First Asp-2078-Gly Mutation Conferring Resistance to Different ACCase Inhibitors in a Polypogon fugax Population from China
Abstract
1. Introduction
2. Results
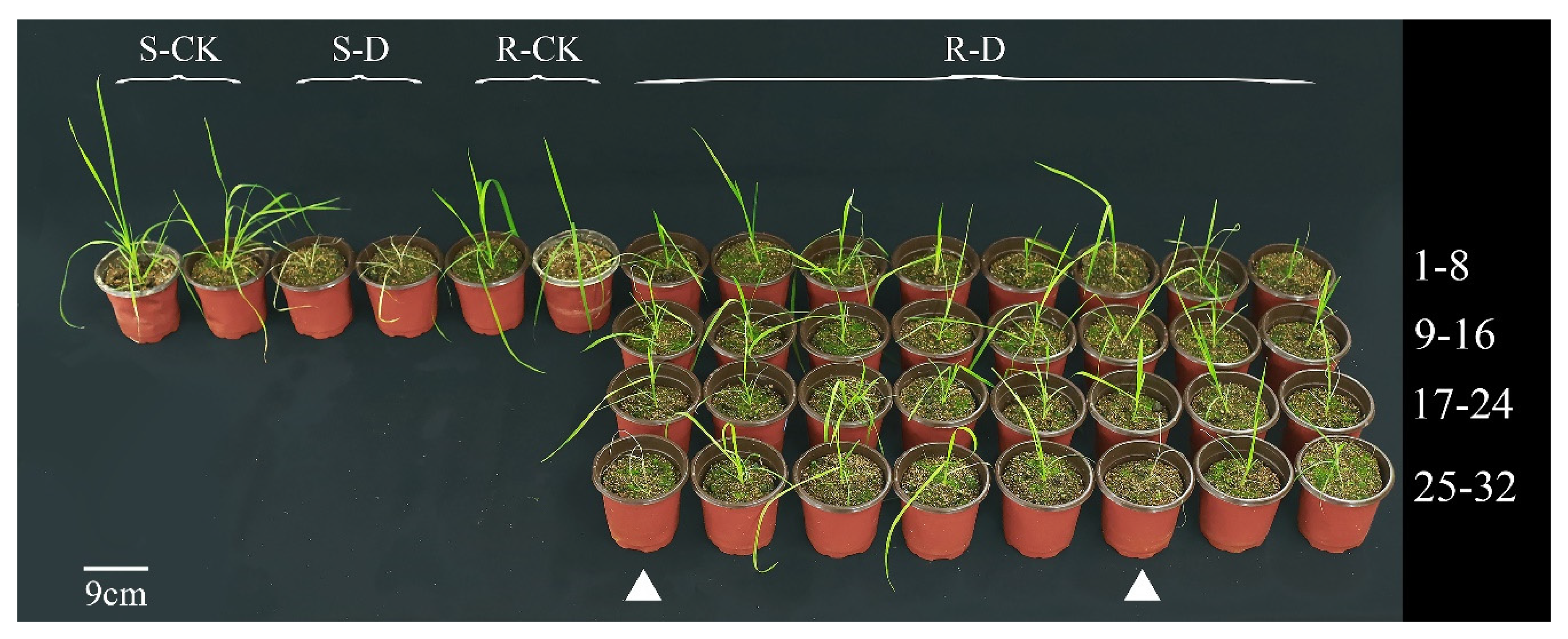
2.1. Quizalofop-p-Ethyl Resistance
2.2. Dose-Response to Other Herbicides
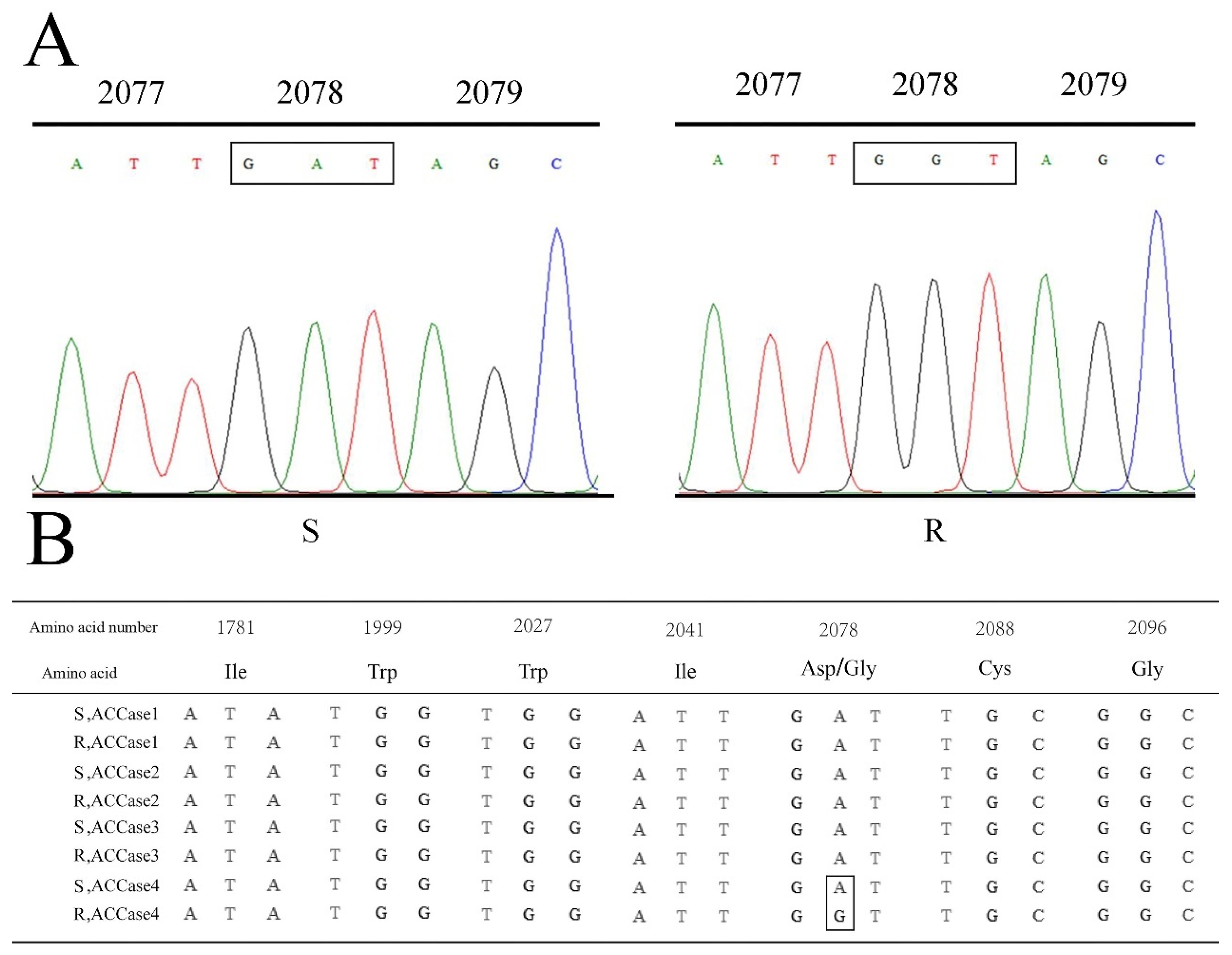
2.3. Identification of ACCase Mutation
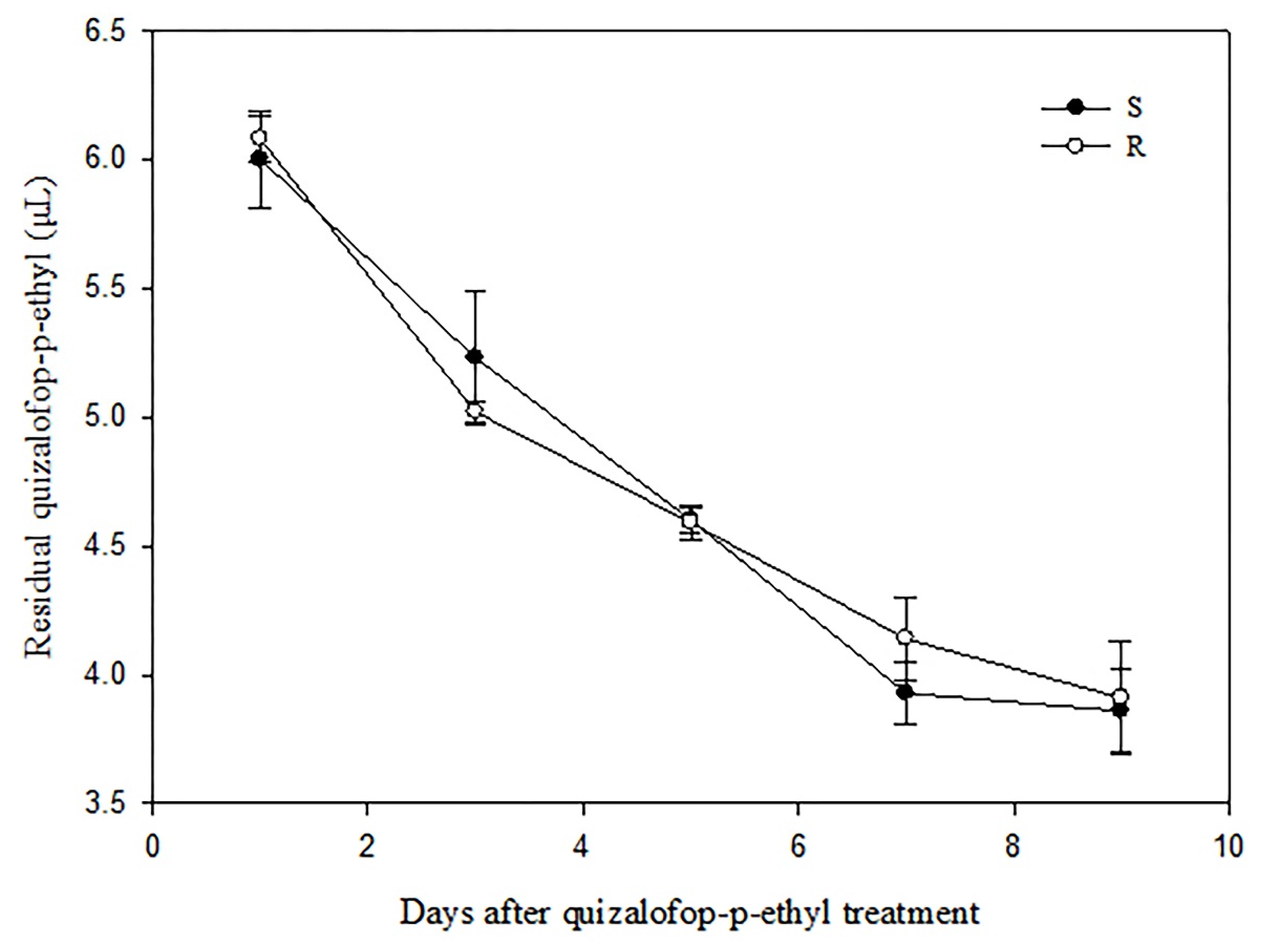
2.4. Determination of Quizalofop-p-Ethyl Residual in P. fugax
2.5. The Correlation between Phenotypes and Genotypes of P. fugax
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Whole-Plant Dose-Response Bioassay
4.3. Identification of the P. fugax Plastidic ACCase Gene Mutation
4.4. Detection of Quizalofop-p-Ethyl Residual in R and S P. fugax Populations
4.5. Development of dCAPS for 2078 Mutation Detection and Genotype Analysis
4.6. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, W.; Harrison, D.K.; Chalupska, D.; Gornicki, P.; O’Donnell C, C.; Adkins, S.W.; Haselkorn, R.; Williams, R.R. Single-site mutations in the carboxyltransferase domain of plastid acetyl-CoA carboxylase confer resistance to grass-specific herbicides. Proc. Natl. Acad. Sci. USA 2007, 104, 3627–3632. [Google Scholar] [CrossRef]
- Troncoso-Ponce, M.A.; Nikovics, K.; Marchive, C.; Lepiniec, L.; Baud, S. New insights on the organization and regulation of the fatty acid biosynthetic network in the model higher plant Arabidopsis thaliana. Biochimie 2016, 120, 3–8. [Google Scholar] [CrossRef] [PubMed]
- HRAC. Herbicide Resistance Action Committee. 2022. Available online: http://www.hracglobal.com (accessed on 10 November 2022).
