Root-Zone Restriction Regulates Soil Factors and Bacterial Community Assembly of Grapevine
Abstract
1. Introduction
2. Results
2.1. Biochemical Dynamics of Root Zone Restriction Approach on Grapevine Fruit
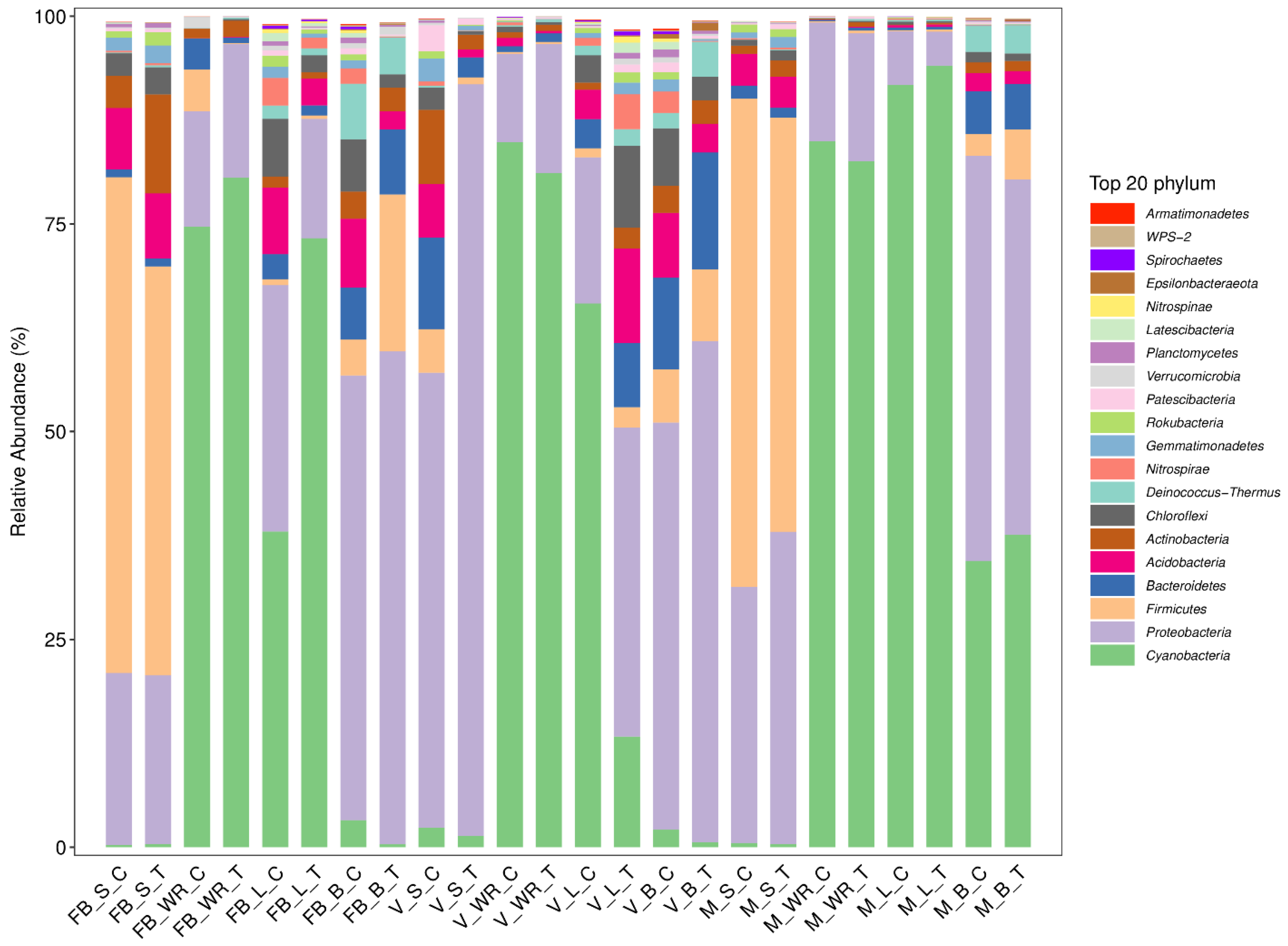
2.2. Bacterial Taxonomic Diversity throughout the Grapevine Influenced by Root Zone Cultivation
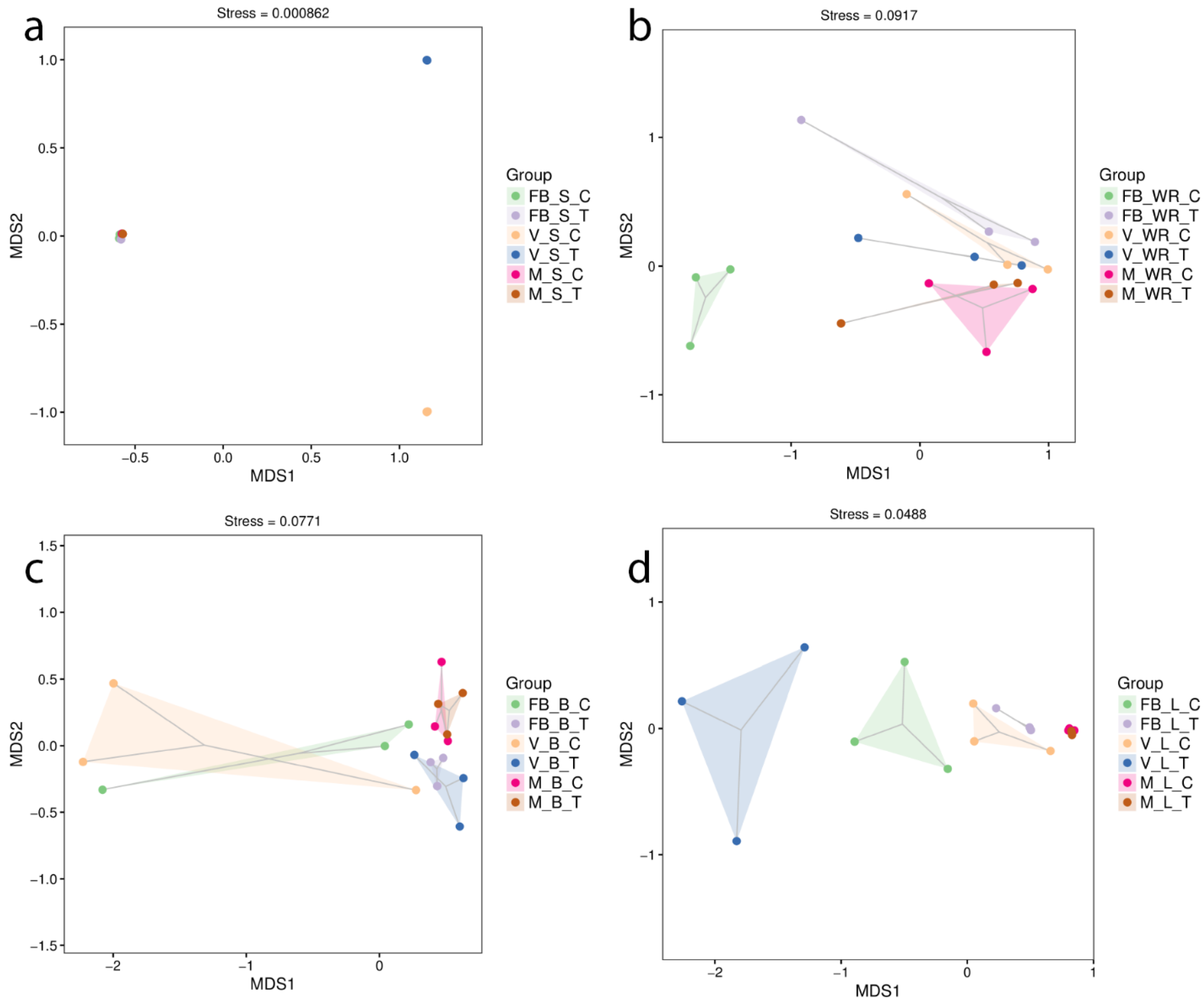
2.3. Impact on Bacterial Community Structure in Plant Organs across Three Growth Stages
2.4. Richness and Diversity of Bacterial Communities Associated with Grapevine Habitats
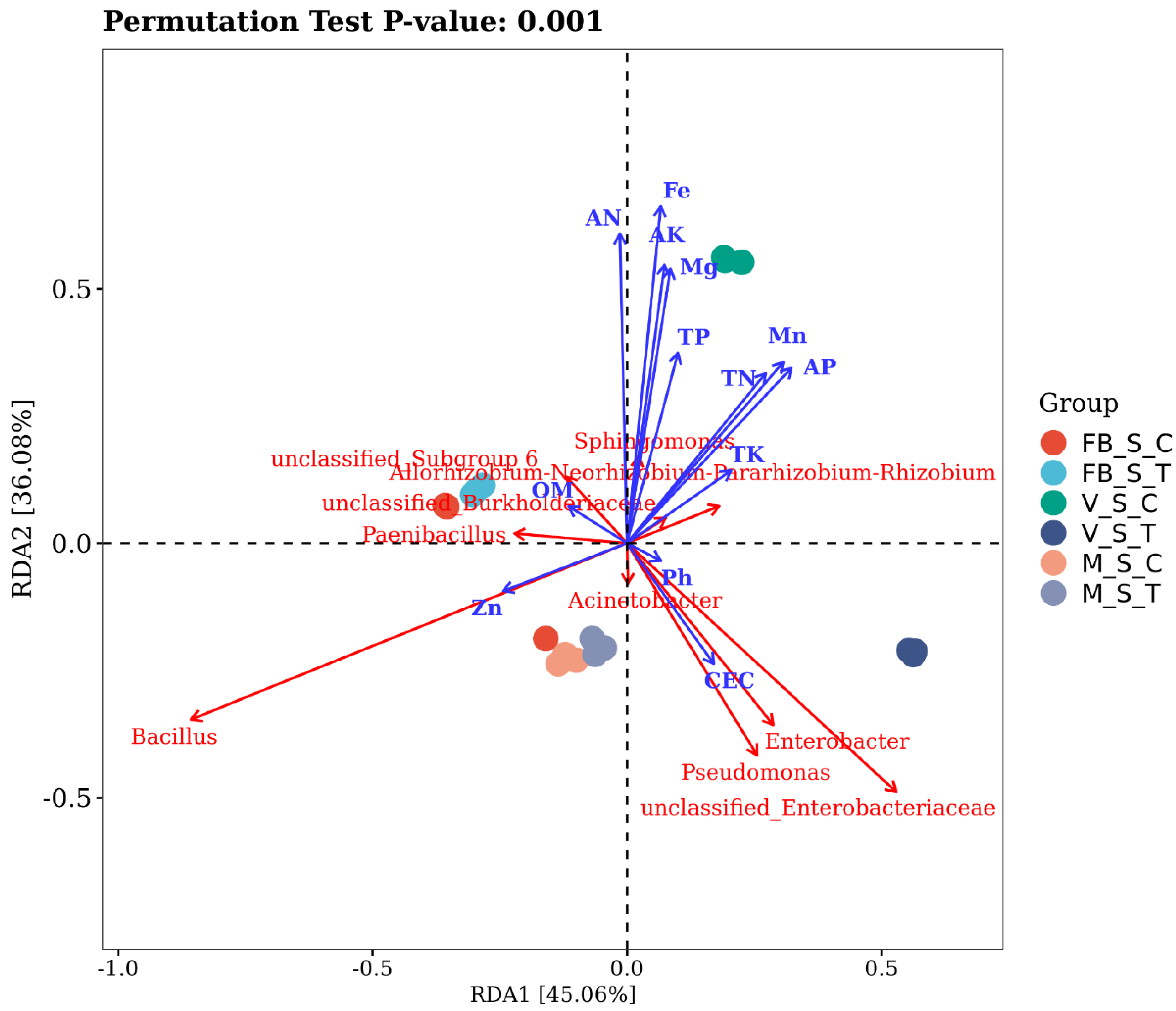
2.5. The Preponderant Influence of Soil Parameters on Bacterial Community Structuring
2.6. Species Difference Analysis and Notable Species
2.7. Metagenome Prediction
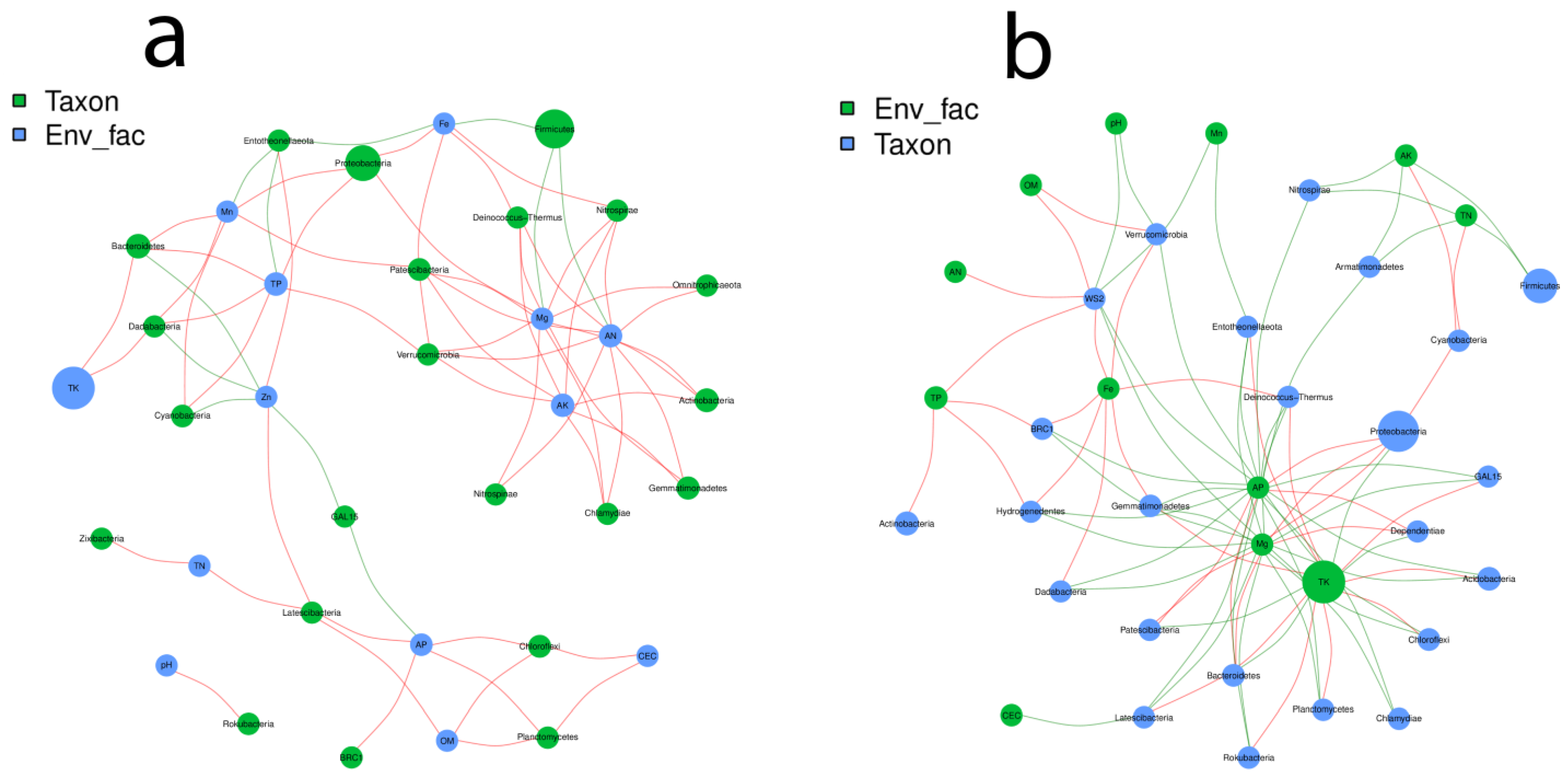
2.8. Microbial Co-Relation Network Analysis
3. Discussion
3.1. Root-Zone Restriction Influences the Bacterial Structuring across Successional Stages
3.2. Improved Fruit Quality
3.3. The Bacterial Population at Predisposal of Soil Elements in Root-Restricted Cultivation
3.4. Correlation between Soil and Bacterial Niches
3.5. Root Restriction Regulates the Bacterial Networking in the Grapevine Habitats
4. Materials and Methods
4.1. Plant Material and Sample Collection
4.2. Biochemical Characterization of Grapevine
4.3. Soil Physiochemical Characterization
4.4. Genomic Extraction, 16S rDNA Library Preparation and MiSeq Sequencing
4.5. Computational, Bioinformatics and Statistical Approaches for MiSeq Analysis
4.6. Diversity and Statistical Analysis
4.7. Prediction of Metabolic Functions
4.8. Bacterial Network Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alston, J.M.; Sambucci, O. Grapes in the World Economy. In The Grape Genome; Cantu, D., Walker, M.A., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 1–24. ISBN 978-3-030-18601-2. [Google Scholar]
- Wang, S.; Okamoto, G.; Hirano, K.; Lu, J.; Zhang, C. Effects of Restricted Rooting Volume on Vine Growth and Berry Development of Kyoho Grapevines. Am. J. Enol. Vitic. 2001, 52, 248–253. [Google Scholar] [CrossRef]
- Xie, Z.; Forney, C.; Xu, W.; Wang, S. Effects of Root Restriction on Ultrastructure of Phloem Tissues in Grape Berry. HortScience 2009, 44, 1334–1339. [Google Scholar] [CrossRef]
- Li, J.; Li, D.; Liu, B.; Wang, R.; Yan, Y.; Li, G.; Wang, L.; Ma, C.; Xu, W.; Zhao, L.; et al. Effects of Root Restriction on Phytohormone Levels in Different Growth Stages and Grapevine Organs. Sci. Rep. 2022, 12, 1323. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, C.; Wang, F.; Niu, R.; Zhu, W.; Wu, X. Root Restriction Effects of Nectarines Grown in a Non-Arable Land Greenhouse. Sci. Hortic. 2019, 250, 399–404. [Google Scholar] [CrossRef]
