Chromatin-Remodelling ATPases ISWI and BRM Are Essential for Reproduction in the Destructive Pest Tuta absoluta
Abstract
:1. Introduction
2. Results
2.1. Chromatin-Remodelling Gene Cloning
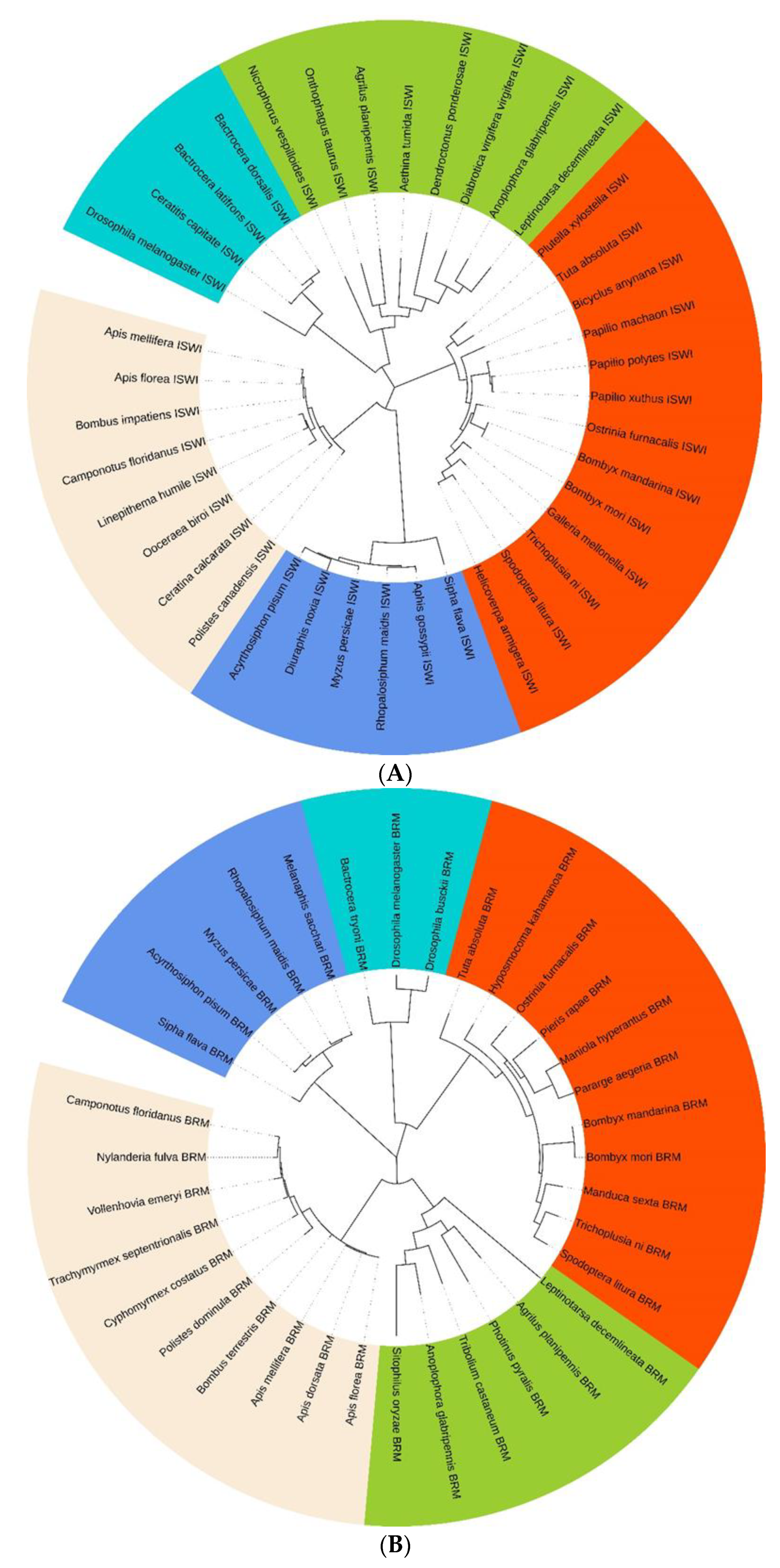
2.2. TaISWI and TaBRM Characterisation
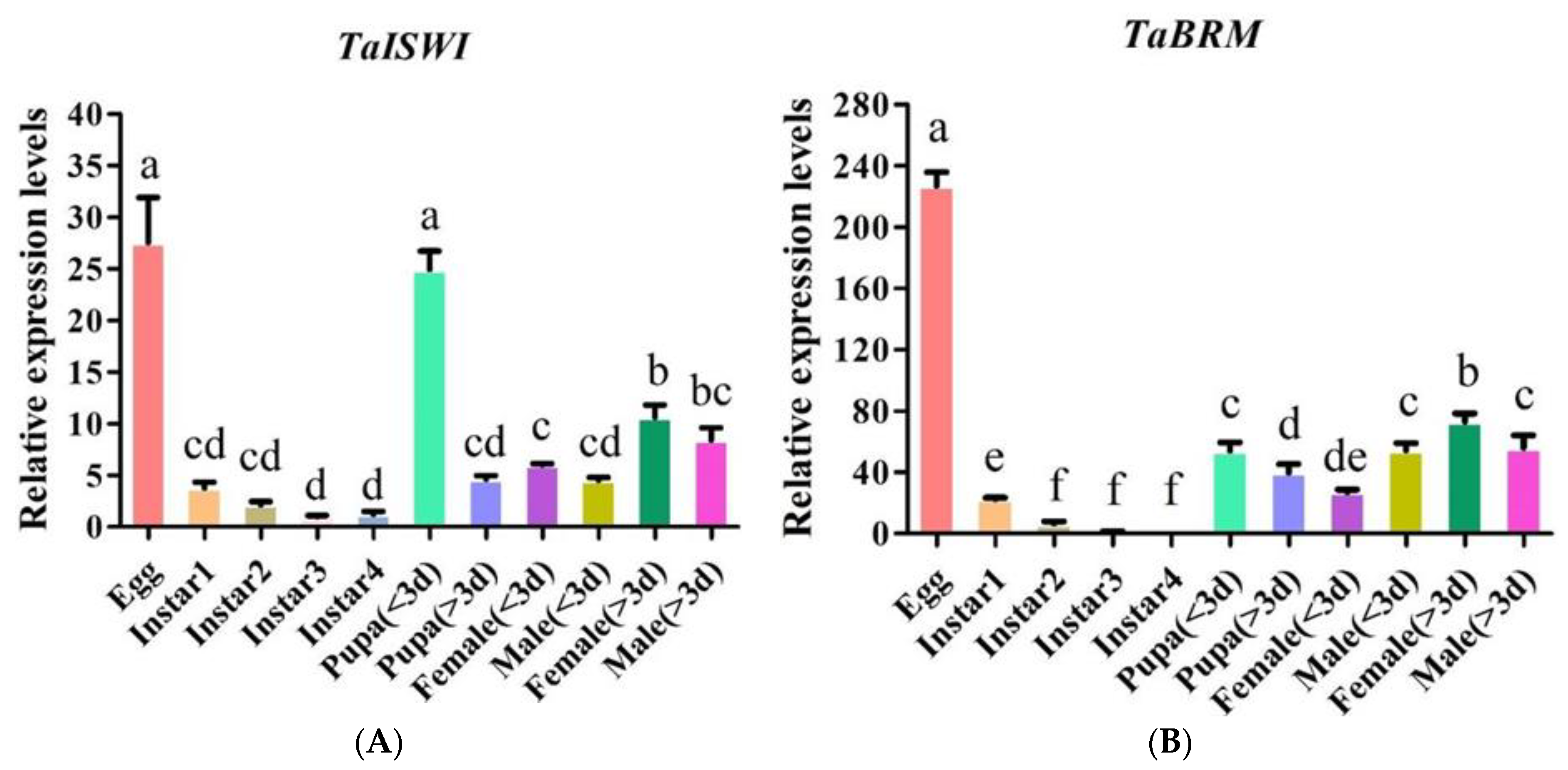
2.3. TaISWI and TaBRM Expression Patterns during Different Life Cycle Stages
2.4. Effects of dsRNA Injection on TaISWI and TaBRM Expression
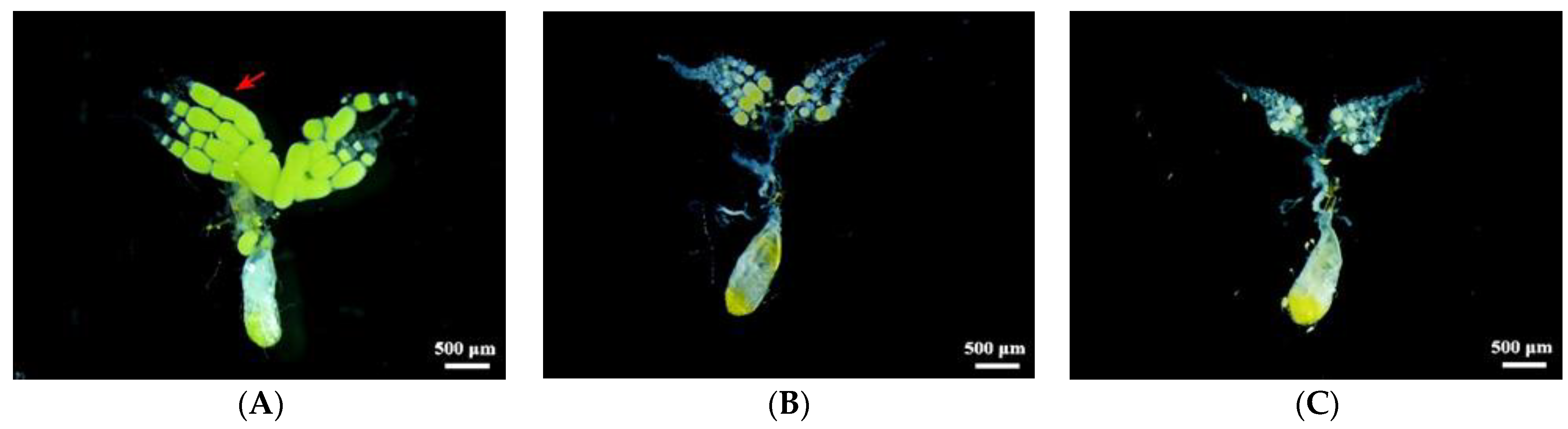
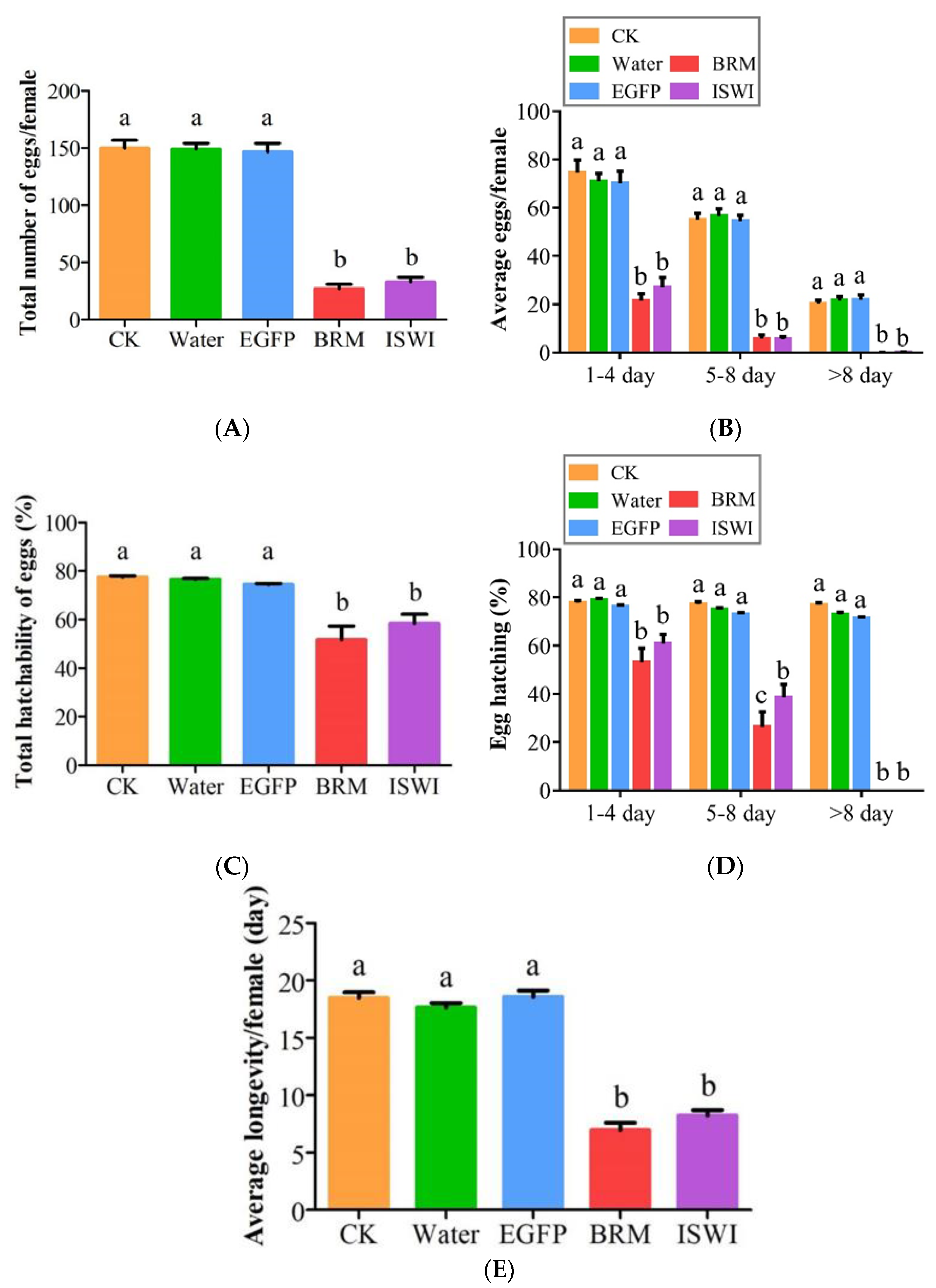
2.5. Effects of dsRNA Injection on Ovarian Development and Fecundity
3. Discussion
4. Materials and Methods
4.1. Insect Rearing
4.2. RNA Isolation and cDNA Synthesis
4.3. Chromatin-Remodelling Gene Cloning
4.4. Gene Sequences Analysis and Phylogenetic Relationships
4.5. RT-qPCR
4.6. Double-Stranded RNA (dsRNA) Synthesis
4.7. dsRNA Injection and Detection
