Claudin Barriers on the Brink: How Conflicting Tissue and Cellular Priorities Drive IBD Pathogenesis
Abstract
1. Barrier Loss in IBD
2. Claudin Expression in Health and Disease
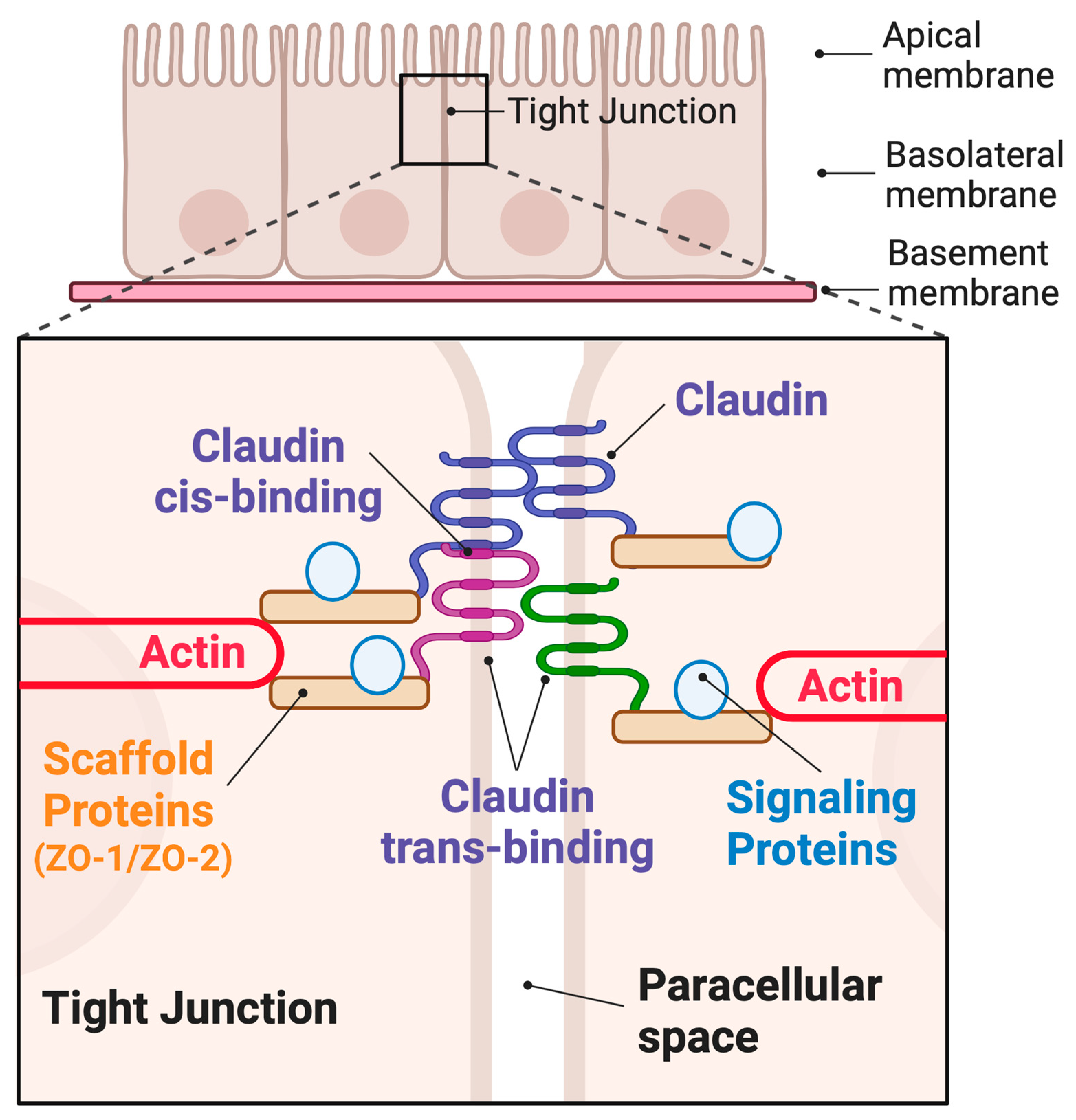
3. Claudin Family Proteins and the Hierarchy of Tight Junction Structure
4. Claudins Exhibit Dynamic Self-Assembly
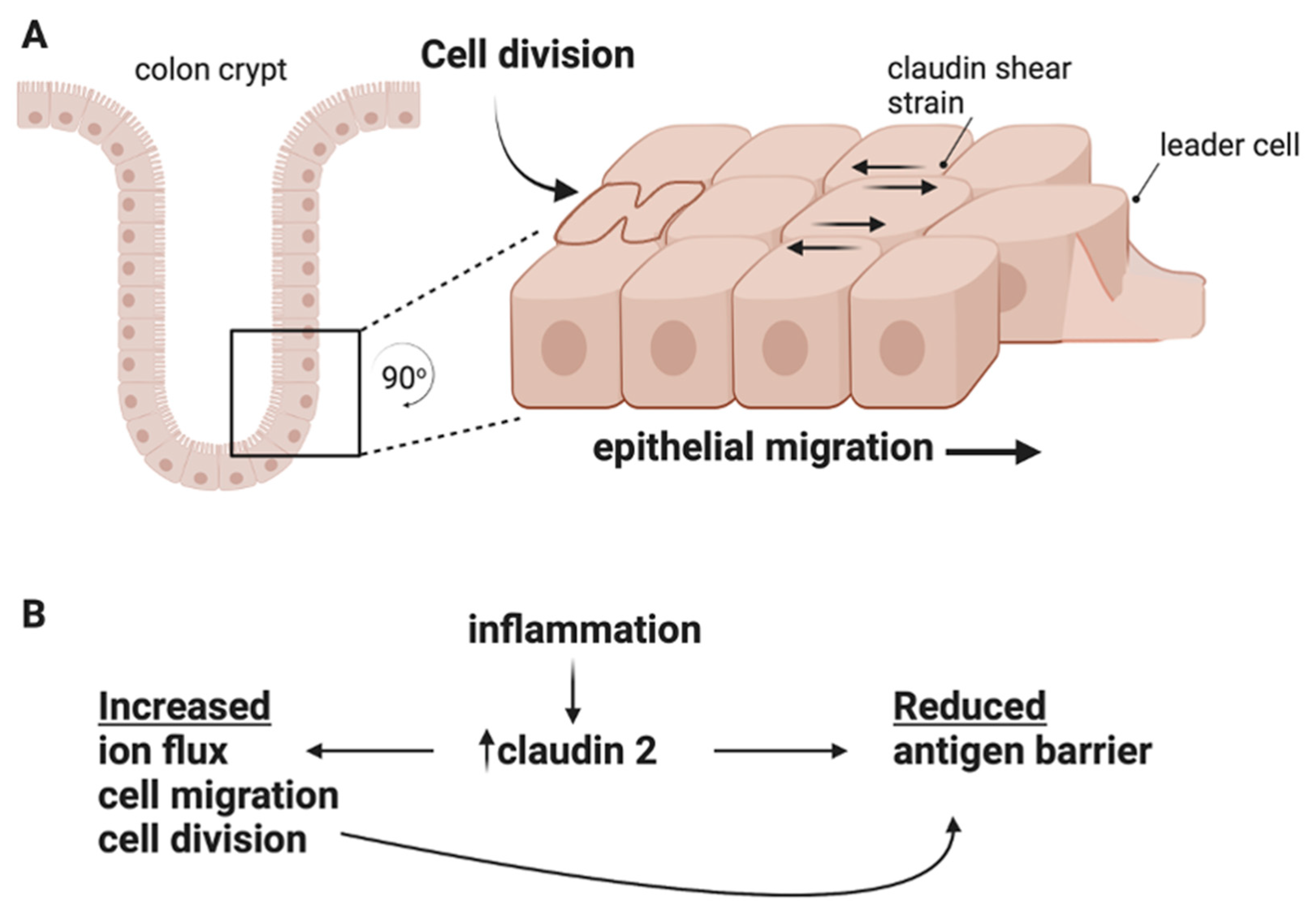
5. Claudins Regulate Cell Proliferation and Cell Migration
6. Claudins Participate in Mechanotransduction
7. Concluding Remarks
Funding
Acknowledgments
Conflicts of Interest
References
- Kobayashi, T.; Siegmund, B.; Le Berre, C.; Wei, S.C.; Ferrante, M.; Shen, B.; Bernstein, C.N.; Danese, S.; Peyrin-Biroulet, L.; Hibi, T. Ulcerative colitis. Nat. Rev. Dis. Primers 2020, 6, 74. [Google Scholar] [CrossRef] [PubMed]
- Roda, G.; Chien Ng, S.; Kotze, P.G.; Argollo, M.; Panaccione, R.; Spinelli, A.; Kaser, A.; Peyrin-Biroulet, L.; Danese, S. Crohn’s disease. Nat. Rev. Dis. Primers 2020, 6, 22. [Google Scholar] [CrossRef] [PubMed]
- Leung, G.; Muise, A.M. Monogenic Intestinal Epithelium Defects and the Development of Inflammatory Bowel Disease. Physiology 2018, 33, 360–369. [Google Scholar] [CrossRef] [PubMed]
- Villablanca, E.J.; Selin, K.; Hedin, C.R.H. Mechanisms of mucosal healing: Treating inflammatory bowel disease without immunosuppression? Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 493–507. [Google Scholar] [CrossRef]
- Odenwald, M.A.; Turner, J.R. Intestinal permeability defects: Is it time to treat? Clin. Gastroenterol. Hepatol. 2013, 11, 1075–1083. [Google Scholar] [CrossRef]
- Weidinger, C.; Krug, S.M.; Voskens, C.; Moschen, A.R.; Atreya, I. Editorial: Loss of Epithelial Barrier Integrity in Inflammatory Diseases: Cellular Mediators and Therapeutic Targets. Front. Med. 2021, 8, 813153. [Google Scholar] [CrossRef]
- Williams, J.M.; Duckworth, C.A.; Watson, A.J.; Frey, M.R.; Miguel, J.C.; Burkitt, M.D.; Sutton, R.; Hughes, K.R.; Hall, L.J.; Caamano, J.H.; et al. A mouse model of pathological small intestinal epithelial cell apoptosis and shedding induced by systemic administration of lipopolysaccharide. Dis. Model. Mech. 2013, 6, 1388–1399. [Google Scholar] [CrossRef]
- Chanez-Paredes, S.D.; Abtahi, S.; Kuo, W.T.; Turner, J.R. Differentiating Between Tight Junction-Dependent and Tight Junction-Independent Intestinal Barrier Loss In Vivo. Methods Mol. Biol. 2021, 2367, 249–271. [Google Scholar] [CrossRef]
- Van Itallie, C.M.; Anderson, J.M. Architecture of tight junctions and principles of molecular composition. Semin. Cell. Dev. Biol. 2014, 36, 157–165. [Google Scholar] [CrossRef]
- Zhu, L.; Han, J.; Li, L.; Wang, Y.; Li, Y.; Zhang, S. Claudin Family Participates in the Pathogenesis of Inflammatory Bowel Diseases and Colitis-Associated Colorectal Cancer. Front. Immunol. 2019, 10, 1441. [Google Scholar] [CrossRef]
- Barkas, F.; Liberopoulos, E.; Kei, A.; Elisaf, M. Electrolyte and acid-base disorders in inflammatory bowel disease. Ann. Gastroenterol. 2013, 26, 23–28. [Google Scholar]
