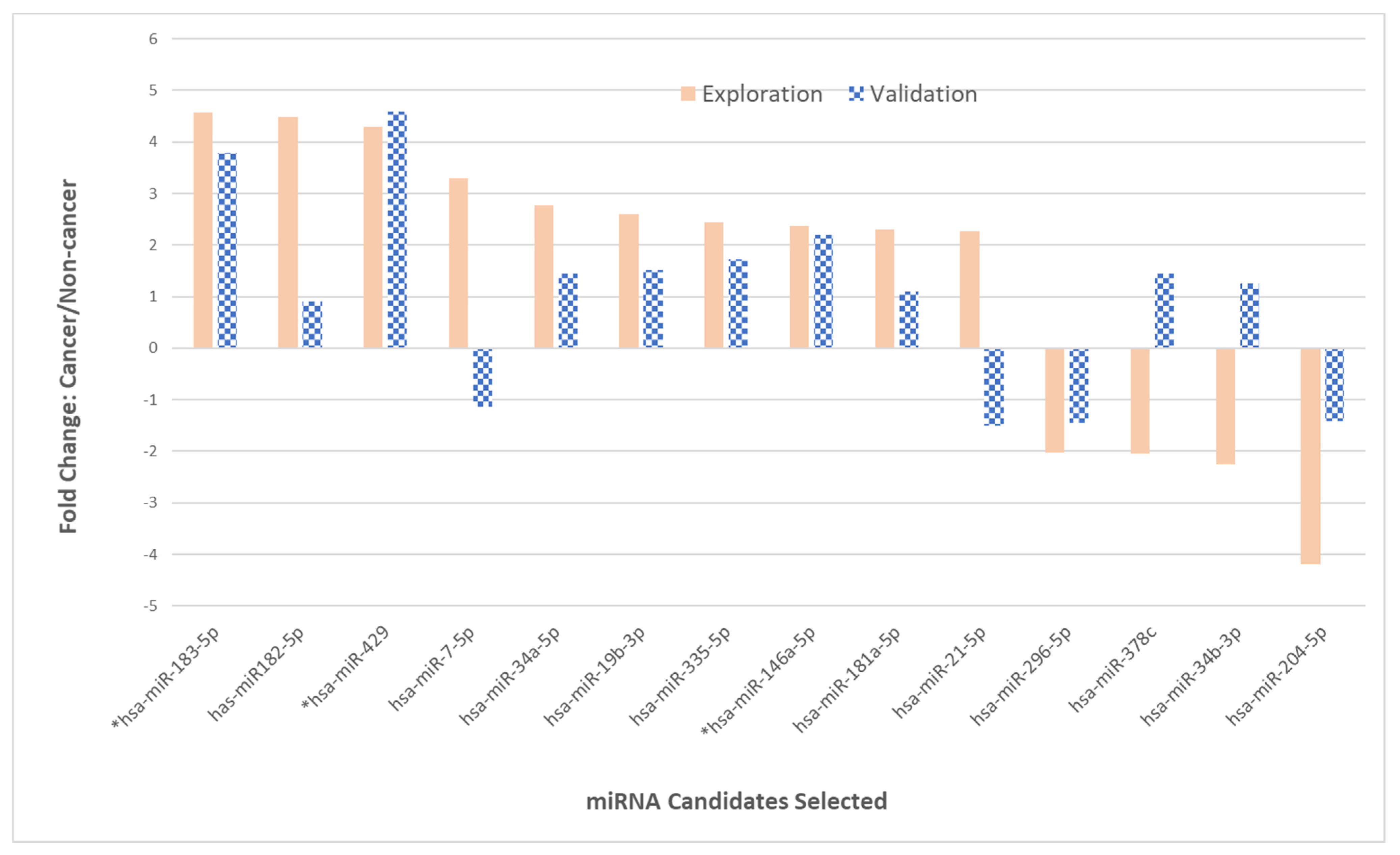
2.3. Phase II—Validation of 14 miRNAs
2.4. Clinical and Demographic Features of Patients in the Study
3. Discussion
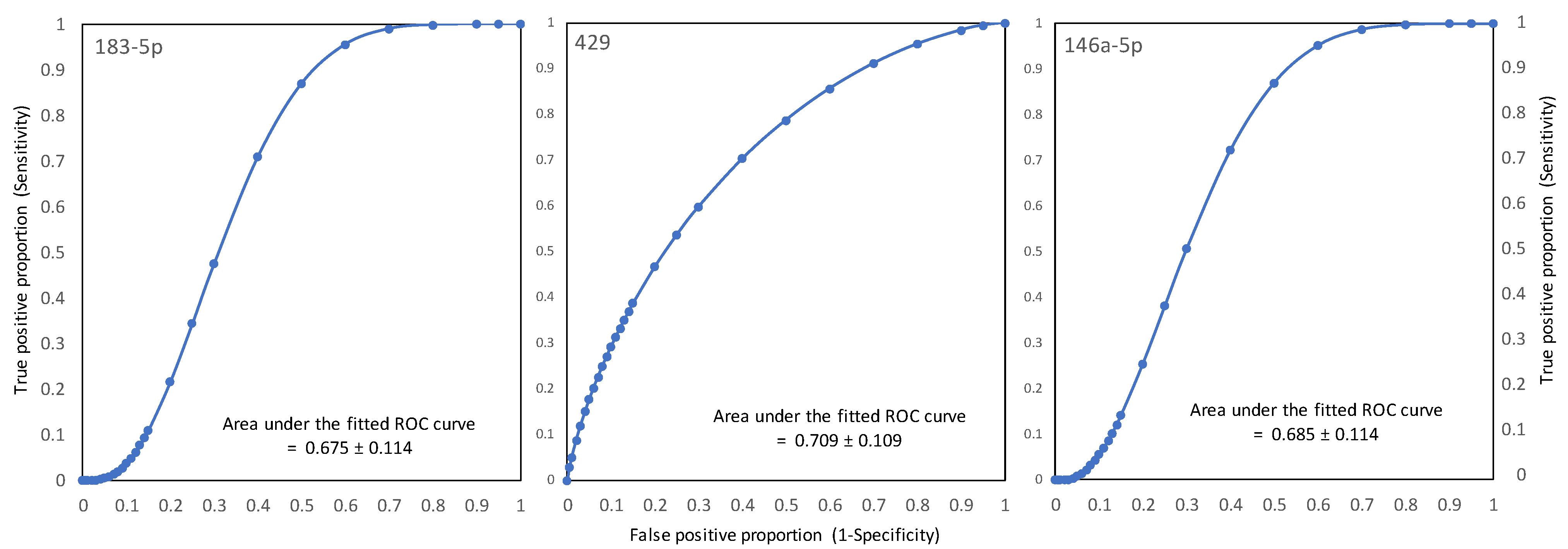
3.1. EC-Relevant miRNAs Identified in Phase I of the Study
3.2. Several Unique miRNAs Identified in EC Endometrial Fluid
3.3. Inconsistency in miRNA Expression between Phase I and Phase II
3.4. Comparing the miRNA Expression Changes Identified in This Study with Those Reported in the Literature
3.5. Clinical Implications
3.6. Strengths and Limitations of Our Study
4. Materials and Methods
4.1. Endometrial Fluid Sample Collection and Preparation
4.2. miRNA Isolation and Custom miRNA qPCR Micro-Array Plate
4.2.1. Total RNA (Including miRNA) Extraction and Quantification
4.2.2. First-Strand DNA Synthesis (Reverse Transcription)
4.2.3. Customized 96-Well miScript miRNA PCR Array Plate and Real-Time qPCR Amplification
4.2.4. Data Analysis and Normalization
4.3. Pathological Diagnosis
4.4. Experimental Design—Phase I (Exploration) and Phase II (Validation)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Key Statistics for Endometrial Cancer. Available online: https://www.cancer.org/cancer/endometrial-cancer/about/key-statistics.html (accessed on 10 November 2022).
- Goldstein, S.R. Modern evaluation of the endometrium. Obs. Gynecol. 2010, 116, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Simon, C.; Crawford, S.C.; Davis, J.A.; Siddiqui, N.A.; Caestecker, L.D.; Gillis, C.R.; Hole, D.; Penney, G. The waiting time paradox: Population based retrospective study of treatment delay and survival of women with endometrial cancer in Scotland. BMJ 2002, 325, 196. [Google Scholar] [CrossRef]
- Elit, L.M.; O’Leary, E.M.; Pond, G.R.; Seow, H.Y. Impact of wait times on survival for women with uterine cancer. J. Clin. Oncol. 2014, 32, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Spencer, C.P.; Whitehead, M.I. Endometrial assessment re-visited. Br. J. Obs. Gynaecol. 1999, 106, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Guido, R.S.; Kanbour-Shakir, A.; Rulin, M.C.; Christopherson, A.W. Pipelle endometrial sampling. Sensitivity in the detection of endometrial cancer. J. Reprod. Med. 1995, 40, 553–555. [Google Scholar]
- Visser, N.C.M.; Reijinen, C.; Massuger, L.F.A.G.; Nagtegaal, I.D.; Bulten, J.; Pijnenborg, J.M.A. Accuracy of endometrial sampling in endometrial carcinoma: A systematic review and meta-analysis. Obstet. Gynecol. 2017, 130, 803–813. [Google Scholar] [CrossRef]
- Shen, Y.; Yang, W.; Liu, J.; Zhang, Y. Minimally invasive approaches for the early detection of endometrial cancer. Mol. Cancer 2023, 22, 53. [Google Scholar] [CrossRef]