- Heap, I.M. The International Survey of Herbicide Resistant Weeds. 2022. Available online: http://www.weedscience.org (accessed on 10 November 2022).
- Kaundun, S.S. Resistance to acetyl-CoA carboxylase-inhibiting herbicides. Pest Manag. Sci. 2014, 70, 1405–1417. [Google Scholar] [CrossRef]
- Gaines, T.A.; Duke, S.O.; Morran, S.; Rigon, C.A.G.; Tranel, P.J.; Kupper, A.; Dayan, F.E. Mechanisms of evolved herbicide resistance. J. Biol. Chem. 2020, 295, 10307–10330. [Google Scholar]
- Mohamed, I.A.; Li, R.Z.; You, Z.G.; Li, Z.H. Japanese Foxtail (Alopecurus japonicus) Resistance to Fenoxaprop and Pinoxaden in China. Weed Sci. 2012, 60, 167–171. [Google Scholar] [CrossRef]
- Deng, W.; Cai, J.X.; Zhang, J.Y.; Chen, Y.Y.; Chen, Y.R.; Di, Y.J.; Yuan, S.Z. Molecular basis of resistance to ACCase-inhibiting herbicide cyhalofop-butyl in Chinese sprangletop (Leptochloa chinensis (L.) Nees) from China. Pestic. Biochem. Physiol. 2019, 158, 143–148. [Google Scholar] [CrossRef]
- Wang, J.Z.; Peng, Y.J.; Chen, W.; Yu, Q.; Bai, L.Y.; Pan, L. The Ile-2041-Val mutation in the ACCase gene confers resistance to clodinafop-propargyl in American sloughgrass (Beckmannia syzigachne Steud). Pest Manag. Sci. 2021, 77, 2425–2432. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Li, J.; Zhang, T.; Zhang, D.; Dong, L.Y. Cross-resistance patterns to acetyl coenzyme A carboxylase (ACCase) inhibitors associated with different ACCase mutations in Beckmannia syzigachne. Weed Res. 2015, 55, 609–620. [Google Scholar] [CrossRef]
- Saini, R.K.; Kleemann, S.G.L.; Preston, C.; Gill, G.S. Control of clethodim-resistant Lolium rigidum (rigid ryegrass) in triazine-tolerant canola (Brassica napus L.) in southern Australia. Crop Prot. 2015, 78, 99–105. [Google Scholar] [CrossRef]
- Powles, S.B.; Yu, Q. Evolution in Action: Plants Resistant to Herbicides. Annu. Rev. Plant Biol. 2010, 61, 317–347. [Google Scholar] [CrossRef]
- Delye, C.; Jasieniuk, M.; Le Corre, V. Deciphering the evolution of herbicide resistance in weeds. Trends Genet. 2013, 29, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Iwakami, S.; Kamidate, Y.; Yamaguchi, T.; Ishizaka, M.; Endo, M.; Suda, H.; Nagai, K.; Sunohara, Y.; Toki, S.; Uchino, A.; et al. CYP81A P450s are involved in concomitant cross-resistance to acetolactate synthase and acetyl-CoA carboxylase herbicides in Echinochloa phyllopogon. New Phytol. 2019, 221, 2112–2122. [Google Scholar] [CrossRef]
- Cummins, I.; Wortley, D.J.; Sabbadin, F.; He, Z.S.; Coxon, C.R.; Straker, H.E.; Sellars, J.D.; Knight, K.; Edwards, L.; Hughes, D.; et al. Key role for a glutathione transferase in multiple-herbicide resistance in grass weeds. Proc. Natl. Acad. Sci. USA 2013, 110, 5812–5817. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Li, J.; Xu, H.; Dong, L. Factors Affecting Seed Germination and Seedling Emergence of Asia Minor Bluegrass (Polypogon fugax). Weed Sci. 2017, 63, 440–447. [Google Scholar] [CrossRef]