- Ahmad, R.; Chaturvedi, R.; Olivares-Villagomez, D.; Habib, T.; Asim, M.; Shivesh, P.; Polk, D.B.; Wilson, K.T.; Washington, M.K.; Van Kaer, L.; et al. Targeted colonic claudin-2 expression renders resistance to epithelial injury, induces immune suppression, and protects from colitis. Mucosal Immunol. 2014, 7, 1340–1353. [Google Scholar] [CrossRef]
- Capaldo, C.T.; Nusrat, A. Claudin switching: Physiological plasticity of the Tight Junction. Semin. Cell. Dev. Biol. 2015, 42, 22–29. [Google Scholar] [CrossRef]
- Garcia-Hernandez, V.; Quiros, M.; Nusrat, A. Intestinal epithelial claudins: Expression and regulation in homeostasis and inflammation. Ann. N. Y. Acad. Sci. 2017, 1397, 66–79. [Google Scholar] [CrossRef]
- Serigado, J.M.; Foulke-Abel, J.; Hines, W.C.; Hanson, J.A.; In, J.; Kovbasnjuk, O. Ulcerative Colitis: Novel Epithelial Insights Provided by Single Cell RNA Sequencing. Front. Med. 2022, 9, 868508. [Google Scholar] [CrossRef]
- Ahmad, R.; Sorrell, M.F.; Batra, S.K.; Dhawan, P.; Singh, A.B. Gut permeability and mucosal inflammation: Bad, good or context dependent. Mucosal Immunol. 2017, 10, 307–317. [Google Scholar] [CrossRef]
- Barrett, K.E. Claudin-2 pore causes leak that breaches the dam in intestinal inflammation. J. Clin. Invest. 2020, 130, 5100–5101. [Google Scholar] [CrossRef]
- Capaldo, C.T.; Nusrat, A. Cytokine regulation of tight junctions. Biochim. Biophys. Acta 2009, 1788, 864–871. [Google Scholar] [CrossRef]
- Schmitz, H.; Barmeyer, C.; Fromm, M.; Runkel, N.; Foss, H.D.; Bentzel, C.J.; Riecken, E.O.; Schulzke, J.D. Altered tight junction structure contributes to the impaired epithelial barrier function in ulcerative colitis. Gastroenterology 1999, 116, 301–309. [Google Scholar] [CrossRef]
- Zeissig, S.; Burgel, N.; Gunzel, D.; Richter, J.; Mankertz, J.; Wahnschaffe, U.; Kroesen, A.J.; Zeitz, M.; Fromm, M.; Schulzke, J.D. Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active Crohn’s disease. Gut 2007, 56, 61–72. [Google Scholar] [CrossRef]
- Rodriguez, P.; Heyman, M.; Candalh, C.; Blaton, M.A.; Bouchaud, C. Tumour necrosis factor-alpha induces morphological and functional alterations of intestinal HT29 cl.19A cell monolayers. Cytokine 1995, 7, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Li, J. Context-Dependent Roles of Claudins in Tumorigenesis. Front. Oncol. 2021, 11, 676781. [Google Scholar] [CrossRef] [PubMed]
- Seker, M.; Fernandez-Rodriguez, C.; Martinez-Cruz, L.A.; Muller, D. Mouse Models of Human Claudin-Associated Disorders: Benefits and Limitations. Int. J. Mol. Sci. 2019, 20, 5504. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Lu, Z.; Foreman, O.; Tatum, R.; Lu, Q.; Renegar, R.; Cao, J.; Chen, Y.H. Inflammation and disruption of the mucosal architecture in claudin-7-deficient mice. Gastroenterology 2012, 142, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Weber, C.R.; Raleigh, D.R.; Yu, D.; Turner, J.R. Tight junction pore and leak pathways: A dynamic duo. Annu. Rev. Physiol. 2011, 73, 283–309. [Google Scholar] [CrossRef]