- Duque, G.; Manterola, C.; Otzen, T.; Arias, C.; Palacios, D.; Mora, M.; Galindo, B.; Holguin, J.P.; Albarracin, L. Cancer biomarkers in liquid biopsy for early detection of breast cancer: A systematic review. Clin. Med. Insights Oncol. 2022, 16, 1–16. [Google Scholar] [CrossRef]
- Chang, L.; Ni, J.; Zhu, Y.; Pang, B.; Graham, P.; Zhang, H.; Li, Y. Liquid biopsy in ovarian cancer: Recent advances in circulating extracellular vesicle detection for early diagnosis and monitoring progression. Theranostics 2019, 9, 4130. [Google Scholar] [CrossRef]
- Rahmioglu, N.; Fassbender, A.; Vitonis, A.F.; Tworoger, S.S.; Hummelshoj, L.; D’Hooghe, T.M.; Adamson, G.D.; Giudice, L.C.; Becker, C.M.; Zondervan, K.T.; et al. WERF EPHect Working Group. World Endometriosis Research Foundation Endometriosis Phenome and Biobanking Harmonization Project: III. Fluid biospecimen collection, processing, and storage in endometriosis research. Fertil. Steril. 2014, 102, 1233–1243. [Google Scholar] [CrossRef]
- Njoku, K.; Chiasserini, D.; Geary, B.; Pierce, A.; Jones, E.R.; Whetton, A.D.; Crosbie, E.J. Comprehensive Library Generation for Identification and Quantification of Endometrial Cancer Protein Biomarkers in Cervico-Vaginal Fluid. Cancers 2021, 13, 3804. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.D.; Lyons, K.S.; Gerçel-Taylor, C. Shed membrane fragment-associated markers for endometrial and ovarian cancers. Gynecol. Oncol. 2002, 84, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Romero-Cordoba, S.L.; Salido-Guadarrama, I.; Rodriguez-Dorantes, M.; Hidalgo-Miranda, A. miRNA biogenesis: Biological impact in development of cancer. Cancer Biol. Ther. 2014, 15, 444–455. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Ruvkun, G. Molecular biology. Glimpses of a tiny RNA world. Science 2001, 294, 797–799. [Google Scholar] [CrossRef]
- Fasoulakis, Z.; Daskalakis, G.; Diakosavvas, M.; Papapanagiotou, I.; Theodora, M.; Bourazan, A.; Alatzidou, D.; Pagkalos, A.; Kontomanolis, E.N. MicroRNAs Determining Carcinogenesis by Regulating Oncogenes and Tumor Suppressor Genes during Cell Cycle. Microrna 2020, 9, 82–92. [Google Scholar] [CrossRef]
- Yanokura, M.; Banno, K.; Iida, M.; Irie, H.; Umene, K.; Masuda, K.; Kobayashi, Y.; Tominaga, E.; Aoki, D. MicroRNAS in endometrial cancer: Recent advances and potential clinical applications. EXCLI J. 2015, 14, 190–198. [Google Scholar] [CrossRef]
- Banno, K.; Yanokura, M.; Kisu, L.; Yamagami, W.; Susumu, N.; Aoki, D. MicroRNAs in Endometrial Cancer. Int. J. Clin. Oncol. 2013, 18, 186–192. [Google Scholar] [CrossRef]
- Jurcevic, S.; Olsson, B.; Klinga-Levan, K. MicroRNA expression in human endometrial adenocarcinoma. Cancer Cell Int. 2014, 14, 88. [Google Scholar] [CrossRef]