- Xie, Z.S.; Li, B.; Forney, C.F.; Xu, W.P.; Wang, S.P. Changes in Sugar Content and Relative Enzyme Activity in Grape Berry in Response to Root Restriction. Sci. Hortic. 2009, 123, 39–45. [Google Scholar] [CrossRef]
- Jiu, S.; Xu, Y.; Wang, J.; Haider, M.S.; Xu, J.; Wang, L.; Wang, S.; Li, J.; Liu, X.; Sun, W.; et al. Molecular Mechanisms Underlying the Action of Strigolactones Involved in Grapevine Root Development by Interacting with Other Phytohormone Signaling. Sci. Hortic. 2022, 293, 110709. [Google Scholar] [CrossRef]
- White, M.; Tustin, D.; Foote, K.; Campbell, J. GROWTH OF YOUNG SWEET CHERRY TREES IN RESPONSE TO ROOT RESTRICTION USING ROOT CONTROL BAGS. Acta Hortic. 2001, 557, 391–398. [Google Scholar] [CrossRef]
- Zakaria, N.I.; Ismail, M.R.; Awang, Y.; Megat Wahab, P.E.; Berahim, Z. Effect of Root Restriction on the Growth, Photosynthesis Rate, and Source and Sink Relationship of Chilli (Capsicum annuum L.) Grown in Soilless Culture. Biomed Res. Int. 2020, 2020, 2706937. [Google Scholar] [CrossRef]
- Hooijdonk, B.; Woolley, D.; Warrington, I.; Tustin, S. Architectural Development of “Royal Gala” Apple Scions in Response to Rootstock, Root Restriction, and Benzylaminopurine. Acta Hortic. 2006, 727, 561–567. [Google Scholar] [CrossRef]
- Ayarna, A.W.; Tsukagoshi, S.; Nkansah, G.O. Effect of Root Restriction on the Performance of Three-Truss Cultivated Tomato in the Low-Node Pinching Order at High-Density Cultivation System. Horticulturae 2021, 7, 60. [Google Scholar] [CrossRef]
- Wang, B.; He, J.; Duan, C.; Yu, X.; Zhu, L.; Xie, Z.; Zhang, C.; Xu, W.; Wang, S. Root Restriction Affects Anthocyanin Accumulation and Composition in Berry Skin of “Kyoho” Grape (Vitis vinifera L. × Vitis labrusca L.) during Ripening. Sci. Hortic. 2012, 137, 20–28. [Google Scholar] [CrossRef]
- Li, D.; Liu, B.; Wang, Z.; Li, X.; Sun, S.; Ma, C.; Wang, L.; Wang, S. Sugar Accumulation May Be Regulated by a Transcriptional Cascade of ABA-VvGRIP55-VvMYB15-VvSWEET15 in Grape Berries under Root Restriction. Plant Sci. 2022, 322, 111288. [Google Scholar] [CrossRef]
- Li, H.; Gao, Z.; Chen, Q.; Li, Q.; Luo, M.; Wang, J.; Hu, L.; Zahid, M.S.; Wang, L.; Zhao, L.; et al. Grapevine ABA Receptor VvPYL1 Regulates Root Hair Development in Transgenic Arabidopsis. Plant Physiol. Biochem. 2020, 149, 190–200. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, Q.; Liu, J.; Dou, F.; Wang, H.; Song, Y.; Ren, Y.; He, J.; Wang, L.; Zhang, C.; et al. Identification of Grape MiRNA Revealed Vvi-MiR164b Involved in Auxin Induced Root Development. Sci. Hortic. 2022, 295, 110804. [Google Scholar] [CrossRef]