- Hempel, C.; Rosenthal, R.; Fromm, A.; Krug, S.M.; Fromm, M.; Gunzel, D.; Piontek, J. Tight junction channels claudin-10b and claudin-15: Functional mapping of pore-lining residues. Ann. N. Y. Acad. Sci. 2022, 1515, 129–142. [Google Scholar] [CrossRef]
- Gunzel, D.; Yu, A.S. Claudins and the modulation of tight junction permeability. Physiol. Rev. 2013, 93, 525–569. [Google Scholar] [CrossRef]
- Mitchell, L.A.; Overgaard, C.E.; Ward, C.; Margulies, S.S.; Koval, M. Differential effects of claudin-3 and claudin-4 on alveolar epithelial barrier function. Am. J. Physiol. Lung Cell. Mol. Physiol. 2011, 301, L40–L49. [Google Scholar] [CrossRef]
- Liu, X.; Yang, G.; Geng, X.R.; Cao, Y.; Li, N.; Ma, L.; Chen, S.; Yang, P.C.; Liu, Z. Microbial products induce claudin-2 to compromise gut epithelial barrier function. PLoS ONE 2013, 8, e68547. [Google Scholar] [CrossRef]
- Tanaka, H.; Takechi, M.; Kiyonari, H.; Shioi, G.; Tamura, A.; Tsukita, S. Intestinal deletion of Claudin-7 enhances paracellular organic solute flux and initiates colonic inflammation in mice. Gut 2015, 64, 1529–1538. [Google Scholar] [CrossRef]
- Tsai, P.Y.; Zhang, B.; He, W.Q.; Zha, J.M.; Odenwald, M.A.; Singh, G.; Tamura, A.; Shen, L.; Sailer, A.; Yeruva, S.; et al. IL-22 Upregulates Epithelial Claudin-2 to Drive Diarrhea and Enteric Pathogen Clearance. Cell. Host Microbe 2017, 21, 671–681.e674. [Google Scholar] [CrossRef]
- Raju, P.; Shashikanth, N.; Tsai, P.Y.; Pongkorpsakol, P.; Chanez-Paredes, S.; Steinhagen, P.R.; Kuo, W.T.; Singh, G.; Tsukita, S.; Turner, J.R. Inactivation of paracellular cation-selective claudin-2 channels attenuates immune-mediated experimental colitis in mice. J. Clin. Invest. 2020, 130, 5197–5208. [Google Scholar] [CrossRef]
- Furuse, M.; Fujita, K.; Hiiragi, T.; Fujimoto, K.; Tsukita, S. Claudin-1 and -2: Novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J. Cell. Biol. 1998, 141, 1539–1550. [Google Scholar] [CrossRef]
- Gunzel, D.; Fromm, M. Claudins and other tight junction proteins. Compr. Physiol. 2012, 2, 1819–1852. [Google Scholar] [CrossRef]
- Morita, K.; Furuse, M.; Fujimoto, K.; Tsukita, S. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc. Natl. Acad. Sci. USA 1999, 96, 511–516. [Google Scholar] [CrossRef]
- Daugherty, B.L.; Ward, C.; Smith, T.; Ritzenthaler, J.D.; Koval, M. Regulation of heterotypic claudin compatibility. J. Biol. Chem. 2007, 282, 30005–30013. [Google Scholar] [CrossRef]
- Colegio, O.R.; Van Itallie, C.; Rahner, C.; Anderson, J.M. Claudin extracellular domains determine paracellular charge selectivity and resistance but not tight junction fibril architecture. Am. J. Physiol. Cell. Physiol. 2003, 284, C1346–C1354. [Google Scholar] [CrossRef]
- Rosenthal, R.; Gunzel, D.; Krug, S.M.; Schulzke, J.D.; Fromm, M.; Yu, A.S. Claudin-2-mediated cation and water transport share a common pore. Acta Physiol. 2017, 219, 521–536. [Google Scholar] [CrossRef]
