Assembly of the Complete Mitochondrial Genome of Gelsemium elegans Revealed the Existence of Homologous Conformations Generated by a Repeat Mediated Recombination
Abstract
1. Introduction
2. Results
2.1. General Features of G. elegans Mitochondrial Genome
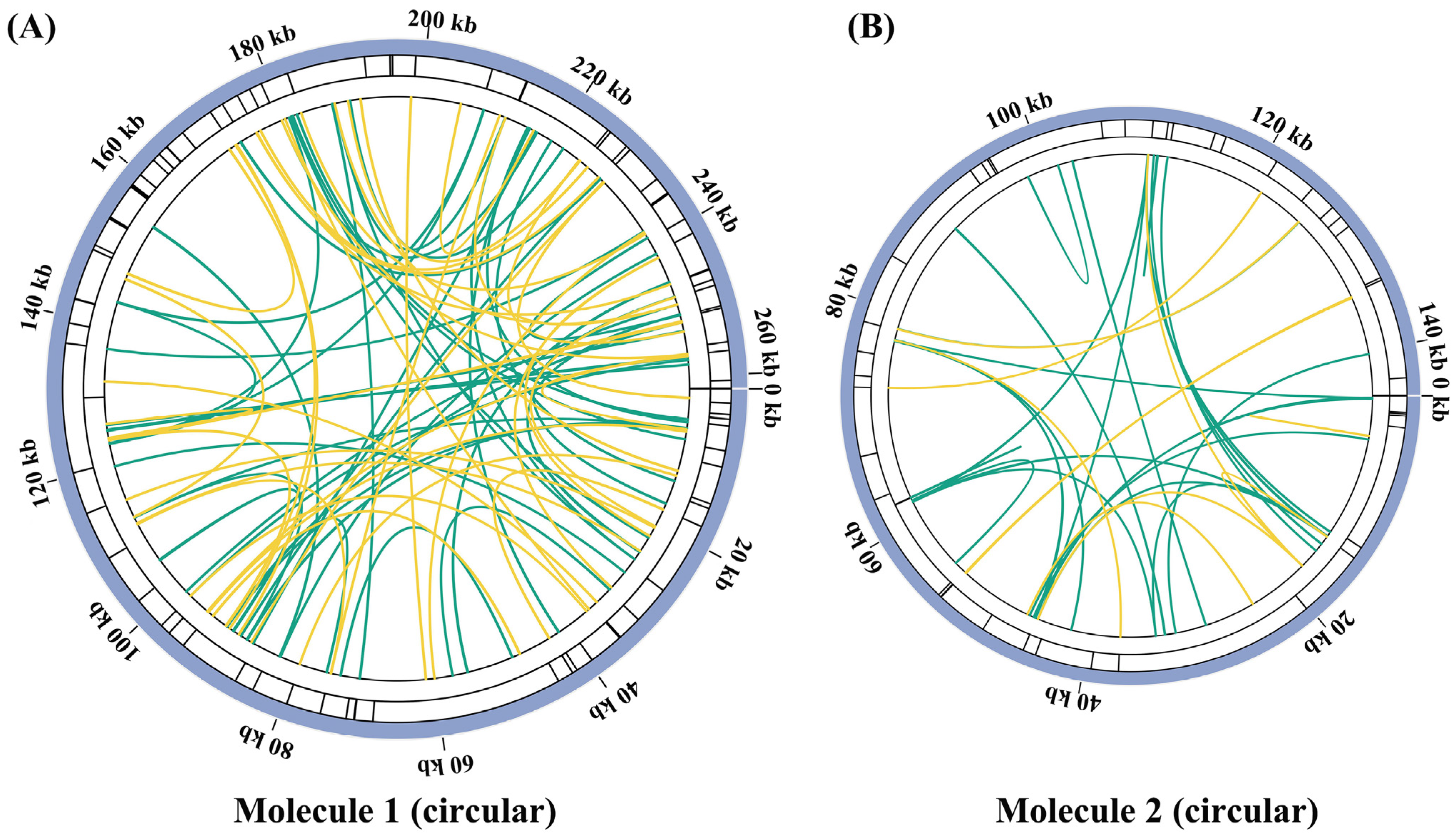
2.2. Repeat Sequence Analysis
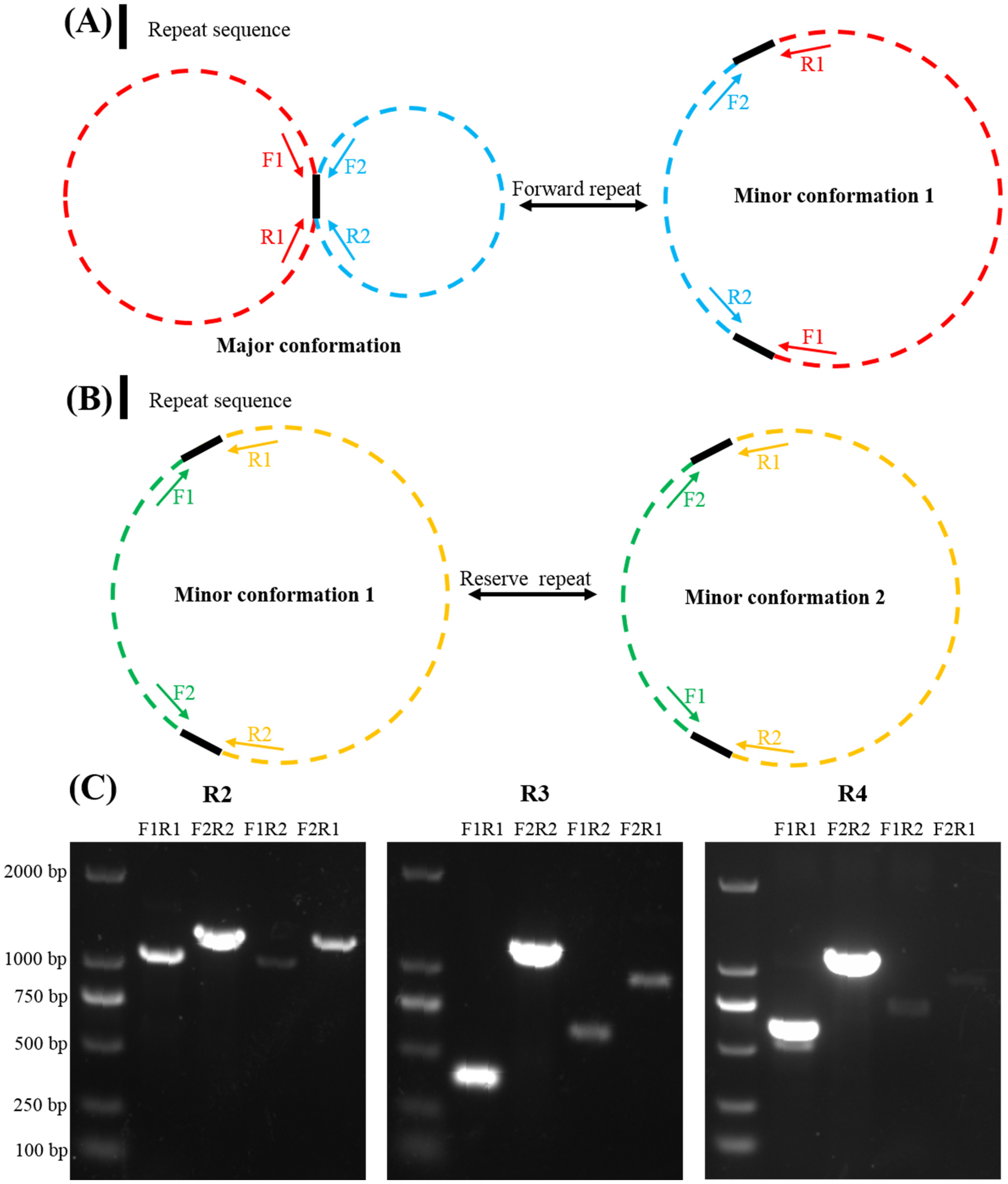
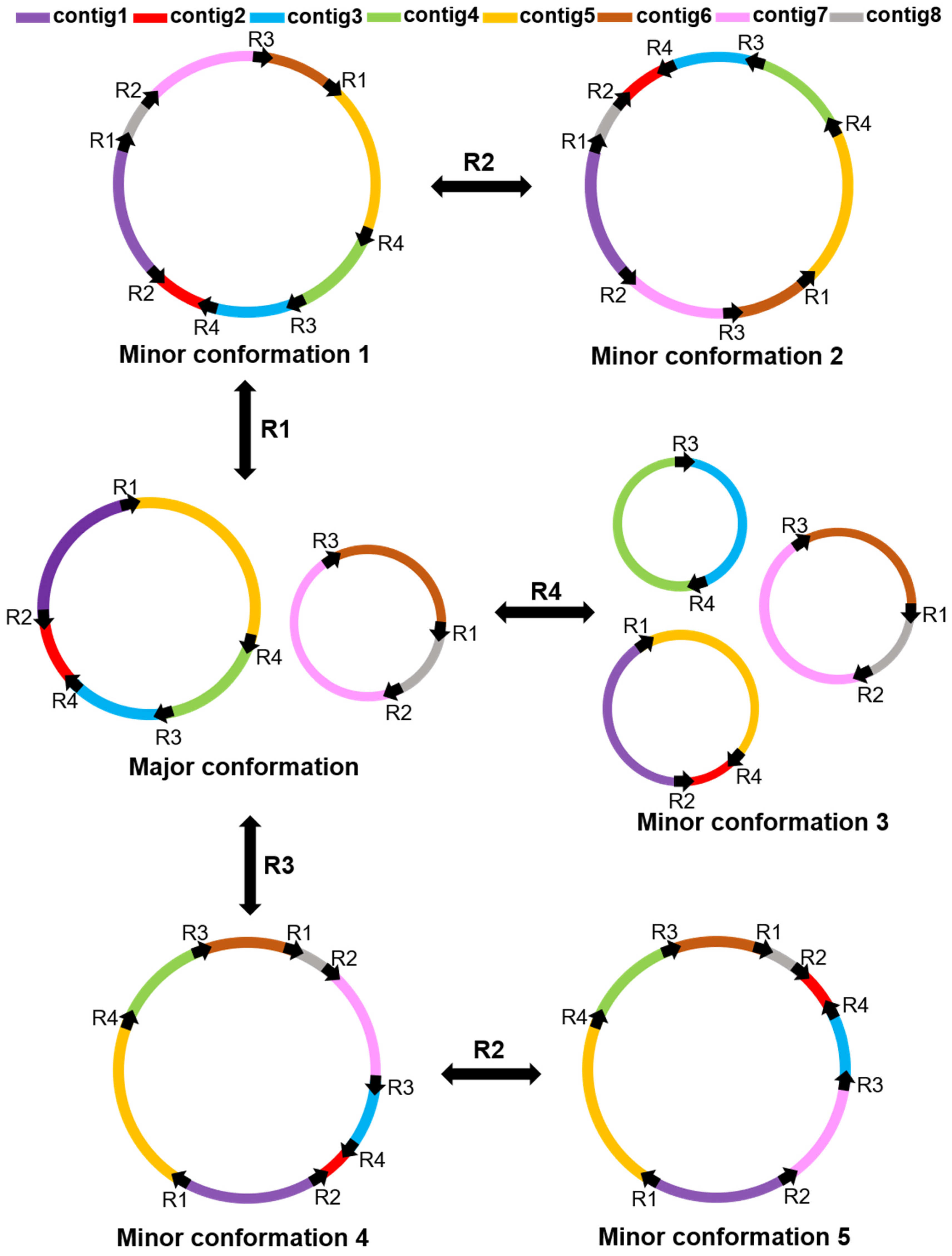
2.3. Repeat-Mediated Homologous Recombination
2.4. Identification of MTPTs
2.5. Phylogenetic Analysis
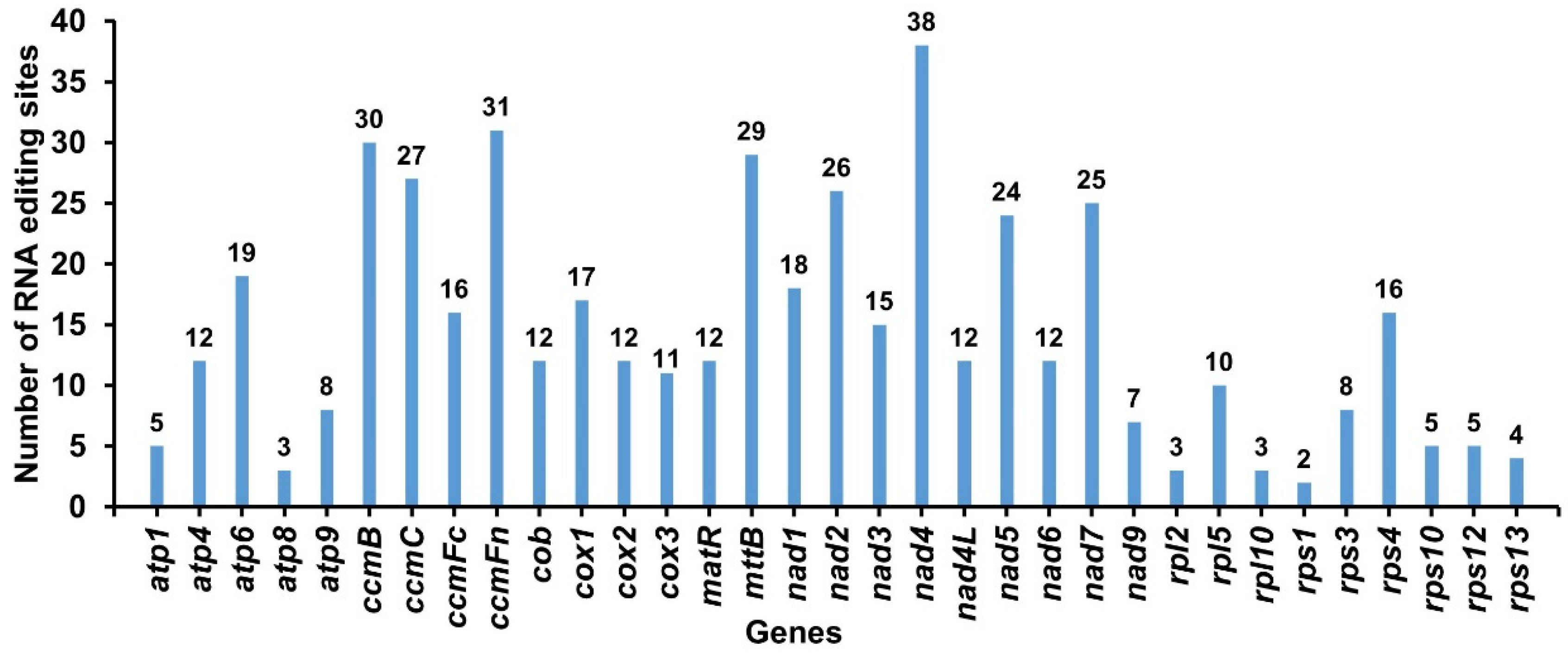
2.6. RNA Editing Events in G. elegans
3. Discussion
3.1. Multiple Circular Conformations in Plant Mitochondrial Genome
3.2. Repeated Sequence-Mediated Homologous Recombination
3.3. Phylogenetic Relationships of Homologous Species
3.4. Prediction of RNA Editing Sites
3.5. Intracellular Sequence Transfer Events in G. elegans Mitogenome
4. Materials and Methods
4.1. Plant Materials, DNA Extraction, and Sequencing
4.2. Genome Assembly, Modification and Annotation
4.3. DNA Repeat Sequence Analysis
4.4. Repeat-Mediated Homologous Recombination Predication and PCR Amplification Validation
4.5. Phylogenetic Analysis and RNA Editing Site Prediction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, Y.S.; Tang, Q.; Cheng, P.; Zhu, M.F.; Zhang, H.; Liu, J.Z.; Zuo, M.T.; Huang, C.Y.; Wu, C.Q.; Sun, Z.L.; et al. Whole-genome sequencing and analysis of the Chinese herbal plant Gelsemium elegans. Acta Pharm. Sin. B 2020, 10, 374–382. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.L.; Qiu, H.Q.; Cheng, Y.; Liu, M.; Chen, M.H.; Que, Y.X.; Que, W.C. Gelsemium elegans Benth: Chemical components, pharmacological effects, and toxicity mechanisms. Molecules 2021, 26, 7145. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Y.W.; Sun, Z.L.; Li, Y.Y.; Chen, X.J. Research progress on chemical constituents and pharmacological effects of Gelsemium elegans Benth. J. Tradit. Chin. Med. 2016, 35, 27. [Google Scholar]
- Wu, J.W. Study on the Promotion of Growth in Pigs by Gelsemiumelegans (Gardncechamp) Benth. Ph.D. Thesis, Fujian Agriculture and Forestry University, Fuzhou, China, 2015. [Google Scholar]