- Chen, W.; Wu, L.; Wang, J.; Yu, Q.; Bai, L.; Pan, L. Quizalofop-p-ethyl resistance in Polypogon fugax involves glutathione S-transferases. Pest Manag. Sci. 2020, 76, 3800–3805. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Ge, L.; Yan, Y.; Bai, S.; Wang, D.; Liu, W.; Wang, J. Trp-1999-Ser mutation of acetyl-CoA carboxylase and cytochrome P450s-involved metabolism confer resistance to fenoxaprop-P-ethyl in Polypogon fugax. Pest Manag. Sci. 2019, 75, 3175–3183. [Google Scholar] [CrossRef]
- Tang, W.; Zhou, F.; Chen, J.; Zhou, X. Resistance to ACCase-inhibiting herbicides in an Asia minor bluegrass (Polypogon fugax) population in China. Pestic. Biochem. Physiol. 2014, 108, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.L.; Luo, K.; Mao, A.X.; Pan, L.; Yan, B.; Wu, J.; Hu, L.F.; Liu, M.; Liu, X.Y.; Bai, L.Y. Endophytes enhance Asia minor bluegrass (Polypogon fugax) resistance to quizalofop-p-ethyl. Plant Soil. 2020, 450, 373–384. [Google Scholar] [CrossRef]
- Dimaano, N.G.; Yamaguchi, T.; Fukunishi, K.; Tominaga, T.; Iwakami, S. Functional characterization of cytochrome P450 CYP81A subfamily to disclose the pattern of cross-resistance in Echinochloa phyllopogon. Plant Mol. Biol. 2020, 102, 403–416. [Google Scholar] [CrossRef]
- Huan, Z.B.; Zhang, H.J.; Hou, Z.; Zhang, S.Y.; Zhang, Y.; Liu, W.T.; Bi, Y.L.; Wang, J.X. Resistance Level and Metabolism of Barnyard-Grass (Echinochloa crusgalli (L.) Beauv.) Populations to Quizalofop-p-ethyl in Heilongjiang Province, China. Agric. Sci. China. 2011, 10, 1914–1922. [Google Scholar] [CrossRef]
- Huan, Z.B.; Xu, Z.; Lv, D.Z.; Wang, J.X. Determination of ACCase Sensitivity and Gene Expression in Quizalofop-Ethyl-Resistant and -Susceptible Barnyardgrass (Echinochloa crus-galli) Biotypes. Weed Sci. 2013, 61, 537–542. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, H.; Xu, H.L.; Gao, Y.; Zhang, W.; Dong, L.Y. Mechanism of Fenoxaprop-P-ethyl Resistance in Italian Ryegrass (Lolium perenne ssp. multiflorum) from China. Weed Sci. 2017, 65, 710–717. [Google Scholar] [CrossRef]
- Xu, H.L.; Zhu, X.D.; Wang, H.C.; Li, J.; Dong, L.Y. Mechanism of resistance to fenoxaprop in Japanese foxtail (Alopecurus japonicus) from China. Pestic. Biochem. Physiol. 2013, 107, 25–31. [Google Scholar] [CrossRef]
- Guo, W.L.; Yuan, G.H.; Liu, W.T.; Bi, Y.L.; Du, L.; Zhang, C.; Li, Q.; Wang, J.X. Multiple resistance to ACCase and AHAS-inhibiting herbicides in shortawn foxtail (Alopecurus aequalis Sobol.) from China. Pestic. Biochem. Physiol. 2015, 124, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.T.; Yu, J.X.; Pan, L.; Wu, X.B.; Dong, L.Y. Target-Site Resistance to Fenoxaprop-P-ethyl in Keng Stiffgrass (Sclerochloa kengiana) from China. Weed Sci. 2017, 65, 452–460. [Google Scholar] [CrossRef]