- Milatz, S.; Krug, S.M.; Rosenthal, R.; Gunzel, D.; Muller, D.; Schulzke, J.D.; Amasheh, S.; Fromm, M. Claudin-3 acts as a sealing component of the tight junction for ions of either charge and uncharged solutes. Biochim. Biophys. Acta 2010, 1798, 2048–2057. [Google Scholar] [CrossRef]
- Zhao, J.; Krystofiak, E.S.; Ballesteros, A.; Cui, R.; Van Itallie, C.M.; Anderson, J.M.; Fenollar-Ferrer, C.; Kachar, B. Multiple claudin-claudin cis interfaces are required for tight junction strand formation and inherent flexibility. Commun. Biol. 2018, 1, 50. [Google Scholar] [CrossRef]
- Suzuki, H.; Tani, K.; Tamura, A.; Tsukita, S.; Fujiyoshi, Y. Model for the architecture of claudin-based paracellular ion channels through tight junctions. J. Mol. Biol. 2015, 427, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Itoh, M.; Furuse, M.; Morita, K.; Kubota, K.; Saitou, M.; Tsukita, S. Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J. Cell. Biol. 1999, 147, 1351–1363. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Zheng, J.J. PDZ domains and their binding partners: Structure, specificity, and modification. Cell. Commun. Signal. 2010, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Tang, V.W. Proteomic and bioinformatic analysis of epithelial tight junction reveals an unexpected cluster of synaptic molecules. Biol. Direct 2006, 1, 37. [Google Scholar] [CrossRef]
- Shashikanth, N.; France, M.M.; Xiao, R.; Haest, X.; Rizzo, H.E.; Yeste, J.; Reiner, J.; Turner, J.R. Tight junction channel regulation by interclaudin interference. Nat. Commun. 2022, 13, 3780. [Google Scholar] [CrossRef]
- Ye, Q.; Phan, T.; Hu, W.S.; Liu, X.; Fan, L.; Tan, W.S.; Zhao, L. Transcriptomic Characterization Reveals Attributes of High Influenza Virus Productivity in MDCK Cells. Viruses 2021, 13, 2200. [Google Scholar] [CrossRef]
- Amasheh, S.; Meiri, N.; Gitter, A.H.; Schoneberg, T.; Mankertz, J.; Schulzke, J.D.; Fromm, M. Claudin-2 expression induces cation-selective channels in tight junctions of epithelial cells. J. Cell. Sci. 2002, 115, 4969–4976. [Google Scholar] [CrossRef]
- Van Itallie, C.M.; Holmes, J.; Bridges, A.; Gookin, J.L.; Coccaro, M.R.; Proctor, W.; Colegio, O.R.; Anderson, J.M. The density of small tight junction pores varies among cell types and is increased by expression of claudin-2. J. Cell. Sci. 2008, 121, 298–305. [Google Scholar] [CrossRef]
- Beumer, J.; Clevers, H. Cell fate specification and differentiation in the adult mammalian intestine. Nat. Rev. Mol. Cell. Biol. 2021, 22, 39–53. [Google Scholar] [CrossRef]
- Dhawan, P.; Ahmad, R.; Chaturvedi, R.; Smith, J.J.; Midha, R.; Mittal, M.K.; Krishnan, M.; Chen, X.; Eschrich, S.; Yeatman, T.J.; et al. Claudin-2 expression increases tumorigenicity of colon cancer cells: Role of epidermal growth factor receptor activation. Oncogene 2011, 30, 3234–3247. [Google Scholar] [CrossRef]