- Cai, J.; Lei, L.S.; Chi, D.B. Antineoplastic effect of koumine in mice bearing H22 solid tumor. J. South. Med. Univ. 2009, 29, 1851. [Google Scholar]
- Xu, Y.; Qiu, H.Q.; Liu, H.; Liu, M.; Huang, Z.Y.; Yang, J.; Su, Y.P.; Yu, C.X. Effects of koumine, an alkaloid of Gelsemium elegans Benth., on inflammatory and neuropathic pain models and possible mechanism with allopregnanolone. Pharmacol. Biochem. Behav. 2012, 101, 504–514. [Google Scholar] [CrossRef]
- Que, W.C.; Wu, Z.Y.; Chen, M.H.; Zhang, B.Q.; You, C.H.; Lin, H.L.; Zhao, Z.C.; Liu, M.B.; Qiu, H.Q.; Cheng, Y. Molecular mechanism of Gelsemium elegans (Gardner and Champ.) Benth. against neuropathic pain based on network pharmacology and experimental evidence. Front. Pharmacol. 2022, 12, 792932. [Google Scholar] [CrossRef]
- Liu, Y.C.; Li, L.; Pi, C.; Sun, Z.L.; Wu, Y.; Liu, Z.Y. Fingerprint analysis of Gelsemium elegans by HPLC followed by the targeted identification of chemical constituents using HPLC coupled with quadrupole-time-off light mass spectrometry. Fitoterapia 2017, 121, 94–105. [Google Scholar] [CrossRef]
- Liu, Y.C.; Xiao, S.; Yang, K.; Ling, L.; Sun, Z.L.; Liu, Z.Y. Comprehensive identification and structural characterization of target components from Gelsemium elegans by high-performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry based on accurate mass databases combined with MS/MS spectra. J. Mass. Spectrom. 2017, 52, 378–396. [Google Scholar]
- Jin, G.L.; Su, Y.P.; Liu, M.; Xu, Y.; Yang, J.; Liao, K.J.; Yu, C.X. Medicinal plants of the genus Gelsemium (Gelsemiaceae, Gentianales)—A review of their phytochemistry, pharmacology, toxicology and traditional use. J. Ethnopharmacol. 2014, 152, 33–52. [Google Scholar] [CrossRef]
- Roger, A.J.; Munoz-Gomez, S.A.; Kamikawa, R. The origin and diversification of mitochondria. Curr. Biol. 2017, 27, R1177–R1192. [Google Scholar] [CrossRef]
- Montilla, E.C.; Hillebrand, S.; Winterhalter, P. Anthocyanins in purple sweet potato (Ipomoea batatas L.) varieties. Fruit Veg. Cereal Sci. Biotechnol. 2011, 5, 19–23. [Google Scholar]
- Birky, C.W., Jr. The inheritance of genes in mitochondria and chloroplasts: Laws, mechanisms, and models. Ann. Rev. Genet. 2001, 35, 125–148. [Google Scholar] [CrossRef]