4.8. Post-RNAi Ovary Development and Fecundity
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guillemaud, T.; Blin, A.; Goff, I.L.; Desneux, N.; Reyes, M.; Tabone, E.; Tsagkarakou, A.; Niño, L.; Lombaert, E. The tomato borer, Tuta absoluta, invading the Mediterranean Basin, originates from a single introduction from Central Chile. Sci. Rep. 2015, 5, 8371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, M.F.; Espul, J.C. Bioecology of the tomato moth (Scrobipalpula absoluta) in Mendoza, Argentine Republic. Rev. Investig. Agrop. 1982, 17, 135–146. [Google Scholar]
- Desneux, N.; Wajnberg, E.; Wyckhuys, K.A.G.; Burgio, G.; Arpaia, S.; Narváez-Vasquez, C.A.; González-Cabrera, J.; Ruescas, D.C.; Tabone, E.; Frandon, J.; et al. Biological invasion of European tomato crops by Tuta absoluta: Ecology, geographic expansion and prospects for biological control. J. Pest Sci. 2010, 83, 197–215. [Google Scholar] [CrossRef]
- Biondi, A.; Guedes, R.N.C.; Wan, F.H.; Desneux, N. Ecology, worldwide spread, and management of the invasive South American tomato pinworm, Tuta absoluta: Past, present, and future. Annu. Rev. Entomol. 2018, 63, 239–258. [Google Scholar] [CrossRef] [PubMed]
- Desneux, N.; Luna, M.G.; Guillemaud, T.; Urbaneja, A. The invasive South American tomato pinworm, Tuta absoluta, continues to spread in Afro-Eurasia and beyond: The new threat to tomato world production. J. Pest Sci. 2011, 84, 403–408. [Google Scholar] [CrossRef]
- Guimapi, R.Y.A.; Mohamed, S.A.; Okeyo, G.O.; Ndjomatchoua, F.T.; Ekesi, S.; Tonnang, H.E.Z. Modeling the risk of invasion and spread of Tuta absoluta in Africa. Ecol. Complex. 2016, 28, 77–93. [Google Scholar] [CrossRef]