- Zhu, L.; Wang, S.; Yang, T.; Zhang, C.; Xu, W. Vine Growth and Nitrogen Metabolism of “Fujiminori” Grapevines in Response to Root Restriction. Sci. Hortic. 2006, 107, 143–149. [Google Scholar] [CrossRef]
- Yu, X.; Wang, B.; Zhang, C.; Xu, W.; He, J.; Zhu, L.; Wang, S. Effect of Root Restriction on Nitrogen Levels and Glutamine Synthetase Activity in “Kyoho” Grapevines. Sci. Hortic. 2012, 137, 156–163. [Google Scholar] [CrossRef]
- Li, D.; Pang, Y.; Li, H.; Guo, D.; Wang, R.; Ma, C.; Xu, W.; Wang, L.; Wang, S. Comparative Analysis of the Gene Expression Profile under Two Cultivation Methods Reveals the Critical Role of ABA in Grape Quality Promotion. Sci. Hortic. 2021, 281, 109924. [Google Scholar] [CrossRef]
- Orozco-Mosqueda, M.D.C.; Santoyo, G. Plant-Microbial Endophytes Interactions: Scrutinizing Their Beneficial Mechanisms from Genomic Explorations. Curr. Plant Biol. 2021, 25, 100189. [Google Scholar] [CrossRef]
- Compant, S.; Clément, C.; Sessitsch, A. Plant Growth-Promoting Bacteria in the Rhizo- and Endosphere of Plants: Their Role, Colonization, Mechanisms Involved and Prospects for Utilization. Soil Biol. Biochem. 2010, 42, 669–678. [Google Scholar] [CrossRef]
- Lugtenberg, B.; Kamilova, F. Plant-Growth-Promoting Rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Cambon, M.C.; Vacher, C.; Mitter, B.; Samad, A.; Sessitsch, A. The Plant Endosphere World—Bacterial Life within Plants. Environ. Microbiol. 2021, 23, 1812–1829. [Google Scholar] [CrossRef]
- Garbeva, P.; van Veen, J.A.; van Elsas, J.D. MICROBIAL DIVERSITY IN SOIL: Selection of Microbial Populations by Plant and Soil Type and Implications for Disease Suppressiveness. Annu. Rev. Phytopathol. 2004, 42, 243–270. [Google Scholar] [CrossRef]
- Gómez-Aparicio, L.; Domínguez-Begines, J.; Villa-Sanabria, E.; García, L.V.; Muñoz-Pajares, A.J. Tree Decline and Mortality Following Pathogen Invasion Alters the Diversity, Composition and Network Structure of the Soil Microbiome. Soil Biol. Biochem. 2022, 166, 108560. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; van Themaat, E.V.L.; Schulze-Lefert, P. Structure and Functions of the Bacterial Microbiota of Plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef]
- Vorholt, J.A. Microbial Life in the Phyllosphere. Nat. Rev. Microbiol. 2012, 10, 828–840. [Google Scholar] [CrossRef]