- Wolfe, K.H.; Li, W.H.; Sharp, P.M. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. USA 1987, 84, 9054–9058. [Google Scholar] [CrossRef]
- Wu, Z.Q.; Liao, X.Z.; Zhang, X.N.; Tembrock, L.R.; Broz, A. Genomic architectural variation of plant mitochondria—A review of multichromosomal structuring. J. Syst. Evol. 2022, 60, 160–168. [Google Scholar] [CrossRef]
- Kozik, A.; Rowan, B.A.; Lavelle, D.; Berke, L.; Schranz, M.E.; Michelmore, R.W.; Christensen, A.C. The alternative reality of plant mitochondrial DNA: One ring does not rule them all. PLoS Genet. 2019, 15, e1008373. [Google Scholar] [CrossRef]
- Sloan, D.B. One ring to rule them all? Genome sequencing provides new insights into the ‘master circle’ model of plant mitochondrial DNA structure. New Phytol. 2013, 200, 978–985. [Google Scholar] [CrossRef]
- Wu, Z.Q.; Cuthbert, J.M.; Taylor, D.R.; Sloan, D.B. The massive mitochondrial genome of the angiosperm Silene noctiflora is evolving by gain or loss of entire chromosomes. Proc. Natl. Acad. Sci. USA 2015, 112, 10185–10191. [Google Scholar] [CrossRef]
- Zou, Y.; Zhu, W.D.; Sloan, D.B.; Wu, Z.Q. Long-read sequencing characterizes mitochondrial and plastid genome variants in Arabidopsis msh1 mutants. Plant J. 2022, 112, 738–755. [Google Scholar] [CrossRef]
- Bi, C.W.; Paterson, A.H.; Wang, X.L.; Xu, Y.Q.; Wu, D.Y.; Qu, Y.S.; Jiang, A.N.; Ye, Q.L.; Ye, N. Analysis of the complete mitochondrial genome sequence of the diploid cotton Gossypium raimondii by comparative genomics approaches. BioMed Res. Int. 2019, 2019, 9691253. [Google Scholar] [CrossRef]
- Millar, A.H.; Heazlewood, J.L.; Kristensen, B.K.; Braun, H.P.; Møller, I.M. The plant mitochondrial proteome. Trends Plant Sci. 2005, 10, 36–43. [Google Scholar] [CrossRef]
- Shtolz, N.; Mishmar, D. The mitochondrial genome–on selective constraints and signatures at the organism, cell, and single mitochondrion levels. Front. Ecol. Evol. 2019, 7, 342. [Google Scholar] [CrossRef]