- Zhang, G.-F.; Ma, D.-Y.; Wang, Y.-S.; Gao, Y.-H.; Liu, W.-X.; Zhang, R.; Fu, W.-J.; Xian, X.-Q.; Wang, J.; Kuang, M.; et al. First report of the South American tomato leafminer, Tuta absoluta (Meyrick), in China. J. Integr. Agr. 2020, 19, 1912–1917. [Google Scholar] [CrossRef]
- Li, X.W.; Li, D.; Zhang, Z.J.; Huang, J.; Zhang, J.M.; Hafeez, M.; Wang, L.K.; Guo, W.C.; Lu, Y.B. Supercooling capacity and cold tolerance of the South American tomato pinworm, Tuta absoluta, a newly invaded pest in China. J. Pest Sci. 2021, 94, 845–858. [Google Scholar] [CrossRef]
- Campos, M.R.; Biondi, A.; Adiga, A.; Guedes, R.N.C.; Desneux, N. From the Western Palaearctic region to beyond: Tuta absoluta ten years after invading Europe. J. Pest Sci. 2017, 90, 787–796. [Google Scholar] [CrossRef]
- FAO (Food and Agriculture Organization). Online Statistical Database: Food and Agriculture Data. FAOSTAT. 2019. Available online: http://www.fao.org/faostat (accessed on 6 February 2021).