- Reinhold-Hurek, B.; Bünger, W.; Burbano, C.S.; Sabale, M.; Hurek, T. Roots Shaping Their Microbiome: Global Hotspots for Microbial Activity. Annu. Rev. Phytopathol. 2015, 53, 403–424. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Ver Loren van Themaat, E.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing Structure and Assembly Cues for Arabidopsis Root-Inhabiting Bacterial Microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Vieira, S.; Sikorski, J.; Dietz, S.; Herz, K.; Schrumpf, M.; Bruelheide, H.; Scheel, D.; Friedrich, M.W.; Overmann, J. Drivers of the Composition of Active Rhizosphere Bacterial Communities in Temperate Grasslands. ISME J. 2020, 14, 463–475. [Google Scholar] [CrossRef]
- Bodenhausen, N.; Horton, M.W.; Bergelson, J. Bacterial Communities Associated with the Leaves and the Roots of Arabidopsis Thaliana. PLoS ONE 2013, 8, e56329. [Google Scholar] [CrossRef]
- Tkacz, A.; Cheema, J.; Chandra, G.; Grant, A.; Poole, P.S. Stability and Succession of the Rhizosphere Microbiota Depends upon Plant Type and Soil Composition. ISME J. 2015, 9, 2349–2359. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Badri, D.V.; Vivanco, J.M. Rhizosphere Microbiome Assemblage Is Affected by Plant Development. ISME J. 2014, 8, 790–803. [Google Scholar] [CrossRef] [PubMed]
- Vink, S.N.; Chrysargyris, A.; Tzortzakis, N.; Salles, J.F. Bacterial Community Dynamics Varies with Soil Management and Irrigation Practices in Grapevines (Vitis vinifera L.). Appl. Soil Ecol. 2021, 158, 103807. [Google Scholar] [CrossRef]
- Dombrowski, N.; Schlaeppi, K.; Agler, M.T.; Hacquard, S.; Kemen, E.; Garrido-Oter, R.; Wunder, J.; Coupland, G.; Schulze-Lefert, P. Root Microbiota Dynamics of Perennial Arabis Alpina Are Dependent on Soil Residence Time but Independent of Flowering Time. ISME J. 2017, 11, 43–55. [Google Scholar] [CrossRef]
- Heck, K.L.; van Belle, G.; Simberloff, D. Explicit Calculation of the Rarefaction Diversity Measurement and the Determination of Sufficient Sample Size. Ecology 1975, 56, 1459–1461. [Google Scholar] [CrossRef]
- Kemp, P.F.; Aller, J.Y. Bacterial Diversity in Aquatic and Other Environments: What 16S RDNA Libraries Can Tell Us. FEMS Microbiol. Ecol. 2004, 47, 161–177. [Google Scholar] [CrossRef]
- Chen, S.; Waghmode, T.R.; Sun, R.; Kuramae, E.E.; Hu, C.; Liu, B. Root-Associated Microbiomes of Wheat under the Combined Effect of Plant Development and Nitrogen Fertilization. Microbiome 2019, 7, 136. [Google Scholar] [CrossRef]
- Gobbi, A.; Acedo, A.; Imam, N.; Santini, R.G.; Ortiz-Álvarez, R.; Ellegaard-Jensen, L.; Belda, I.; Hansen, L.H. A Global Microbiome Survey of Vineyard Soils Highlights the Microbial Dimension of Viticultural Terroirs. Commun. Biol. 2022, 5, 241. [Google Scholar] [CrossRef]