- Buchert, M.; Papin, M.; Bonnans, C.; Darido, C.; Raye, W.S.; Garambois, V.; Pelegrin, A.; Bourgaux, J.F.; Pannequin, J.; Joubert, D.; et al. Symplekin promotes tumorigenicity by up-regulating claudin-2 expression. Proc. Natl. Acad. Sci. USA 2010, 107, 2628–2633. [Google Scholar] [CrossRef]
- Lima, W.R.; Parreira, K.S.; Devuyst, O.; Caplanusi, A.; N’Kuli, F.; Marien, B.; Van Der Smissen, P.; Alves, P.M.; Verroust, P.; Christensen, E.I.; et al. ZONAB promotes proliferation and represses differentiation of proximal tubule epithelial cells. J. Am. Soc. Nephrol. 2010, 21, 478–488. [Google Scholar] [CrossRef]
- Tamura, A.; Hayashi, H.; Imasato, M.; Yamazaki, Y.; Hagiwara, A.; Wada, M.; Noda, T.; Watanabe, M.; Suzuki, Y.; Tsukita, S. Loss of claudin-15, but not claudin-2, causes Na+ deficiency and glucose malabsorption in mouse small intestine. Gastroenterology 2011, 140, 913–923. [Google Scholar] [CrossRef]
- Lechuga, S.; Braga-Neto, M.B.; Naydenov, N.G.; Rieder, F.; Ivanov, A.I. Understanding disruption of the gut barrier during inflammation: Should we abandon traditional epithelial cell lines and switch to intestinal organoids? Front. Immunol. 2023, 14, 1108289. [Google Scholar] [CrossRef]
- Ahmad, R.; Kumar, B.; Chen, Z.; Chen, X.; Muller, D.; Lele, S.M.; Washington, M.K.; Batra, S.K.; Dhawan, P.; Singh, A.B. Loss of claudin-3 expression induces IL6/gp130/Stat3 signaling to promote colon cancer malignancy by hyperactivating Wnt/beta-catenin signaling. Oncogene 2017, 36, 6592–6604. [Google Scholar] [CrossRef]
- de Souza, W.F.; Fortunato-Miranda, N.; Robbs, B.K.; de Araujo, W.M.; de-Freitas-Junior, J.C.; Bastos, L.G.; Viola, J.P.; Morgado-Diaz, J.A. Claudin-3 overexpression increases the malignant potential of colorectal cancer cells: Roles of ERK1/2 and PI3K-Akt as modulators of EGFR signaling. PLoS ONE 2013, 8, e74994. [Google Scholar] [CrossRef]
- Lechuga, S.; Ivanov, A.I. Disruption of the epithelial barrier during intestinal inflammation: Quest for new molecules and mechanisms. Biochim. Biophys. Acta Mol. Cell. Res. 2017, 1864, 1183–1194. [Google Scholar] [CrossRef]
- Venugopal, S.; Anwer, S.; Szaszi, K. Claudin-2: Roles beyond Permeability Functions. Int. J. Mol. Sci. 2019, 20, 5655. [Google Scholar] [CrossRef]
- Geoffroy, M.; Kleinclauss, A.; Kuntz, S.; Grillier-Vuissoz, I. Claudin 1 inhibits cell migration and increases intercellular adhesion in triple-negative breast cancer cell line. Mol. Biol. Rep. 2020, 47, 7643–7653. [Google Scholar] [CrossRef]
- Kim, H.; Kim, S.H.; Hwang, D.; An, J.; Chung, H.S.; Yang, E.G.; Kim, S.Y. Extracellular pyruvate kinase M2 facilitates cell migration by upregulating claudin-1 expression in colon cancer cells. Biochem. Cell. Biol. 2020, 98, 219–226. [Google Scholar] [CrossRef]
- Sasaki, H.; Matsui, C.; Furuse, K.; Mimori-Kiyosue, Y.; Furuse, M.; Tsukita, S. Dynamic behavior of paired claudin strands within apposing plasma membranes. Proc. Natl. Acad. Sci. USA 2003, 100, 3971–3976. [Google Scholar] [CrossRef] [PubMed]