- Gras, D.E.; Mansilla, N.; Rodríguez, C.; Welchen, E.; Gonzalez, D.H. Arabidopsis thaliana SURFEIT1-like genes link mitochondrial function to early plant development and hormonal growth responses. Plant J. Cell Mol. Biol. 2020, 103, 690–704. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.H.; Wu, J.J.; Peng, Y.Q.; Hu, S.H.; Yan, H.S.; Xiao, F.; Zhou, X.; Gao, J.; Wang, R.H.; Xu, L.; et al. The complete mitochondrial genome of heartbreak grass Gelsemium elegans (Gardner & Champ.) Benth. (Gelsemiaceae). Mitochondrial DNA Part B Resour. 2019, 4, 3721–3722. [Google Scholar]
- Chase, C.D. Cytoplasmic male sterility: A window to the world of plant mitochondrial–nuclear interactions. Trends Genet. 2007, 23, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.W.; Zheng, X.M.; Li, C.L.; Xie, X.R.; Chen, Y.L.; Chen, L.T.; Zhao, X.C.; Zheng, H.Q.; Zhou, J.J.; Ye, S.; et al. Multi-step formation, evolution, and functionalization of new cytoplasmic male sterility genes in the plant mitochondrial genomes. Cell Res. 2017, 27, 130–146. [Google Scholar] [CrossRef]
- Toriyama, K. Molecular basis of cytoplasmic male sterility and fertility restoration in rice. Plant Biotechnol. 2021, 38, 285–295. [Google Scholar] [CrossRef]
- Melonek, J.; Small, I. Triticeae genome sequences reveal huge expansions of gene families implicated in fertility restoration. Curr. Opin. Plant Biol. 2022, 66, 102166. [Google Scholar] [CrossRef]
- Sun, T.; Bentolila, S.; Hanson, M.R. The unexpected diversity of plant organelle RNA editosomes. Trends Plant Sci. 2016, 21, 962–973. [Google Scholar] [CrossRef]
- Gualberto, J.M.; Mileshina, D.; Wallet, C.; Niazi, A.K.; Weber-Lotfi, F.; Dietrich, A. The plant mitochondrial genome: Dynamics and maintenance. Biochimie 2014, 100, 107–120. [Google Scholar] [CrossRef]
- Yang, Z.J.; Ni, Y.; Lin, Z.B.; Yang, L.B.; Chen, G.T.; Nijiati, N.; Hu, Y.Z.; Chen, X.Y. De novo assembly of the complete mitochondrial genome of sweet potato (Ipomoea batatas [L.] Lam) revealed the existence of homologous conformations generated by the repeat-mediated recombination. BMC Plant Biol. 2022, 22, 285. [Google Scholar] [CrossRef]