- Akköprü, A.; Akat, Ş.; Özaktan, H.; Gül, A.; Akbaba, M. The Long-Term Colonization Dynamics of Endophytic Bacteria in Cucumber Plants, and Their Effects on Yield, Fruit Quality and Angular Leaf Spot Disease. Sci. Hortic. 2021, 282, 110005. [Google Scholar] [CrossRef]
- Rho, H.; Van Epps, V.; Kim, S.H.; Doty, S.L. Endophytes Increased Fruit Quality with Higher Soluble Sugar Production in Honeycrisp Apple (Malus pumila). Microorganisms 2020, 8, 699. [Google Scholar] [CrossRef] [PubMed]
- Vives-Peris, V.; de Ollas, C.; Gómez-Cadenas, A.; Pérez-Clemente, R.M. Root Exudates: From Plant to Rhizosphere and Beyond. Plant Cell Rep. 2020, 39, 3–17. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Badri, D.V.; Bakker, M.G.; Sugiyama, A.; Manter, D.K.; Vivanco, J.M. Root Exudation of Phytochemicals in Arabidopsis Follows Specific Patterns That Are Developmentally Programmed and Correlate with Soil Microbial Functions. PLoS ONE 2013, 8, e55731. [Google Scholar] [CrossRef]
- Sugiyama, A.; Ueda, Y.; Zushi, T.; Takase, H.; Yazaki, K. Changes in the Bacterial Community of Soybean Rhizospheres during Growth in the Field. PLoS ONE 2014, 9, e100709. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.A.; Van Der Lelie, D.; Zarraonaindia, I. Microbial Terroir for Wine Grapes. Proc. Natl. Acad. Sci. USA 2014, 111, 5–6. [Google Scholar] [CrossRef]
- Compant, S.; Kaplan, H.; Sessitsch, A.; Nowak, J.; Ait Barka, E.; Clément, C. Endophytic Colonization of Vitis vinifera L. by Burkholderia Phytofirmans Strain PsJN: From the Rhizosphere to Inflorescence Tissues. FEMS Microbiol. Ecol. 2008, 63, 84–93. [Google Scholar] [CrossRef]
- Van Overbeek, L.; Van Elsas, J.D. Effects of Plant Genotype and Growth Stage on the Structure of Bacterial Communities Associated with Potato (Solanum tuberosum L.). FEMS Microbiol. Ecol. 2008, 64, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The Hidden World within Plants: Ecological and Evolutionary Considerations for Defining Functioning of Microbial Endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef]
- Jenkins, S.N.; Waite, I.S.; Blackburn, A.; Husband, R.; Rushton, S.P.; Manning, D.C.; O’Donnell, A.G. Actinobacterial Community Dynamics in Long Term Managed Grasslands. Antonie Van Leeuwenhoek 2009, 95, 319–334. [Google Scholar] [CrossRef]
- Barata, A.; Malfeito-Ferreira, M.; Loureiro, V. The Microbial Ecology of Wine Grape Berries. Int. J. Food Microbiol. 2012, 153, 243–259. [Google Scholar] [CrossRef]
- Zarraonaindia, I.; Owens, S.M.; Weisenhorn, P.; West, K.; Hampton-Marcell, J.; Lax, S.; Bokulich, N.A.; Mills, D.A.; Martin, G.; Taghavi, S.; et al. The Soil Microbiome Influences Grapevine-Associated Microbiota. MBio 2015, 6, e02527-14. [Google Scholar] [CrossRef]