- Bagaria, M.; Shields, E.; Bakkum-Gamez, J.N. Novel approaches to early detection of endometrial cancer. Curr. Opin. Obstet. Gynecol. 2017, 29, 40–46. [Google Scholar] [CrossRef]
- Boren, T.; Xiong, Y.; Hakam, A.; Wenham, R.; Apte, S.; Wei, Z.; Kamath, S.; Chen, D.T.; Dressman, H.; Lancaster, J.M. MicroRNAs and their target messenger RNAs associated with endometrial carcinogenesis. Gynecol. Oncol. 2008, 110, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Kolanska, K.; Bendifallah, S.; Canlorbe, G.; Mekinian, A.; Touboul, C.; Aractingi, S.; Chabbert-Buffet, N.; Daraï, E. Role of miRNAs in Normal Endometrium and in Endometrial Disorders: Comprehensive Review. J. Clin. Med. 2021, 10, 3457. [Google Scholar] [CrossRef] [PubMed]
- Donkers, H.; Bekkers, R.; Galaal, K. Diagnostic Value of microRNA panel in endometrial cancer: A systemic review. Oncotarget 2020, 11, 2010–2023. [Google Scholar] [CrossRef] [PubMed]
- Favier, A.; Rocher, G.; Larsen, A.K.; Delangle, R.; Uzan, C.; Sabbah, M.; Castela, M.; Duval, A.; Mehats, C.; Canlorbe, G. MicroRNA as epigenetic modifiers in endometrial cancer: A systematic review. Cancers 2020, 13, 1137. [Google Scholar] [CrossRef]
- Klicka, K.; Grzywa, T.M.; Linke, A.; Mielniczuk, A.; Wlodarski, P.K. The role of miRNA in the regulation of endometrial cancer invasiveness and metastasis—A systematic review. Cancers 2021, 13, 3393. [Google Scholar] [CrossRef]
- Yoneyama, K.; Ishibashi, O.; Kawase, R.; Kurose, K.; Takeshita, T. miR-200a, miR-200b and miR-429 are onco-miRs that target the PTEN gene in endometrioid endometrial carcinoma. Antican. Res. 2015, 35, 1401–1410. [Google Scholar]
- Jurcevic, S.; Klinga-Levan, K.; Olsson, B.; Ejeskär, K. Verification of microRNA expression in human endometrial adenocarcinoma. BMC Cancer 2016, 16, 261. [Google Scholar] [CrossRef]
- Wilczynski, M.; Danielska, J.; Dzieniecka, M.; Szymanska, B.; Wojciechowski, M.; Malinowski, A. Prognostic and clinical significance of miRNA-205 in endometrioid endometrial cancer. PLoS ONE. 2016, 11, e0164687. [Google Scholar] [CrossRef]
- Donkers, H.; Hirschfeld, M.; Weiß, D.; Erbes, T.; Jaeger, M.; Pijnenborg, J.M.A.; Bekkers, R.; Galaaal, K. ENITEC-consortium. Usefulness of miRNA detection in the diagnosis of endometrial cancer. Acta Obstet. Gynecol. Scand. 2021, 100, 1148–1154. [Google Scholar] [CrossRef]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef]
- Bevacqua, E.; Ammirato, S.; Cione, E.; Curcio, R.; Dolce, V.; Tucci, P. The potential of microRNA as non-invasive prostate cancer biomarkers: A systematic literature review based on a machine learning approach. Cancers 2022, 14, 5418. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sun, H.; Wang, X.; Yu, Q.; Li, S.; Yu, X.; Gong, W. Increased exosomal microRNA-21 and microRNA-146a levels in the cervicovaginal lavage specimens of patients with cervical cancer. Int. J. Mol. Sci. 2014, 15, 758–773. [Google Scholar] [CrossRef] [PubMed]