- Yu, Q.; Ahmad-Hamdani, M.S.; Han, H.; Christoffers, M.J.; Powles, S.B. Herbicide resistance-endowing ACCase gene mutations in hexaploid wild oat (Avena fatua): Insights into resistance evolution in a hexaploid species. Heredity 2013, 110, 220–231. [Google Scholar] [CrossRef]
- Xu, Y.; Xu, L.; Li, X.; Zheng, M. Investigation of resistant level to tribenuron-methyl, diversity and regional difference of the resistant mutations on acetolactate synthase (ALS) isozymes in Descurainia sophia L. from China. Pestic. Biochem. Physiol. 2020, 169, 104653. [Google Scholar] [CrossRef]
- Tanigaki, S.; Uchino, A.; Okawa, S.; Miura, C.; Hamamura, K.; Matsuo, M.; Yoshino, N.; Ueno, N.; Toyama, Y.; Fukumi, N.; et al. Gene expression shapes the patterns of parallel evolution of herbicide resistance in the agricultural weed Monochoria vaginalis. New Phytol. 2021, 232, 928–940. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Xu, H.; Zhang, T.; Bai, C.; Dong, L. Fenoxaprop-P-ethyl resistance conferred by cytochrome P450s and target site mutation in Alopecurus japonicus. Pest Manag. Sci. 2018, 74, 1694–1703. [Google Scholar] [CrossRef]
- Yu, Q.; Collavo, A.; Zheng, M.-Q.; Owen, M.; Sattin, M.; Powles, S.B. Diversity of acetyl-coenzyme a carboxylase mutations in resistant Lolium populations: Evaluation using clethodim. Plant Physiol. 2007, 145, 547–558. [Google Scholar] [CrossRef]
- Delye, C.; Matejicek, A.; Michel, S. Cross-resistance patterns to ACCase-inhibiting herbicides conferred by mutant ACCase isoforms in Alopecurus myosuroides Huds. (black-grass), re-examined at the recommended herbicide field rate. Pest Manag. Sci. 2008, 64, 1179–1186. [Google Scholar] [CrossRef] [PubMed]
- Collavo, A.; Panozzo, S.; Lucchesi, G.; Scarabel, L.; Sattin, M. Characterisation and management of Phalaris paradoxa resistant to ACCase-inhibitors. Crop Prot. 2011, 30, 293–299. [Google Scholar] [CrossRef]
- Cruz-Hipolito, H.; Osuna, M.D.; Dominguez-Valenzuela, J.A.; Espinoza, N.; De Prado, R. Mechanism of Resistance to ACCase-Inhibiting Herbicides in Wild Oat (Avena fatua) from Latin America. J. Agric. Food Chem. 2011, 59, 7261–7267. [Google Scholar] [CrossRef]
- Tang, W.; Zhou, F.; Zhang, Y.; Chen, J. Resistance of American sloughgrass (Bechmannia syzigachne) populations to ACCase-inhibiting herbicides involves three different target site mutations from China. Pestic. Biochem. Physiol. 2015, 124, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; He, Z.; Liu, T.; Li, J.; Dong, L. A novel mutation Asp-2078-Glu in ACCase confers resistance to ACCase herbicides in barnyardgrass (Echinochloa crus-galli). Pestic. Biochem. Physiol. 2020, 168, 104634. [Google Scholar] [CrossRef]
- Zhu, X.L.; Ge-Fei, H.; Zhan, C.G.; Yang, G.F. Computational simulations of the interactions between acetyl-coenzyme-A carboxylase and clodinafop: Resistance mechanism due to active and nonactive site mutations. J. Chem. Inf. Model. 2009, 49, 1936–1943. [Google Scholar] [CrossRef] [PubMed]