- Furuse, M.; Sasaki, H.; Fujimoto, K.; Tsukita, S. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J. Cell. Biol. 1998, 143, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Otani, T.; Nguyen, T.P.; Tokuda, S.; Sugihara, K.; Sugawara, T.; Furuse, K.; Miura, T.; Ebnet, K.; Furuse, M. Claudins and JAM-A coordinately regulate tight junction formation and epithelial polarity. J. Cell. Biol. 2019, 218, 3372–3396. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, S.; Nguyen, T.P.; Furuse, K.; Fukazawa, Y.; Otani, T.; Furuse, M. Tight junction formation by a claudin mutant lacking the COOH-terminal PDZ domain-binding motif. Ann. N. Y. Acad. Sci. 2022, 1516, 85–94. [Google Scholar] [CrossRef]
- Gonschior, H.; Schmied, C.; Van der Veen, R.E.; Eichhorst, J.; Himmerkus, N.; Piontek, J.; Gunzel, D.; Bleich, M.; Furuse, M.; Haucke, V.; et al. Nanoscale segregation of channel and barrier claudins enables paracellular ion flux. Nat. Commun. 2022, 13, 4985. [Google Scholar] [CrossRef]
- Capaldo, C.T.; Farkas, A.E.; Hilgarth, R.S.; Krug, S.M.; Wolf, M.F.; Benedik, J.K.; Fromm, M.; Koval, M.; Parkos, C.; Nusrat, A. Proinflammatory cytokine-induced tight junction remodeling through dynamic self-assembly of claudins. Mol. Biol. Cell. 2014, 25, 2710–2719. [Google Scholar] [CrossRef]
- Kirschner, M.; Gerhart, J.; Mitchison, T. Molecular “vitalism”. Cell 2000, 100, 79–88. [Google Scholar] [CrossRef]
- Twiss, F.; Oldenkamp, M.; Hiemstra, A.; Zhou, H.; Matheron, L.; Mohammed, S.; de Rooij, J. HGF signaling regulates Claudin-3 dynamics through its C-terminal tyrosine residues. Tissue Barriers 2013, 1, e27425. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Tokumasu, R.; Kimura, H.; Tsukita, S. Role of claudin species-specific dynamics in reconstitution and remodeling of the zonula occludens. Mol. Biol. Cell. 2011, 22, 1495–1504. [Google Scholar] [CrossRef]
- Shen, L.; Weber, C.R.; Turner, J.R. The tight junction protein complex undergoes rapid and continuous molecular remodeling at steady state. J. Cell. Biol. 2008, 181, 683–695. [Google Scholar] [CrossRef]
- Van Itallie, C.M.; Tietgens, A.J.; Anderson, J.M. Visualizing the dynamic coupling of claudin strands to the actin cytoskeleton through ZO-1. Mol. Biol. Cell. 2017, 28, 524–534. [Google Scholar] [CrossRef]
- Higashi, T.; Saito, A.C.; Fukazawa, Y.; Furuse, M.; Higashi, A.Y.; Ono, M.; Chiba, H. EpCAM proteolysis and release of complexed claudin-7 repair and maintain the tight junction barrier. J. Cell. Biol. 2023, 222, e202204079. [Google Scholar] [CrossRef]
- Fanning, A.S.; Van Itallie, C.M.; Anderson, J.M. Zonula occludens-1 and -2 regulate apical cell structure and the zonula adherens cytoskeleton in polarized epithelia. Mol. Biol. Cell. 2012, 23, 577–590. [Google Scholar] [CrossRef]
- Kalyoncu, S.; Keskin, O.; Gursoy, A. Interaction prediction and classification of PDZ domains. BMC Bioinform. 2010, 11, 357. [Google Scholar] [CrossRef]