- Ibañez-Perez, J.; Díaz-Nuñez, M.; Clos-García, M.; Lainz, L.; Iglesias, M.; Díez-Zapirain, M.; Rabanal, A.; Bárcena, L.; González, M.; Lozano, J.J.; et al. microRNA-based signatures obtained from endometrial fluid identify implantative endometrium. Hum. Reprod. 2022, 37, 2375–2391. [Google Scholar] [CrossRef]
- von Grothusen, C.; Frisendahl, C.; Modhukur, V.; Lalitkumar, P.G.; Peters, M.; Faridani, O.R.; Salumets, A.; Boggavarapu, N.R.; Gemzell-Danielsson, K. Uterine fluid microRNAs are dysregulated in women with recurrent implantation failure. Hum. Reprod. 2022, 37, 734–746. [Google Scholar] [CrossRef] [PubMed]
- Grasso, A.; Navarro, R.; Balaguer, N.; Moreno, I.; Alama, P.; Jimenez, J.; Simón, C.; Vilella, F. Endometrial Liquid Biopsy Provides a miRNA Roadmap of the Secretory Phase of the Human Endometrium. J. Clin. Endocrinol. Metab. 2020, 105, 877–889. [Google Scholar] [CrossRef]
- Campoy, I.; Lanau, L.; Altadill, T.; Sequeiros, T.; Cabrera, S.; Cubo-Abert, M.; Pérez-Benavente, A.; Garcia, A.; Borrós, S.; Santamaria, A.; et al. Exosome-like vesicles in uterine aspirates: A comparison of ultracentrifugation-based isolation protocols. J. Trans. Med. 2016, 14, 180. [Google Scholar] [CrossRef]
- Ng, Y.H.; Rome, S.; Jalabert, A.; Forterre, A.; Singh, H.; Hincks, C.L.; Salamonsen, L.A. Endometrial exosomes/microvesicles in the uterine microenvironment: A new paradigm for embryo-endometrial cross talk at implantation. PLoS ONE 2013, 8, e58502. [Google Scholar] [CrossRef]
- Colas, E.; Perez, C.; Cabrera, S.; Pedrola, N.; Monge, M.; Castellvi, J.; Eyzaguirre, F.; Gregorio, J.; Ruiz, A.; Llaurado, M.; et al. Molecular markers of endometrial carcinoma detected in uterine aspirates. Int. J. Cancer 2011, 129, 2435–2444. [Google Scholar] [CrossRef]
- Hunter, M.P.; Ismail, N.; Zhang, X.; Aguda, B.D.; Lee, E.J.; Yu, L.; Xiao, T.; Schafer, J.; Lee, M.-L.T.; Schmittgen, T.D.; et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE 2008, 3, e3694. [Google Scholar] [CrossRef]
- Banno, K.; Nogami, Y.; Kisu, I.; Yanokura, M.; Umene, K.; Masuda, K.; Kobayashi, Y.; Yamagami, W.; Susumu, N.; Aoki, D. Candidate biomarkers for genetic and clinicopathological diagnosis of endometrial cancer. Int. J. Mol. Sci. 2013, 14, 12123–12137. [Google Scholar] [CrossRef]
- Banno, K.; Kisu, I.; Yanokura, M.; Tsuji, K.; Masuda, K.; Ueki, A.; Kobayashi, Y.; Yamagami, W.; Nomura, H.; Tominaga, E.; et al. Biomarkers in endometrial cancer: Possible clinical applications (Review). Oncol. Lett. 2012, 3, 1175–1180. [Google Scholar] [CrossRef] [PubMed]