- Wang, S.B.; Li, D.W.; Yao, X.H.; Song, Q.W.; Wang, Z.P.; Zhang, Q.; Zhong, C.H.; Liu, Y.F.; Huang, H.W. Evolution and diversification of kiwifruit mitogenomes through extensive whole-genome rearrangement and mosaic loss of intergenic sequences in a highly variable region. Genome Biol. Evol. 2019, 11, 1192–1206. [Google Scholar] [CrossRef]
- Li, J.L.; Xu, Y.C.; Shan, Y.Y.; Pei, X.Y.; Yong, S.Y.; Liu, C.; Yu, J. Assembly of the complete mitochondrial genome of an endemic plant, Scutellaria tsinyunensis, revealed the existence of two conformations generated by a repeat-mediated recombination. Planta 2021, 254, 36. [Google Scholar] [CrossRef]
- Alverson, A.J.; Rice, D.W.; Dickinson, S.; Barry, K.; Palmer, J.D. Origins and recombination of the bacterial-sized multichromosomal mitochondrial genome of cucumber. Plant Cell 2011, 23, 2499–2513. [Google Scholar] [CrossRef]
- Yu, R.X.; Sun, C.Y.; Zhong, Y.; Liu, Y.; Sanchez-Puerta, M.V.; Mower, J.P.; Zhou, R.C. The minicircular and extremely heteroplasmic mitogenome of the holoparasitic plant Rhopalocnemis phalloides. Curr. Biol. 2022, 32, 470–479. [Google Scholar] [CrossRef]
- Guo, W.; Zhu, A.; Fan, W.; Mower, J.P. Complete mitochondrial genomes from the ferns Ophioglossum californicum and Psilotum nudum are highly repetitive with the largest organellar introns. New Phytol. 2017, 213, 391–403. [Google Scholar] [CrossRef]
- Dong, S.; Zhao, C.; Chen, F.; Liu, Y.; Zhang, S.; Wu, H.; Zhang, L.; Liu, Y. The complete mitochondrial genome of the early flowering plant Nymphaea colorata is highly repetitive with low recombination. BMC Genom. 2018, 19, 614. [Google Scholar] [CrossRef]
- Fang, B.; Li, J.L.; Zhao, Q.; Liang, Y.P.; Yu, J. Assembly of the complete mitochondrial genome of Chinese plum (Prunus salicina): Characterization of genome recombination and RNA editing sites. Genes 2021, 12, 1970. [Google Scholar] [CrossRef]
- Morgante, M.; Hanafey, M.; Powell, W. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat. Genet. 2002, 30, 194–200. [Google Scholar] [CrossRef]
- Liu, Y.C.; Liu, S.; Liu, D.C.; Wei, Y.X.; Liu, C.; Yang, Y.M.; Tao, C.G.; Liu, W.S. Exploiting EST databases for the development and characterization of EST-SSR markers in blueberry (Vaccinium) and their cross-species transferability in Vaccinium spp. Sci. Hortic. 2014, 176, 319–329. [Google Scholar] [CrossRef]