- Marasco, R.; Rolli, E.; Fusi, M.; Cherif, A.; Abou-Hadid, A.; El-Bahairy, U.; Borin, S.; Sorlini, C.; Daffonchio, D. Plant Growth Promotion Potential Is Equally Represented in Diverse Grapevine Root-Associated Bacterial Communities from Different Biopedoclimatic Environments. Biomed Res. Int. 2013, 2013, 491091. [Google Scholar] [CrossRef]
- Samad, A.; Trognitz, F.; Compant, S.; Antonielli, L.; Sessitsch, A. Shared and Host-Specific Microbiome Diversity and Functioning of Grapevine and Accompanying Weed Plants. Environ. Microbiol. 2017, 19, 1407–1424. [Google Scholar] [CrossRef] [PubMed]
- Burns, K.N.; Kluepfel, D.A.; Strauss, S.L.; Bokulich, N.A.; Cantu, D.; Steenwerth, K.L. Vineyard Soil Bacterial Diversity and Composition Revealed by 16S RRNA Genes: Differentiation by Geographic Features. Soil Biol. Biochem. 2015, 91, 232–247. [Google Scholar] [CrossRef]
- Compant, S.; Mitter, B.; Colli-Mull, J.G.; Gangl, H.; Sessitsch, A. Endophytes of Grapevine Flowers, Berries, and Seeds: Identification of Cultivable Bacteria, Comparison with Other Plant Parts, and Visualization of Niches of Colonization. Microb. Ecol. 2011, 62, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Lareen, A.; Burton, F.; Schäfer, P. Plant Root-Microbe Communication in Shaping Root Microbiomes. Plant Mol. Biol. 2016, 90, 575–587. [Google Scholar] [CrossRef] [PubMed]
- Fahrbach, M.; Kuever, J.; Remesch, M.; Huber, B.E.; Kämpfer, P.; Dott, W.; Hollender, J. Steroidobacter Denitrificans Gen. Nov., Sp. Nov., a Steroidal Hormone-Degrading Gammaproteobacterium. Int. J. Syst. Evol. Microbiol. 2008, 58, 2215–2223. [Google Scholar] [CrossRef]
- Holguin, J.; Collins, S.L.; McLaren, J.R. Belowground Responses to Altered Precipitation Regimes in Two Semi-Arid Grasslands. Soil Biol. Biochem. 2022, 171, 108725. [Google Scholar] [CrossRef]
- Compant, S.; Van Der Heijden, M.G.A.; Sessitsch, A. Climate Change Effects on Beneficial Plant-Microorganism Interactions. FEMS Microbiol. Ecol. 2010, 73, 197–214. [Google Scholar] [CrossRef]
- Li, H.; Gao, Z.; Zahid, M.S.; Li, D.; Javed, H.U.; Wang, L.; Song, S.; Zhao, L.; Xu, W.; Zhang, C.; et al. Small RNA Sequencing Analysis of MiRNA Expression Reveals Novel Insihts into Root Formation under Root Restriction Cultivation in Grapevine (Vitis vinifera L.). Int. J. Mol. Sci. 2020, 21, 3513. [Google Scholar] [CrossRef]
- Bever, J.D.; Platt, T.G.; Morton, E.R. Microbial Population and Community Dynamics on Plant Roots and Their Feedbacks on Plant Communities. Annu. Rev. Microbiol. 2012, 66, 265–283. [Google Scholar] [CrossRef]