- Supernat, A.; Łapińska-Szumczyk, S.; Majewska, H.; Gulczyński, J.; Biernat, W.; Wydra, D.; Zaczek, A.J. A multimarker qPCR platform for the characterisation of endometrial cancer. Oncol. Rep. 2014, 31, 1003–1013. [Google Scholar] [CrossRef] [PubMed]
- Grushko, T.A.; Filiaci, V.L.; Mundt, A.J.; Ridderstråle, K.; Olopade, O.I.; Fleming, G.F. Gynecologic Oncology Group. An exploratory analysis of HER-2 amplification and overexpression in advanced endometrial carcinoma: A Gynecologic Oncology Group study. Gynecol. Oncol. 2008, 108, 3–9. [Google Scholar] [CrossRef]
- miRBase. Available online: http://www.mirbase.org (accessed on 4 November 2015).
- Yan, H.; Sun, B.M.; Zhang, Y.Y.; Li, Y.J.; Huang, C.X.; Feng, F.Z.; Li, C. Upregulation of miR-183-5p is responsible for the promotion of apoptosis and inhibition of the epithelial-mesenchymal transition, proliferation, invasion and migration of human endometrial cancer cells by downregulating Ezrin. Int. J. Mol. Med. 2018, 42, 2469–2480. [Google Scholar] [CrossRef]
- Iacona, J.R.; Lutz, C.S. miR-146a-5p: Expression, regulation, and functions in cancer. Wiley Interdiscip. Rev. RNA 2019, 10, e1533. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.X.; Zhao, W.; Mao, L.W.; Wang, Y.L.; Xia, L.Q.; Cao, M.; Shen, J.; Chen, J. Long non-coding RNA NIFK-AS1 inhibits M2 polarization of macrophages in endometrial cancer through targeting miR-146a. Int. J. Biochem. Cell Biol. 2018, 104, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Zhao, N.; Zhao, N.; Hu, X.; He, X.; Xu, Y.; Chen, J.; Chen, W.; Liu, X.; Zhou, Z.; et al. Tumor-Suppressive and Oncogenic Roles of microRNA-149-5p in Human Cancers. Int. J. Mol. Sci. 2022, 23, 10823. [Google Scholar] [CrossRef] [PubMed]
- Ren, F.J.; Yao, Y.; Cai, X.Y.; Cai, Y.T.; Su, Q.; Fang, G.Y. MiR-149-5p: An Important miRNA Regulated by Competing Endogenous RNAs in Diverse Human Cancers. Front. Oncol. 2021, 11, 743077. [Google Scholar] [CrossRef]
- Liu, Y.; Chang, Y.; Cai, Y. Hsa_circ_0061140 promotes endometrial carcinoma progression via regulating miR-149-5p/STAT3. Gene 2020, 745, 144625. [Google Scholar] [CrossRef]
- Wang, C.; Li, Q.; He, Y. MicroRNA-21-5p promotes epithelial to mesenchymal transition by targeting SRY-box 17 in endometrial cancer. Oncol. Rep. 2020, 43, 1897–1905. [Google Scholar] [CrossRef]
- Dong, M.; Xie, Y.; Xu, Y. miR-7-5p regulates the proliferation and migration of colorectal cancer cells by negatively regulating the expression of Krüppel-like factor 4. Oncol. Lett. 2019, 17, 3241–3246. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Huang, W.; Huang, H.; Wang, L.; Wang, M.; Zhang, T.; Fang, X.; Xia, X. miR-182-5p and miR-96-5p target PIAS1 and mediate the negative feedback regulatory loop between PIAS1 and STAT3 in endometrial cancer. DNA Cell Biol. 2021, 40, 618–628. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Zhu, Z.; Song, L.; He, Z. Transferred by exosomes-derived MiR-19b-3p targets PTEN to regulate esophageal cancer cell apoptosis, migration, and invasion. Biosci. Rep. 2020, 40, BSR20201858. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Zhao, Y.; Chen, L.; Sun, Y.; Fan, S. miR-335-5p Inhibits Progression of Uterine Leiomyoma by Targeting ARGLU1. Comput. Math. Methods Med. 2022, 2022, 2329576. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.; Li, L.X.; Bian, X.Y.; Shi, X.J.; Lu, L.L.; Zhou, H.X.; Pan, T.J.; Zhou, J.; Fan, J.; Wu, W.Z. miR-296-5p suppresses EMT of hepatocellular carcinoma via attenuating NRG1/ERBB2/ERBB3 signaling. J. Exp. Clin. Cancer Res. 2018, 37, 294. [Google Scholar] [CrossRef]
- Li, S.J.; Yang, F.X.; Wang, M.Y.; Cao, W.J.; Yang, Z. miR-378 functions as an onco-miRNA by targeting the ST7L/Wnt/β-catenin pathway in cervical cancer. Int. J. Mol. Med. 2017, 40, 1047–1056. [Google Scholar] [CrossRef]
- Zeng, M.; Zhu, L.; Li, L.; Kang, C. miR-378 suppresses the proliferation, migration and invasion of colon cancer cells by inhibiting SDAD1. Cell. Mol. Biol. Lett. 2017, 22, 12. [Google Scholar] [CrossRef]
- Hiroki, E.; Suzuki, F.; Akahira, J.; Nagase, S.; Ito, K.; Sugawara, H.; Miki, Y.; Suzuki, T.; Sasano, H.; Yaegashi, N. MicroRNA-34b functions as a potential tumor suppressor in endometrial serous adenocarcinoma. Int. J. Cancer 2011, 131, e395–e404. [Google Scholar] [CrossRef]
- Bao, W.; Wang, H.H.; Tian, F.J.; He, X.Y.; Qiu, M.T.; Wang, J.Y.; Zhang, H.J.; Wang, L.H.; Wan, X.P. A TrkB-STAT3-miR-204-5p regulatory circuitry controls proliferation and invasion of endometrial carcinoma cells. Mol. Cancer 2013, 12, 155. [Google Scholar] [CrossRef]
- Cao, S.; Li, N.; Liao, X. miR-362-3p acts as a tumor suppressor by targeting SERBP1 in ovarian cancer. J. Ovarian Res. 2021, 14, 23. [Google Scholar] [CrossRef]
- Kang, H.; Kim, C.; Lee, H.; Rho, J.G.; Seo, J.W.; Nam, J.W.; Song, W.K.; Nam, S.W.; Kim, W.; Lee, E.K. Downregulation of microRNA-362-3p and microRNA-329 promotes tumor progression in human breast cancer. Cell Death Differ. 2016, 23, 484–495. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhang, X.; Sun, Y.; Shi, J.; Jiang, D.; Wang, J.; Liu, Y.; Hu, C.; Pan, J.; Zheng, L.; et al. MicroRNA-362-3p Inhibits Migration and Invasion via Targeting BCAP31 in Cervical Cancer. Front. Mol. Biosci. 2020, 7, 107. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Zhang, L.; Yang, H.; Luo, K.; Qing, C. Long non-coding RNA NEAT1 promotes ovarian cancer cell invasion and migration by interacting with miR-1321 and regulating tight junction protein 3 expression. Mol. Med. Rep. 2020, 22, 3429–3439. [Google Scholar] [CrossRef]