- Zhang, G.; Xian, X.; Zhang, Y.; Liu, W.; Liu, H.; Feng, X.; Ma, D.; Wang, Y.; Gao, Y.; Zhang, R.; et al. Outbreak of the South American tomato leafminer, Tuta absoluta, in the Chinese mainland: Geographic and potential host range expansion. Pest Manag. Sci. 2021, 77, 5475–5488. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.M.; Li, X.W.; He, T.J.; Li, P.J.; Liu, Y.; Zhou, S.X.; Wu, Q.C.; Chen, T.T.; Lu, Y.B.; Hou, Y.M. Comparative biochemical and transcriptome analyses in tomato and eggplant reveal their differential responses to Tuta absoluta infestation. Genomics 2021, 113, 2108–2121. [Google Scholar] [CrossRef] [PubMed]
- Urbaneja, A.; González-Cabrera, J.; Arnó, J.; Gabarra, R. Prospects for the biological control of Tuta absoluta in tomatoes of the Mediterranean basin. Pest Manag. Sci. 2012, 68, 1215–1222. [Google Scholar] [CrossRef] [PubMed]
- Mansour, R.; Brévault, T.; Chailleux, A.; Cherif, A.; Grissa-Lebdi, K.; Haddi, K.; Mohamed, S.A.; Nofemela, R.; Oke, A.; Sylla, S.; et al. Occurrence, biology, natural enemies and management of Tuta absoluta in Africa. Entomol. Gen. 2018, 38, 83–112. [Google Scholar] [CrossRef]
- Biondi, A.; Zappalà, L.; Stark, J.D.; Desneux, N. Do biopesticides affect the demographic traits of a parasitoid wasp and its biocontrol services through sublethal effects? PLoS ONE 2013, 8, e76548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Passos, L.C.; Soares, M.A.; Collares, L.J.; Malagoli, I.; Desneux, N.; Carvalho, G.A. Lethal, sublethal and transgenerational effects of insecticides on Macrolophus basicornis, predator of Tuta absoluta. Entomol. Gen. 2018, 38, 127–143. [Google Scholar] [CrossRef]
- Garzia, G.T.; Siscaro, G.; Biondi, A.; Zappalà, L. Tuta absoluta, a South American pest of tomato now in the EPPO region: Biology, distribution and damage. EPPO Bull. 2012, 42, 205–210. [Google Scholar] [CrossRef]
- Siqueira, H.Á.A.; Guedes, R.N.C.; Picanço, M.C. Insecticide resistance in populations of Tuta absoluta (Lepidoptera: Gelechiidae). Agric. For. Entomol. 2000, 2, 147–153. [Google Scholar] [CrossRef]
- Roditakis, E.; Vasakis, E.; Grispou, M.; Stavrakaki, M.; Nauen, R.; Gravouil, M.; Bassi, A. First report of Tuta absoluta resistance to diamide insecticides. J. Pest Sci. 2015, 88, 9–16. [Google Scholar] [CrossRef]
- Roditakis, E.; Steinbach, D.; Moritz, G.; Vasakis, E.; Stavrakaki, M.; Ilias, A.; García-Vidal, L.; Martínez-Aguirre, M.D.R.; Bielza, P.; Morou, E.; et al. Ryanodine receptor point mutations confer diamide insecticide resistance in tomato leafminer, Tuta absoluta (Lepidoptera: Gelechiidae). Insect Biochem. Mol. Biol. 2017, 80, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Langa, T.P.; Dantas, K.C.T.; Pereira, D.L.; Oliveira, M.; Ribeiro, L.M.S.; Siqueira, H.A.A. Basis and monitoring of methoxyfenozide resistance in the South American tomato pinworm Tuta absoluta. J. Pest Sci. 2021, 95, 351–364. [Google Scholar] [CrossRef]
- Sohrabi, F.; Nouryazdan, H.R.; Gharati, B.; Saeidi, Z. Evaluation of ten tomato cultivars for resistance against tomato leaf miner, Tuta absoluta (Meyrick) (Lepidoptera: Gelechiidae) under field infestation conditions. Entomol. Gen. 2016, 36, 163–175. [Google Scholar] [CrossRef]