- Pérez-Jaramillo, J.E.; Mendes, R.; Raaijmakers, J.M. Impact of Plant Domestication on Rhizosphere Microbiome Assembly and Functions. Plant Mol. Biol. 2016, 90, 635–644. [Google Scholar] [CrossRef]
- Grayston, S.J.; Wang, S.; Campbell, C.D.; Edwards, A.C. Selective Influence of Plant Species on Microbial Diversity in the Rhizosphere. Soil Biol. Biochem. 1998, 30, 369–378. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The Rhizosphere Microbiome and Plant Health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Gantar, M.; Kerby, N.W.; Rowell, P.; Obreht, Z.; Scrimgeour, C. Colonization of Wheat (Triticum vulgare L.) by Ng-Fixing Cyanobacteria. New Phytol. 1991, 118, 485–492. [Google Scholar] [CrossRef]
- Singh, P.; Santoni, S.; Weber, A.; This, P.; Péros, J.P. Understanding the Phyllosphere Microbiome Assemblage in Grape Species (Vitaceae) with Amplicon Sequence Data Structures. Sci. Rep. 2019, 9, 14294. [Google Scholar] [CrossRef] [PubMed]
- Ren, F.; Dong, W.; Yan, D.H. Endophytic Bacterial Communities of Jingbai Pear Trees in North China Analyzed with Illumina Sequencing of 16S RDNA. Arch. Microbiol. 2019, 201, 199–208. [Google Scholar] [CrossRef]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; del Rio, T.G.; et al. Defining the Core Arabidopsis thaliana Root Microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef]
- Weinert, N.; Piceno, Y.; Ding, G.C.; Meincke, R.; Heuer, H.; Berg, G.; Schloter, M.; Andersen, G.; Smalla, K. PhyloChip Hybridization Uncovered an Enormous Bacterial Diversity in the Rhizosphere of Different Potato Cultivars: Many Common and Few Cultivar-Dependent Taxa. FEMS Microbiol. Ecol. 2011, 75, 497–506. [Google Scholar] [CrossRef]
- Zhong, Y.; Yan, W.; Shangguan, Z. Impact of Long-Term N Additions upon Coupling between Soil Microbial Community Structure and Activity, and Nutrient-Use Efficiencies. Soil Biol. Biochem. 2015, 91, 151–159. [Google Scholar] [CrossRef]
- Vepštaitė-Monstavičė, I.; Lukša, J.; Stanevičienė, R.; Strazdaitė-Žielienė, Ž.; Yurchenko, V.; Serva, S.; Servienė, E. Distribution of Apple and Blackcurrant Microbiota in Lithuania and the Czech Republic. Microbiol. Res. 2018, 206, 1–8. [Google Scholar] [CrossRef]
- Enya, J.; Shinohara, H.; Yoshida, S.; Tsukiboshi, T.; Negishi, H.; Suyama, K.; Tsushima, S. Culturable Leaf-Associated Bacteria on Tomato Plants and Their Potential as Biological Control Agents. Microb. Ecol. 2007, 53, 524–536. [Google Scholar] [CrossRef]