- Tehranchian, P.; Norsworthy, J.K.; Korres, N.E.; McElroy, S.; Chen, S.; Scott, R.C. Resistance to aryloxyphenoxypropionate herbicides in Amazon sprangletop: Confirmation, control, and molecular basis of resistance. Pestic. Biochem. Physiol. 2016, 133, 79–84. [Google Scholar] [CrossRef]
- Chen, G.; Wang, L.; Xu, H.; Wu, X.; Pan, L.; Dong, L. Cross-resistance Patterns to Acetyl-CoA Carboxylase Inhibitors Associated with Different Mutations in Japanese Foxtail (Alopecurus japonicus). Weed Sci. 2017, 65, 444–451. [Google Scholar] [CrossRef]
- Beckie, H.J.; Tardif, F.J. Herbicide cross resistance in weeds. Crop Prot. 2012, 35, 15–28. [Google Scholar]
- Guo, W.; Liu, W.; Li, L.; Yuan, G.; Du, L.; Wang, J. Molecular basis for resistance to fenoxaprop in shortawn foxtail (Alopecurus aequalis) from China. Weed Sci. 2015, 63, 416–424. [Google Scholar] [CrossRef]
- Scarabel, L.; Panozzo, S.; Varotto, S.; Sattin, M. Allelic variation of the ACCase gene and response to ACCase-inhibiting herbicides in pinoxaden-resistant Lolium spp. Pest Manag. Sci. 2011, 67, 932–941. [Google Scholar] [CrossRef] [PubMed]
- Iwakami, S.; Uchino, A.; Watanabe, H.; Yamasue, Y.; Inamura, T. Isolation and expression of genes for acetolactate synthase and acetyl-CoA carboxylase in Echinochloa phyllopogon, a polyploid weed species. Pest Manag. Sci. 2012, 68, 1098–1106. [Google Scholar] [CrossRef] [PubMed]
- Ghanizadeh, H.; Buddenhagen, C.E.; Harrington, K.C.; Griffiths, A.G.; Ngow, Z. Pinoxaden resistance in Lolium perenne L. is due to both target-site and non-target-site mechanisms. Pestic. Biochem. Physiol. 2022, 184, 105103. [Google Scholar] [CrossRef]
- Han, H.; Yu, Q.; Owen, M.J.; Cawthray, G.R.; Powles, S.B. Widespread occurrence of both metabolic and target-site herbicide resistance mechanisms in Lolium rigidum populations. Pest Manag. Sci. 2016, 72, 255–263. [Google Scholar] [CrossRef]
- Zhao, N.; Yan, Y.Y.; Ge, L.A.; Zhu, B.L.; Liu, W.T.; Wang, J.X. Target site mutations and cytochrome P450s confer resistance to fenoxaprop-P-ethyl and mesosulfuron-methyl in Alopecurus aequalis. Pest Manag. Sci. 2019, 75, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Si, C.; Liu, M.J.; Qiu, L.H.; Zheng, M.Q. Investigation of resistance levels and mechanisms to nicosulfuron conferred by non-target-site mechanisms in large crabgrass (Digitaria sanguinalis L.) from China. Pestic. Biochem. Physiol. 2017, 141, 84–89. [Google Scholar] [CrossRef]

| Population | Treatment † | GR50 (g a.i.ha−1) (SE) a | RI |
|---|---|---|---|
| R | Q | 102.05 (1.19) | 10.89 |
| Q + N | 95.41 (1.31) | 10.18 | |
| Q + M | 96.86 (1.37) | 10.34 | |
| Q + P | 100.20 (3.43) | 10.69 | |
| S | Q | 9.37 (0.22) | 1.00 |
| Q + N | 9.59 (0.56) | 1.02 | |
| Q + M | 8.80 (0.23) | 0.94 | |
| Q + P | 9.85 (0.70) | 1.05 |