- Cheng, Y.; He, X.; Priyadarshani, S.V.G.N.; Wang, Y.; Ye, L.; Shi, C.; Ye, K.; Zhou, Q.; Luo, Z.; Deng, F.; et al. Assembly and comparative analysis of the complete mitochondrial genome of Suaeda glauca. BMC Genom. 2021, 22, 167. [Google Scholar] [CrossRef]
- Christensen, A.C. Plant mitochondrial genome evolution can be explained by DNA repair mechanisms. Genome Biol. Evol. 2013, 5, 1079–1886. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.R.; Keeling, P.J. Mitochondrial and plastid genome architecture: Reoccurring themes, but significant differences at the extremes. Proc. Natl. Acad. Sci. USA 2015, 112, 10177–10184. [Google Scholar] [CrossRef] [PubMed]
- Hong, Z.; Liao, X.Z.; Ye, Y.J.; Zhang, N.N.; Yang, Z.J.; Zhu, W.D.; Gao, W.; Sharbrough, J.; Tembrock, L.R.; Xu, D.P.; et al. A complete mitochondrial genome for fragrant Chinese rosewood (Dalbergia odorifera, Fabaceae) with comparative analyses of genome structure and intergenomic sequence transfers. BMC Genom. 2021, 22, 672. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.F.; Zhu, G.N.; Li, R.; Yan, S.J.; Fu, D.Q.; Zhu, B.Z.; Tian, H.Q.; Luo, Y.B.; Zhu, H.Q. The RNA editing factor SlORRM4 is required for normal fruit ripening in tomato. Plant Physiol. 2017, 175, 1690–1702. [Google Scholar] [CrossRef] [PubMed]
- He, P.; Xiao, G.H.; Liu, H.; Zhang, L.H.; Zhao, L.; Tang, M.J.; Huang, S.; An, Y.J.; Yu, J.N. Two pivotal RNA editing sites in the mitochondrial atp1 mRNA are required for ATP synthase to produce sufficient ATP for cotton fiber cell elongation. New Phytol. 2018, 218, 167–182. [Google Scholar] [CrossRef] [PubMed]
- Notsu, Y.; Masood, S.; Nishikawa, T.; Kubo, N.; Akiduki, G.; Nakazono, M.; Hirai, A.; Kadowaki, K. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: Frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol. Genet. Genom. 2002, 268, 434–445. [Google Scholar] [CrossRef]
- Wang, X.C.; Chen, H.; Yang, D.; Liu, C. Diversity of mitochondrial plastid DNAs (MTPTs) in seed plants. Mitochondrial DNA Part A 2018, 29, 635–642. [Google Scholar] [CrossRef]