- Campolo, O.; Cherif, A.; Ricupero, M.; Siscaro, G.; Grissa-Lebdi, K.; Russo, A.; Cucci, L.M.; Di Pietro, P.; Satriano, C.; Desneux, N.; et al. Citrus peel essential oil nanoformulations to control the tomato borer, Tuta absoluta: Chemical properties and biological activity. Sci. Rep. 2017, 7, 13036. [Google Scholar] [CrossRef] [PubMed]
- Majidiani, S.; Pourabad, R.F.; Laudani, F.; Campolo, O.; Zappalà, L.; Rahmani, S.; Mohammadi, S.A.; Palmeri, V. RNAi in Tuta absoluta management: Effects of injection and root delivery of dsRNAs. J. Pest Sci. 2019, 92, 1409–1419. [Google Scholar] [CrossRef]
- Bolognesi, R.; Ramaseshadri, P.; Anderson, J.; Bachman, P.; Clinton, W.; Flannagan, R.; Ilagan, O.; Lawrence, C.; Levine, S.; Moar, W.; et al. Characterizing the Mechanism of action of double-stranded RNA activity against western corn rootworm (Diabrotica virgifera virgifera LeConte). PLoS ONE 2012, 7, e47534. [Google Scholar] [CrossRef] [PubMed]
- Scott, J.G.; Michel, K.; Bartholomay, L.C.; Siegfried, B.D.; Hunter, W.B.; Smagghe, G.; Zhu, K.Y.; Douglas, A.E. Towards the elements of successful insect RNAi. J. Insect Physiol. 2013, 59, 1212–1221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vélez, A.M.; Fishilevich, E. The mysteries of insect RNAi: A focus on dsRNA uptake and transport. Pestic. Biochem. Physiol. 2018, 151, 25–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, P.; Bayram, Y.; Shaltiel-Harpaz, L.; Sohrabi, F.; Saji, A.; Esenali, U.T.; Jalilov, A.; Ali, A.; Shashank, P.R.; Ismoilov, K.; et al. Tuta absoluta continues to disperse in Asia: Damage, ongoing management and future challenges. J. Pest Sci. 2019, 92, 1317–1327. [Google Scholar] [CrossRef]
- Laudani, F.; Strano, C.P.; Edwards, M.G.; Malacrinò, A.; Campolo, O.; El Halim, H.M.A.; Gatehouse, A.M.; Palmeri, V. RNAi-mediated gene silencing in Rhynchophorus ferrugineus (Oliver) (Coleoptera: Curculionidae). Open Life Sci. 2017, 12, 214–222. [Google Scholar] [CrossRef]
- Zhang, J.; Khan, S.A.; Heckel, D.G.; Bock, R. Next generation insect-resistant plants: RNAi-mediated crop protection. Trends Biotechnol. 2017, 35, 871–882. [Google Scholar] [CrossRef] [PubMed]
- Corona, D.F.; Tamkun, J.W. Multiple roles for ISWI in transcription, chromosome organization and DNA replication. Biochim. Biophys. Acta (BBA) Gene Struct. Expr. 2004, 1677, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Hopfner, K.P.; Gerhold, C.B.; Lakomek, K.; Wollmann, P. Swi2/Snf2 remodelers: Hybrid views on hybrid molecular machines. Curr. Opin Struct. Biol. 2012, 22, 225–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hota, S.K.; Bruneau, B.G. ATP-dependent chromatin remodeling during mammalian development. Development 2016, 143, 2882–2897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clapier, C.R.; Cairns, B.R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009, 78, 273–304. [Google Scholar] [CrossRef] [PubMed]