| Herbicide | Population | GR50 (g a.i.ha−1) (SE) a | RI |
|---|---|---|---|
| Clodinafop-propargyl | R | 42.73 (0.20) | 9.07 |
| S | 4.71 (0.13) | ||
| Haloxyfop-R-methyl | R | 48.20 (3.55) | 13.65 |
| S | 3.53 (0.44) | ||
| Sethoxydim | R | 72.15 (3.79) | 8.82 |
| S | 8.18 (1.60) | ||
| Pinoxaden | R | 33.88 (2.62) | 5.46 |
| S | 6.20 (0.66) | ||
| Fenoxaprop-P-ethyl | R | 97.53 (2.68) | 18.40 |
| S | 5.30 (0.50) | ||
| Metamifop | R | 156.58 (4.18) | 10.77 |
| S | 14.54 (1.17) | ||
| Clethodim | R | 23.36 (0.39) | 6.54 |
| S | 3.57 (0.12) | ||
| cyhalofop-butyl | R | 114.82 (4.27) | 10.00 |
| S | 11.48 (1.89) | ||
| Mesosulfuron-methyl | R | 9.54 (2.45) | 1.31 |
| S | 7.27 (1.24) | ||
| pyroxsulam | R | 7.82 (2.55) | 0.88 |
| S | 8.91 (2.67) |
| Group † | Herbicide | Formulation ‡ | Company | Treated Doses (g a.i. ha−1) a | |
|---|---|---|---|---|---|
| ACCase | APP | quizalofop-p-ethyl Clodinafop-propargyl | 10% EC 15% ME | Tianrun Hansi | 0, 6.56, 13.13, 26.25, 52.5, 105, 210 0, 5.63, 11.25, 22.5, 45, 90, 180 |
| Haloxyfop-R-methyl | 108 g L−1 EC | Flag | 0, 2, 4, 8, 16, 32, 64 | ||
| Fenoxaprop-P-ethyl | 69 g L−1 EW | Bayer | 0, 7.76, 15.53, 31.05, 62.1, 124.2, 248.4 | ||
| Metamifop | 10% EC | FMC | 0, 12.5, 25,50, 100, 200, 400 | ||
| cyhalofop-butyl | 100 g L−1 EC | Jiahui | 0, 12.5, 25,50, 100, 200, 400 | ||
| CHD | Clethodim | 240 g L−1 EC | Cynda | 0, 3.1, 6.1, 12.2, 24.3, 48.6, 97.2 | |
| Sethoxydim | 12.5% EC | Cynda | 0, 19.5, 39, 78, 156, 312, 624 | ||
| DEN | Pinoxaden | 5% EC | Syngenta | 0, 5.6, 11.3, 22.5, 45, 90, 180 | |
| ALS | Mesosulfuron-methyl | 30 g L−1 OD | FMC | 0, 1.41, 2.81, 5.63, 11.25, 22.5, 45 | |
| pyroxsulam | 4% OD | Dow | 0, 1.13, 2.25, 4.5, 9, 18, 36 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mo, B.; Chen, W.; He, S.; Liu, H.; Bai, L.; Pan, L. First Asp-2078-Gly Mutation Conferring Resistance to Different ACCase Inhibitors in a Polypogon fugax Population from China. Int. J. Mol. Sci. 2023, 24, 528. https://doi.org/10.3390/ijms24010528
Mo B, Chen W, He S, Liu H, Bai L, Pan L. First Asp-2078-Gly Mutation Conferring Resistance to Different ACCase Inhibitors in a Polypogon fugax Population from China. International Journal of Molecular Sciences. 2023; 24(1):528. https://doi.org/10.3390/ijms24010528
Chicago/Turabian StyleMo, Bocheng, Wen Chen, Sifen He, Haozhe Liu, Lianyang Bai, and Lang Pan. 2023. "First Asp-2078-Gly Mutation Conferring Resistance to Different ACCase Inhibitors in a Polypogon fugax Population from China" International Journal of Molecular Sciences 24, no. 1: 528. https://doi.org/10.3390/ijms24010528
APA StyleMo, B., Chen, W., He, S., Liu, H., Bai, L., & Pan, L. (2023). First Asp-2078-Gly Mutation Conferring Resistance to Different ACCase Inhibitors in a Polypogon fugax Population from China. International Journal of Molecular Sciences, 24(1), 528. https://doi.org/10.3390/ijms24010528