- Kitazaki, K.; Kubo, T.; Kagami, H.; Matsumoto, T.; Fujita, A.; Matsuhira, H.; Matsunaga, M.; Mikami, T. A horizontally transferred tRNA(Cys) gene in the sugar beet mitochondrial genome: Evidence that the gene is present in diverse angiosperms and its transcript is aminoacylated. Plant J. Cell Mol. Biol. 2011, 68, 262–272. [Google Scholar] [CrossRef]
- Arseneau, J.R.; Steeves, R.; Laflamme, M. Modified low-salt CTAB extraction of high-quality DNA from contaminant-rich tissues. Mol. Ecol. Resour. 2017, 17, 686–693. [Google Scholar] [CrossRef]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; de Pamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Chen, Y.; Ye, W.; Zhang, Y.; Xu, Y. High speed BLASTN: An accelerated MegaBLAST search tool. Nucleic Acids Res. 2015, 43, 7762–7768. [Google Scholar] [CrossRef]
- Shi, L.; Chen, H.; Jiang, M.; Wang, L.; Wu, X.; Huang, L.; Liu, C. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 2019, 47, W65–W73. [Google Scholar] [CrossRef]
- Lewis, S.E.; Searle, S.; Harris, N.; Gibson, M.; Iyer, V.; Richter, J.; Wiel, C.; Bayraktaroglu, L.; Birney, E.; Crosby, M.; et al. Apollo: A sequence annotation editor. Genome Biol. 2002, 3, research0082.1. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Stefan, K.; Choudhuri, J.V.; Enno, O.; Chris, S.; Jens, S.; Robert, G. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 22, 4633–4642. [Google Scholar]
- Zhang, H.; Meltzer, P.; Davis, S. RCircos: An R package for Circos 2D track plots. BMC Bioinform. 2013, 14, 244. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Mower, J.P. The PREP suite: Predictive RNA editors for plant mitochondrial genes, chloroplast genes and us-er-defined alignments. Nucleic Acids Res. 2009, 37, W253–W259. [Google Scholar] [CrossRef]

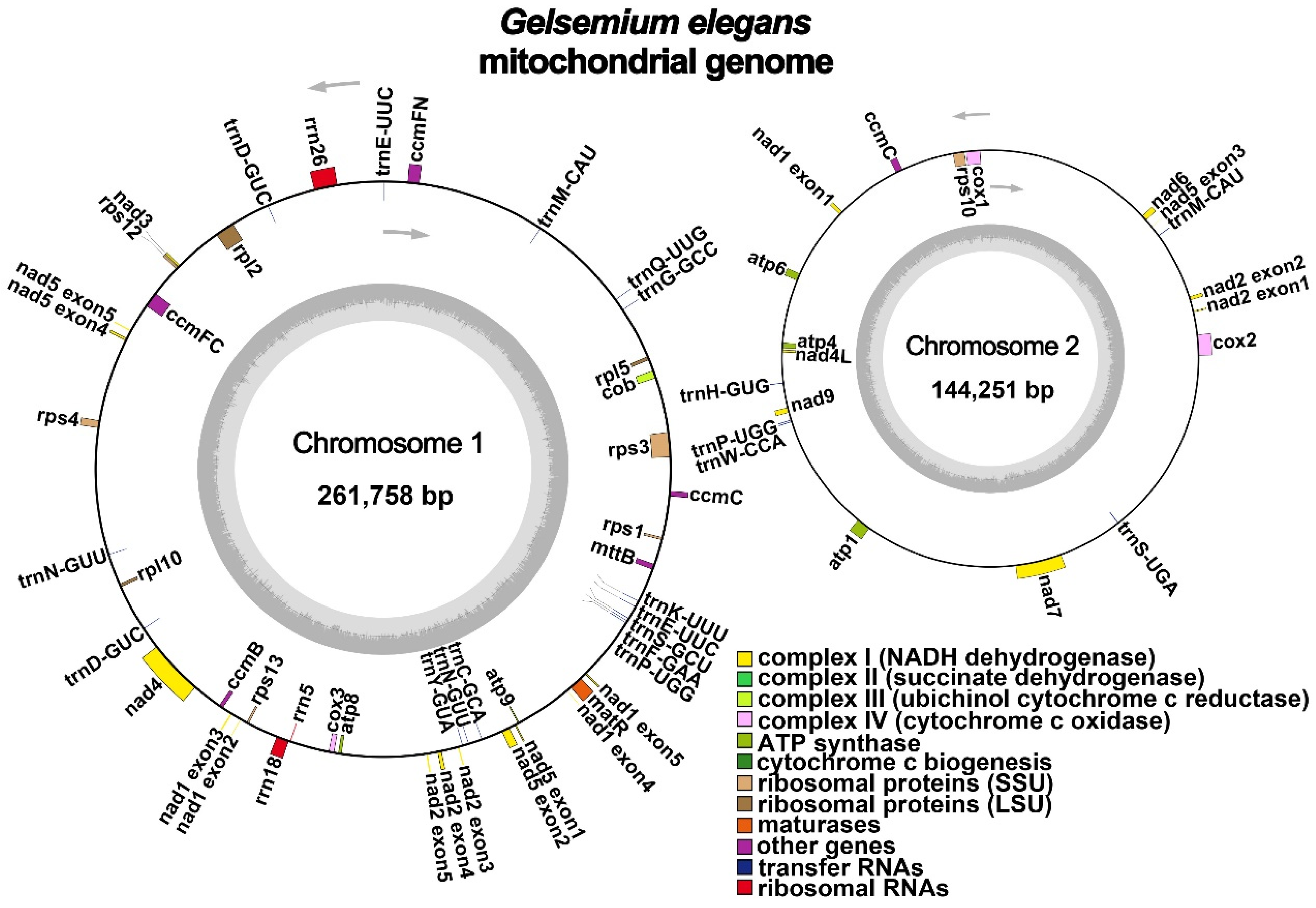

| Group of Genes | Name of Genes |
|---|---|
| ATP synthase | atp1, atp4, atp6, atp8, atp9 |
| NADH dehydrogenase | nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7, nad9 |
| Cytochrome c biogenesis | cob |
| Ubiquinol cytochrome c reductase | ccmB, ccmC (×2), ccmFC, ccmFN |
| Cytochrome c oxidase | cox1, cox2, cox3 |
| Maturases | matR |
| Transport membrane protein | mttB |