- Khajuria, C.; Vélez, A.M.; Rangasamy, M.; Wang, H.; Fishilevich, E.; Frey, M.L.F.; Carneiro, N.P.; Gandra, P.; Narva, K.E.; Siegfried, B.D. Parental RNA interference of genes involved in embryonic development of the western corn rootworm, Diabrotica virgifera virgifera LeConte. Insect Biochem. Mol. Biol. 2015, 63, 54–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fishilevich, E.; Vélez, A.M.; Khajuria, C.; Frey, M.L.; Hamm, R.L.; Wang, H.; Schulenberg, G.A.; Bowling, A.J.; Pence, H.E.; Gandra, P.; et al. Use of chromatin remodeling ATPases as RNAi targets for parental control of western corn rootworm (Diabrotica virgifera virgifera) and Neotropical brown stink bug (Euschistus heros). Insect Biochem. Mol. Biol. 2016, 71, 58–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basnet, S.; Kamble, S.T. Knockdown of the chromatin remodeling gene brahma by RNA interference reduces reproductive fitness and lifespan in common bed bug (Hemiptera: Cimicidae). J. Med. Entomol. 2018, 55, 534–539. [Google Scholar] [CrossRef] [PubMed]
- Basnet, S.; Kamble, S.T. Silencing of four genes involved in chromatin remodeling by RNA interference adversely affects fecundity of bed bugs (Hemiptera: Cimicidae). J. Med. Entomol. 2018, 55, 1440–1445. [Google Scholar] [CrossRef] [PubMed]
- Boyer, L.A.; Latek, R.R.; Peterson, C.L. The SANT domain: A unique histone-tail-binding module? Nat. Rev. Mol. Cell Biol. 2004, 5, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Mathew, R.P.; Mishra, R.K.; Pandey, S.M. ISWI chromatin remodeling complexes: Composition and regulation perspectives. J. Sci. Res. 2018, 62, 133–145. [Google Scholar]
- Ji, S.-X.; Wang, X.-D.; Shen, X.-N.; Liang, L.; Liu, W.-X.; Wan, F.-H.; Lü, Z.-C. Using RNA interference to reveal the function of chromatin remodeling factor ISWI in temperature tolerance in Bemisia tabaci Middle East-Asia Minor 1 cryptic species. Insects 2020, 11, 113. [Google Scholar] [CrossRef] [Green Version]
- Grüne, T.; Brzeski, J.; Eberharter, A.; Clapier, C.R.; Corona, D.F.; Becker, P.B.; Müller, C.W. Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI. Mol. Cell 2003, 12, 449–460. [Google Scholar] [CrossRef]
- Mueller-Planitz, F.; Klinker, H.; Ludwigsen, J.; Becker, P.B. The ATPase domain of ISWI is an autonomous nucleosome remodeling machine. Nat. Struct. Mol. Biol. 2013, 20, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Tamkun, J.W.; Deuring, R.; Scott, M.P.; Kissinger, M.; Pattatucci, A.M.; Kaufman, T.C.; Kennison, J.A. Brahma: A regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell 1992, 68, 561–572. [Google Scholar] [CrossRef]
- Elfring, L.K.; Daniel, C.; Papoulas, O.; Deuring, R.; Sarte, M.; Moseley, S.; Beek, S.J.; Waldrip, W.R.; Daubresse, G.; DePace, A.; et al. Genetic analysis of brahma: The Drosophila homolog of the yeast chromatin remodeling factor SWI2/SNF2. Genetics 1998, 148, 251–265. [Google Scholar] [CrossRef] [PubMed]
- Hannon, G.J. RNA interference. Nature 2002, 418, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Brizuela, B.J.; Elfring, L.; Ballard, J.; Tamkun, J.W.; Kennison, J.A. Genetic analysis of the brahma gene of Drosophila melanogaster and polytene chromosome subdivisions 72AB. Genetics 1994, 137, 803–813. [Google Scholar] [CrossRef]
- Carrera, I.; Zavadil, J.; Treisman, J.E. Two subunits specific to the PBAP chromatin remodeling complex have distinct and redundant functions during Drosophila development. Mol. Cell. Biol. 2008, 28, 5238–5250. [Google Scholar] [CrossRef] [Green Version]
- Su, H.A.; Bai, X.; Zeng, T.; Lu, Y.Y.; Qi, Y.X. Identification, characterization and expression analysis of transient receptor potential channel genes in the oriental fruit fly, Bactrocera dorsalis. BMC Genom. 2018, 19, 674. [Google Scholar] [CrossRef] [Green Version]