- He, S.N.; Guan, S.H.; Wu, M.Y.; Li, W.; Xu, M.D.; Tao, M. Down-regulated hsa_circ_0067934 facilitated the progression of gastric cancer by sponging hsa-mir-4705 to downgrade the expression of BMPR1B. Transl. Cancer Res. 2019, 8, 2691–2703. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.-J.; Zhou, H.; Xiao, H.-X.; Li, Y.; Zhou, T. MiR-378 is an independent prognostic factor and inhibits cell growth and invasion in colorectal cancer. BMC Cancer 2014, 14, 109. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Fan, Y.; Xu, W.; Chen, J.; Meng, Y.; Fang, D.; Wang, J. Exploration of miR-1202 and miR-196a in human endometrial cancer based on high throughout gene screening analysis. Oncol. Rep. 2017, 37, 3493–3501. [Google Scholar] [CrossRef] [PubMed]
- Sladkevicius, P.; Installe, A.; van Den Bosch, T.; Timmerman, D.; Benacerraf, B.; Jokubkiene, L.; Di Legge, A.; Votino, A.; Zannoni, L.; De Moor, B.; et al. International Endometrial Tumor Analysis (IETA) Terminology in Women with Postmenopausal Bleeding and Sonographic Endometrial Thickness ≥4.5 mm: Agreement and Reliability Study. Ultrasound Obstet. Gynecol. 2018, 51, 259–268. [Google Scholar] [CrossRef]
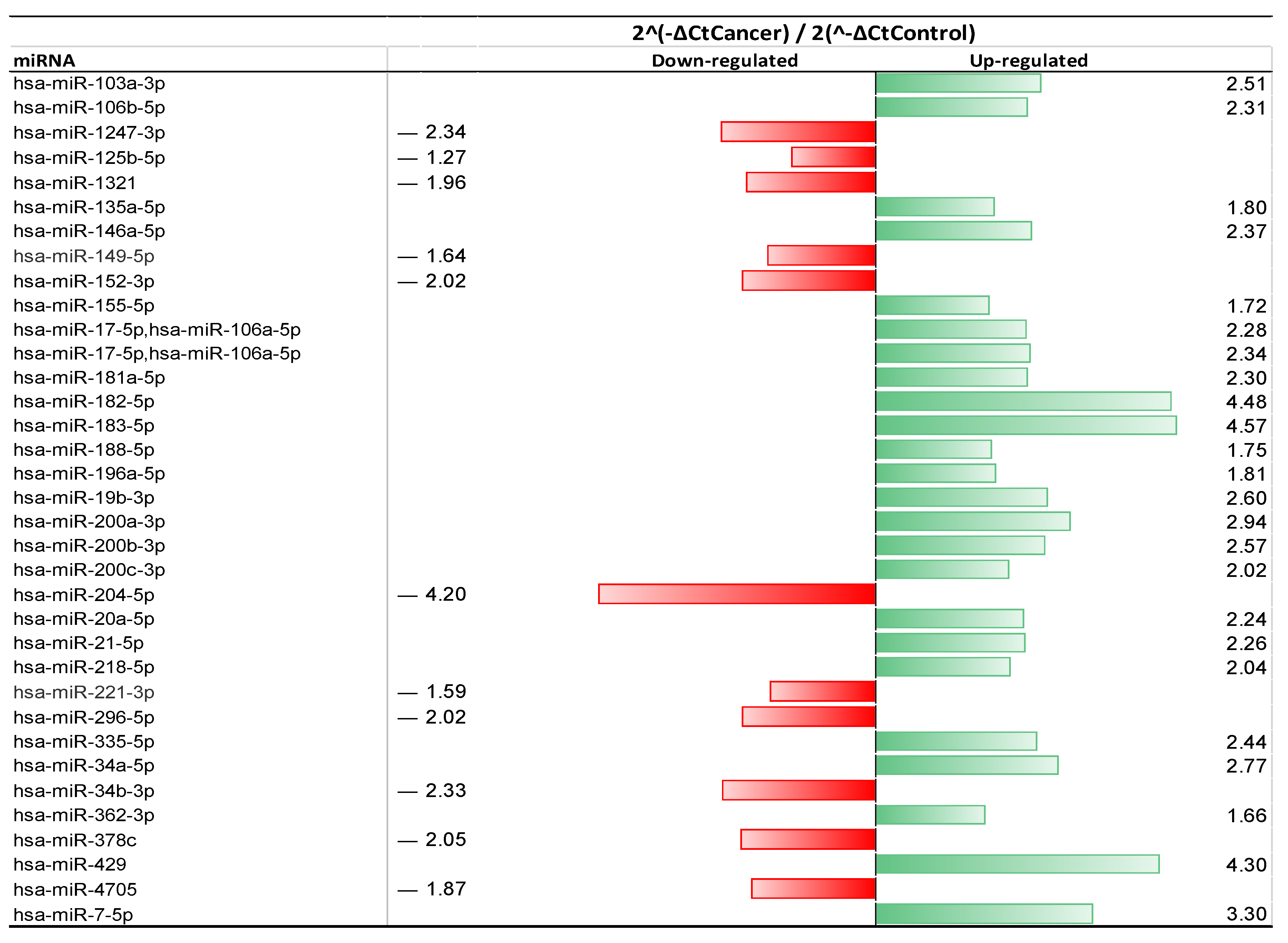
| >2-Fold Change | ||
| Up-Regulated | Down-Regulated | |
| Matched miRNA | miR-182-5p | miR-204-5p |
| miR-183-5p | miR-152-3p | |
| miR-429 | miR-1247-3p | |
| miR-200a-3p | miR-34b-3p | |
| miR-200b-3p | miR-296-5p | |
| miR-200c-3p | ||
| miR-106b-5p | ||
| miR-34a-5p | ||
| miR-20a-5p | ||
| miR-7-5p | ||
| miR-218-5p | ||
| miR-17-5p | ||
| miR-106a-5p | ||
| miR-21-5p | ||
| miR-181a-5p | ||
| miR-103a-3p | ||
| miR-19b-3p | ||
| miR-335-5p | ||
| miR-146a-5p | ||
| Unmatched miRNA | miR-378c | |
| <2-Fold Change | ||
| Up-regulated | Down-regulated | |
| Matched miRNA | miR-155-5p | miR-221-3p |
| miR-188-5p | miR-149-5p | |
| miR-196a-5p | miR-125b-5p | |
| miR-135a-5p | ||
| Unmatched miRNA | miR-362-3p | miR-4705 |
| miR-1321 |
| 2−ΔCtCancer/2−ΔCtControl | ||||
|---|---|---|---|---|
| Count | Functional Change Identified | miRNA Selected | Phase I | Phase II |
| 1 | Up-regulated | * hsa-miR-183-5p | 4.57 | 3.79 |
| 2 | hsa-miR-182-5p | 4.48 | 1.34 | |
| 3 | * hsa-miR-429 | 4.30 | 4.59 | |
| 4 | hsa-miR-7-5p | 3.30 | −1.14 | |
| 5 | hsa-miR-34a-5p | 2.77 | 1.45 | |
| 6 | hsa-miR-19b-3p | 2.60 | 1.51 | |
| 7 | hsa-miR-335-5p | 2.44 | 1.73 | |
| 8 | * hsa-miR-146a-5p | 2.37 | 2.19 | |
| 9 | has-miR-181a-5p | 2.30 | 1.10 | |
| 10 | hsa-miR-21-5p | 2.26 | −1.50 | |
| 11 | Down-regulated | hsa-miR-296-5p | −2.02 | −1.45 |
| 12 | hsa-miR-378c | −2.05 | 1.45 | |
| 13 | hsa-miR-34b-3p | −2.25 | 1.25 | |
| 14 | hsa-miR-204-5p | −4.20 | −1.41 | |