- Nomme, J.; Antanasijevic, A.; Caffrey, M.; Van Itallie, C.M.; Anderson, J.M.; Fanning, A.S.; Lavie, A. Structural Basis of a Key Factor Regulating the Affinity between the Zonula Occludens First PDZ Domain and Claudins. J. Biol. Chem. 2015, 290, 16595–16606. [Google Scholar] [CrossRef]
- Darido, C.; Buchert, M.; Pannequin, J.; Bastide, P.; Zalzali, H.; Mantamadiotis, T.; Bourgaux, J.F.; Garambois, V.; Jay, P.; Blache, P.; et al. Defective claudin-7 regulation by Tcf-4 and Sox-9 disrupts the polarity and increases the tumorigenicity of colorectal cancer cells. Cancer Res. 2008, 68, 4258–4268. [Google Scholar] [CrossRef]
- Takehara, M.; Nishimura, T.; Mima, S.; Hoshino, T.; Mizushima, T. Effect of claudin expression on paracellular permeability, migration and invasion of colonic cancer cells. Biol. Pharm. Bull. 2009, 32, 825–831. [Google Scholar] [CrossRef]
- Kim, D.H.; Lu, Q.; Chen, Y.H. Claudin-7 modulates cell-matrix adhesion that controls cell migration, invasion and attachment of human HCC827 lung cancer cells. Oncol. Lett. 2019, 17, 2890–2896. [Google Scholar] [CrossRef]
- Lei, N.; Cheng, Y.; Wan, J.; Blasig, R.; Li, A.; Bai, Y.; Haseloff, R.F.; Blasig, I.E.; Zhu, L.; Qin, Z. Claudin-3 inhibits tumor-induced lymphangiogenesis via regulating the PI3K signaling pathway in lymphatic endothelial cells. Sci. Rep. 2022, 12, 17440. [Google Scholar] [CrossRef]
- Krndija, D.; El Marjou, F.; Guirao, B.; Richon, S.; Leroy, O.; Bellaiche, Y.; Hannezo, E.; Matic Vignjevic, D. Active cell migration is critical for steady-state epithelial turnover in the gut. Science 2019, 365, 705–710. [Google Scholar] [CrossRef]
- Mayor, R.; Etienne-Manneville, S. The front and rear of collective cell migration. Nat. Rev. Mol. Cell. Biol. 2016, 17, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Belardi, B.; Hamkins-Indik, T.; Harris, A.R.; Kim, J.; Xu, K.; Fletcher, D.A. A Weak Link with Actin Organizes Tight Junctions to Control Epithelial Permeability. Dev. Cell. 2020, 54, 792–804.e797. [Google Scholar] [CrossRef] [PubMed]
- Marchant, C.L.; Malmi-Kakkada, A.N.; Espina, J.A.; Barriga, E.H. Cell clusters softening triggers collective cell migration in vivo. Nat. Mater. 2022, 21, 1314–1323. [Google Scholar] [CrossRef]
- Baumholtz, A.I.; Simard, A.; Nikolopoulou, E.; Oosenbrug, M.; Collins, M.M.; Piontek, A.; Krause, G.; Piontek, J.; Greene, N.D.E.; Ryan, A.K. Claudins are essential for cell shape changes and convergent extension movements during neural tube closure. Dev. Biol. 2017, 428, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Skamrahl, M.; Pang, H.; Ferle, M.; Gottwald, J.; Rubeling, A.; Maraspini, R.; Honigmann, A.; Oswald, T.A.; Janshoff, A. Tight Junction ZO Proteins Maintain Tissue Fluidity, Ensuring Efficient Collective Cell Migration. Adv. Sci. 2021, 8, e2100478. [Google Scholar] [CrossRef]
- Angulo-Urarte, A.; van der Wal, T.; Huveneers, S. Cell-cell junctions as sensors and transducers of mechanical forces. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183316. [Google Scholar] [CrossRef]