| Large subunit of ribosome | rpl2, rpl5, rpl10 |
| Small subunit of ribosome | rps1, rps3, rps4, rps10, rps12 (×2), rps13 |
| Ribosome RNA | rrn5, rrn18, rrn26 |
| Transfer RNA | trnC-GCA, trnD-GUC (×2), trnE-UUC (×2), trnF-GAA, trnG-GCC, trnH-GUG, trnK-UUU, trnM-CAU (×2), trnN-GUU (×2), trnP-UGG (×2), trnQ-UUG, trnS-GCU, trnS-UGA, trnW-CCA, trnY-GUA |
| Repeat Name | Repeat 1 | Repeat 2 | Repeat 3 | Repeat 4 |
|---|---|---|---|---|
| Identities (%) | 100 | 100 | 98.65 | 91.24 |
| Length (bp) | 9816 | 360 | 211 | 137 |
| Position-1 | 251,943–261,758 | 190,267–190,626 | 125,011–125,232 | 82,546–82,676 |
| Position-2 | 38,228–48,043 | 60,101–59,742 | 142,600–142,820 | 155,253–155,389 |
| E-value | 0 | 0 | 6.90 × 10−108 | 1.63 × 10−44 |
| Type | Forward | Reserve | Forward | Forward |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
You, C.; Cui, T.; Zhang, C.; Zang, S.; Su, Y.; Que, Y. Assembly of the Complete Mitochondrial Genome of Gelsemium elegans Revealed the Existence of Homologous Conformations Generated by a Repeat Mediated Recombination. Int. J. Mol. Sci. 2023, 24, 527. https://doi.org/10.3390/ijms24010527
You C, Cui T, Zhang C, Zang S, Su Y, Que Y. Assembly of the Complete Mitochondrial Genome of Gelsemium elegans Revealed the Existence of Homologous Conformations Generated by a Repeat Mediated Recombination. International Journal of Molecular Sciences. 2023; 24(1):527. https://doi.org/10.3390/ijms24010527
Chicago/Turabian StyleYou, Chuihuai, Tianzhen Cui, Chang Zhang, Shoujian Zang, Yachun Su, and Youxiong Que. 2023. "Assembly of the Complete Mitochondrial Genome of Gelsemium elegans Revealed the Existence of Homologous Conformations Generated by a Repeat Mediated Recombination" International Journal of Molecular Sciences 24, no. 1: 527. https://doi.org/10.3390/ijms24010527
APA StyleYou, C., Cui, T., Zhang, C., Zang, S., Su, Y., & Que, Y. (2023). Assembly of the Complete Mitochondrial Genome of Gelsemium elegans Revealed the Existence of Homologous Conformations Generated by a Repeat Mediated Recombination. International Journal of Molecular Sciences, 24(1), 527. https://doi.org/10.3390/ijms24010527