- Bain, S.A.; Marshall, H.; de la Filia, A.G.; Laetsch, D.R.; Husnik, F.; Ross, L. Sex-specific expression and DNA methylation in a species with extreme sexual dimorphism and paternal genome elimination. Mol. Ecol. 2021, 30, 5687–5703. [Google Scholar] [CrossRef]
- Whelan, S.; Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camargo, R.A.; Barbosa, G.O.; Possignolo, I.P.; Peres, L.E.P.; Lam, E.; Lima, J.E.; Figueira, A.; Marques-Souza, H. RNA interference as a gene silencing tool to control Tuta absoluta in tomato (Solanum lycopersicum). Peer J. 2016, 4, e2673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.D.; Lin, Z.K.; Ji, S.X.; Bi, S.Y.; Liu, W.X.; Zhang, G.F.; Wan, F.H.; Lü, Z.C. Molecular characterization of TRPA subfamily genes and function in temperature preference in Tuta absoluta (Meyrick) (Lepidoptera: Gelechiidae). Int. J. Mol. Sci. 2021, 22, 7157. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Primer Name | Primer Sequence (5′ to 3′) |
|---|---|---|
| Primers for cDNA cloning | ||
| BRM | BRM-F1 | AACCATCACGCTAACGC |
| BRM-R1 | GCTGGAACATGAAGGGAT | |
| BRM-F2 | ATCATCAAGTGCGACAT | |
| BRM-R2 | CCGACAGGACTCTACCG | |
| BRM-F3 | CGGAAGCCAACCTACT | |
| BRM-R3 | CCTGCCCTTTCACTAA | |
| ISWI | ISWI-F1 | GCTAATGTATTTGGATAATTT |
| ISWI-R1 | AGTATGGCGAGTTTCCC | |
| ISWI-F2 | CGTCTCAAATCTGAAGTAG | |
| ISWI-R2 | CCCAATGTCTTTCTGTAGT | |
| ISWI-F3 | CAGAAGTTGGAGAGCCTA | |
| ISWI-R3 | TCTACCGCTCATAACCG | |
| Primers for qPCR | ||
| BRM | BRM-QF | CAACGGAAAACTCAAGGAATACCA |
| BRM-QR | GCACCCAGTTTGATAGCGTACTG | |
| ISWI | ISWI-QF | GCTGATGAGATGGGTCTGGGT |
| ISWI-QR | CACACTGCTCTTAGGGAAGGACA | |
| RpL5 | RpL5-QF | CAGTCGTCGAGCCAGCAACA |
| RpL5-QR | TCCCGCATTGAAGGAGACCA | |
| Primers for dsRNA synthesis | ||
| BRM | BRM-DF | TAATACGACTCACTATAGGGGAACCATCACGCTAACG |
| BRM-DR | TAATACGACTCACTATAGGGGGATTCGTGGACCGTA | |
| ISWI | ISWI-DF | TAATACGACTCACTATAGGGTATGGATGTAGGGGACG |
| ISWI-DR | TAATACGACTCACTATAGGGCCAGTTGGTTAGGGTTG | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, S.-X.; Wu, Q.-W.; Bi, S.-Y.; Wang, X.-D.; Zhang, G.-F.; Wan, F.-H.; Lü, Z.-C.; Liu, W.-X. Chromatin-Remodelling ATPases ISWI and BRM Are Essential for Reproduction in the Destructive Pest Tuta absoluta. Int. J. Mol. Sci. 2022, 23, 3267. https://doi.org/10.3390/ijms23063267
Ji S-X, Wu Q-W, Bi S-Y, Wang X-D, Zhang G-F, Wan F-H, Lü Z-C, Liu W-X. Chromatin-Remodelling ATPases ISWI and BRM Are Essential for Reproduction in the Destructive Pest Tuta absoluta. International Journal of Molecular Sciences. 2022; 23(6):3267. https://doi.org/10.3390/ijms23063267
Chicago/Turabian StyleJi, Shun-Xia, Qiang-Wen Wu, Si-Yan Bi, Xiao-Di Wang, Gui-Fen Zhang, Fang-Hao Wan, Zhi-Chuang Lü, and Wan-Xue Liu. 2022. "Chromatin-Remodelling ATPases ISWI and BRM Are Essential for Reproduction in the Destructive Pest Tuta absoluta" International Journal of Molecular Sciences 23, no. 6: 3267. https://doi.org/10.3390/ijms23063267
APA StyleJi, S.-X., Wu, Q.-W., Bi, S.-Y., Wang, X.-D., Zhang, G.-F., Wan, F.-H., Lü, Z.-C., & Liu, W.-X. (2022). Chromatin-Remodelling ATPases ISWI and BRM Are Essential for Reproduction in the Destructive Pest Tuta absoluta. International Journal of Molecular Sciences, 23(6), 3267. https://doi.org/10.3390/ijms23063267