- Lynn, K.S.; Peterson, R.J.; Koval, M. Ruffles and spikes: Control of tight junction morphology and permeability by claudins. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183339. [Google Scholar] [CrossRef]
- Tokuda, S.; Higashi, T.; Furuse, M. ZO-1 knockout by TALEN-mediated gene targeting in MDCK cells: Involvement of ZO-1 in the regulation of cytoskeleton and cell shape. PLoS ONE 2014, 9, e104994. [Google Scholar] [CrossRef]
- Spadaro, D.; Le, S.; Laroche, T.; Mean, I.; Jond, L.; Yan, J.; Citi, S. Tension-Dependent Stretching Activates ZO-1 to Control the Junctional Localization of Its Interactors. Curr. Biol. 2017, 27, 3783–3795.e3788. [Google Scholar] [CrossRef]
- Spadaro, D.; Tapia, R.; Jond, L.; Sudol, M.; Fanning, A.S.; Citi, S. ZO proteins redundantly regulate the transcription factor DbpA/ZONAB. J. Biol. Chem. 2014, 289, 22500–22511. [Google Scholar] [CrossRef]
- Higashi, T.; Arnold, T.R.; Stephenson, R.E.; Dinshaw, K.M.; Miller, A.L. Maintenance of the Epithelial Barrier and Remodeling of Cell-Cell Junctions during Cytokinesis. Curr. Biol. 2016, 26, 1829–1842. [Google Scholar] [CrossRef]
- Monaco, A.; Ovryn, B.; Axis, J.; Amsler, K. The Epithelial Cell Leak Pathway. Int. J. Mol. Sci. 2021, 22, 7677. [Google Scholar] [CrossRef]
- Krug, S.M. Contribution of the tricellular tight junction to paracellular permeability in leaky and tight epithelia. Ann. N. Y. Acad. Sci. 2017, 1397, 219–230. [Google Scholar] [CrossRef]
- Lechuga, S.; Ivanov, A.I. Actin cytoskeleton dynamics during mucosal inflammation: A view from broken epithelial barriers. Curr. Opin. Physiol. 2021, 19, 10–16. [Google Scholar] [CrossRef]
- Kawai, Y.; Hamazaki, Y.; Fujita, H.; Fujita, A.; Sato, T.; Furuse, M.; Fujimoto, T.; Jetten, A.M.; Agata, Y.; Minato, N. Claudin-4 induction by E-protein activity in later stages of CD4/8 double-positive thymocytes to increase positive selection efficiency. Proc. Natl. Acad. Sci. USA 2011, 108, 4075–4080. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Capaldo, C.T. Claudin Barriers on the Brink: How Conflicting Tissue and Cellular Priorities Drive IBD Pathogenesis. Int. J. Mol. Sci. 2023, 24, 8562. https://doi.org/10.3390/ijms24108562
Capaldo CT. Claudin Barriers on the Brink: How Conflicting Tissue and Cellular Priorities Drive IBD Pathogenesis. International Journal of Molecular Sciences. 2023; 24(10):8562. https://doi.org/10.3390/ijms24108562
Chicago/Turabian StyleCapaldo, Christopher T. 2023. "Claudin Barriers on the Brink: How Conflicting Tissue and Cellular Priorities Drive IBD Pathogenesis" International Journal of Molecular Sciences 24, no. 10: 8562. https://doi.org/10.3390/ijms24108562
APA StyleCapaldo, C. T. (2023). Claudin Barriers on the Brink: How Conflicting Tissue and Cellular Priorities Drive IBD Pathogenesis. International Journal of Molecular Sciences, 24(10), 8562. https://doi.org/10.3390/ijms24108562