| Patient-Related Features | Benign (n = 40) * | Cancer (n = 42) * | p ** |
|---|---|---|---|
| Categorical variables, n (%) | |||
| Race | |||
| White | 34 (87.18) | 38 (90.48) | Ref |
| Black | 4 (10.26) | 3 (7.14) | 0.642 |
| Asian | 0 (0) | 1 (2.38) | 0.534 |
| N. American | 1 (2.56) | 0 (0) | 0.480 |
| Ethnicity | |||
| Hispanic | 25 (64.1) | 21 (50) | 0.211 |
| Non-Hispanic | 14 (35.9) | 21 (50) | Ref |
| Contraception | |||
| None | 11 (28.21) | 5 (11.9) | Ref |
| Some form of contraceptives | 23 (58.97) | 6 (14.28) | 0.456 |
| Post-menopause | 5 (12.82) | 29 (69.05) | <0.001 |
| Smoking status | |||
| Former or current smoker | 11 (28.21) | 5 (11.9) | 0.092 |
| Never smoked | 28(73.68) | 37(88.1) | Ref |
| STI | |||
| STI/history of | 3 (7.69) | 2 (4.76) | 0.622 |
| None | 36 (92.31) | 40 (95.24) | Ref |
| Continuous variables, mean ± sd (median, range) | |||
| Age | 45.5 ± 5.6 (45, 36–61) | 58.3±12.3 (57.5, 36–88) | <0.001 |
| BMI | 32.4 ± 5.8 (31.9, 21.7–44.6) | 37.8±8.9 (37.4, 20.5–56) | 0.005 |
| Gravidity | 3.2 ± 1.6 (3, 0–9) | 2.7±1.9 (3, 0–8) | 0.209 |
| Parity | 2.6 ± 1.6 (3, 0–9) | 2.2±1.9 (2, 0–9) | 0.305 |
| Phase I | Phase II | ||||
|---|---|---|---|---|---|
| Histologic Type | Grade | # of Cases | % of Total | # of Cases | % of Total |
| EIN * | - | 4 | 13.33 | 0 | 0 |
| Endometrioid | 25 | 83.33 | 12 | 100 | |
| 1 | 8 | 27 | 10 | 83.33 | |
| 2 | 16 | 53 | 2 | 16.66 | |
| 3 | 1 | 3.33 | 0 | 0 | |
| Rhabdomyosarcoma | 1 | 0 | |||
| 3 | 1 | 3.33 | 0 | 0 | |
| Total | 30 | 100 | 12 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.; Barkley, J.E.; Bhattarai, B.; Firouzi, K.; Monk, B.J.; Coonrod, D.V.; Zenhausern, F. Identification of Endometrial Cancer-Specific microRNA Biomarkers in Endometrial Fluid. Int. J. Mol. Sci. 2023, 24, 8683. https://doi.org/10.3390/ijms24108683
Yang J, Barkley JE, Bhattarai B, Firouzi K, Monk BJ, Coonrod DV, Zenhausern F. Identification of Endometrial Cancer-Specific microRNA Biomarkers in Endometrial Fluid. International Journal of Molecular Sciences. 2023; 24(10):8683. https://doi.org/10.3390/ijms24108683
Chicago/Turabian StyleYang, Jianing, Joel E. Barkley, Bikash Bhattarai, Kameron Firouzi, Bradley J. Monk, Dean V. Coonrod, and Frederic Zenhausern. 2023. "Identification of Endometrial Cancer-Specific microRNA Biomarkers in Endometrial Fluid" International Journal of Molecular Sciences 24, no. 10: 8683. https://doi.org/10.3390/ijms24108683
APA StyleYang, J., Barkley, J. E., Bhattarai, B., Firouzi, K., Monk, B. J., Coonrod, D. V., & Zenhausern, F. (2023). Identification of Endometrial Cancer-Specific microRNA Biomarkers in Endometrial Fluid. International Journal of Molecular Sciences, 24(10), 8683. https://doi.org/10.3390/ijms24108683