- Stefanini, I.; Cavalieri, D. Metagenomic Approaches to Investigate the Contribution of the Vineyard Environment to the Quality of Wine Fermentation: Potentials and Difficulties. Front. Microbiol. 2018, 9, 991. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, E.; Zilber-Rosenberg, I. Microbes Drive Evolution of Animals and Plants: The Hologenome Concept. MBio 2016, 7, e01395-15. [Google Scholar] [CrossRef] [PubMed]
- Layeghifard, M.; Hwang, D.M.; Guttman, D.S. Disentangling Interactions in the Microbiome: A Network Perspective. Trends Microbiol. 2017, 25, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Sgroy, V.; Cassán, F.; Masciarelli, O.; Del Papa, M.F.; Lagares, A.; Luna, V. Isolation and Characterization of Endophytic Plant Growth-Promoting (PGPB) or Stress Homeostasis-Regulating (PSHB) Bacteria Associated to the Halophyte Prosopis Strombulifera. Appl. Microbiol. Biotechnol. 2009, 85, 371–381. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Kucharova, Z.; Davranov, K.; Berg, G.; Makarova, N.; Azarova, T.; Chebotar, V.; Tikhonovich, I.; Kamilova, F.; Validov, S.Z.; et al. Bacteria Able to Control Foot and Root Rot and to Promote Growth of Cucumber in Salinated Soils. Biol. Fertil. Soils 2011, 47, 197–205. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Wirth, S.; Jabborova, D.; Räsänen, L.A.; Liao, H. Coordination between Bradyrhizobium and Pseudomonas Alleviates Salt Stress in Soybean through Altering Root System Architecture. J. Plant Interact. 2017, 12, 100–107. [Google Scholar] [CrossRef]
- Upadhyay, S.K.; Singh, J.S.; Saxena, A.K.; Singh, D.P. Impact of PGPR Inoculation on Growth and Antioxidant Status of Wheat under Saline Conditions. Plant Biol. 2012, 14, 605–611. [Google Scholar] [CrossRef]
- Salomon, M.V.; Bottini, R.; de Souza Filho, G.A.; Cohen, A.C.; Moreno, D.; Gil, M.; Piccoli, P. Bacteria Isolated from Roots and Rhizosphere of Vitis vinifera Retard Water Losses, Induce Abscisic Acid Accumulation and Synthesis of Defense-Related Terpenes in In Vitro Cultured Grapevine. Physiol. Plant. 2014, 151, 359–374. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, P.; Chen, D.; Howell, K. From the Vineyard to the Winery: How Microbial Ecology Drives Regional Distinctiveness of Wine. Front. Microbiol. 2019, 10, 2679. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Collins, T.; Masarweh, C.; Allen, G.; Heymann, H.; Ebeler, S.E.; Mills, D.A. Fermentation Behavior Suggest Microbial Contribution to Regional. MBio 2016, 7, e00631-16. [Google Scholar] [CrossRef]
- Knight, S.; Goddard, M.R. Quantifying Separation and Similarity in a Saccharomyces Cerevisiae Metapopulation. ISME J. 2015, 9, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Zahid, M.S.; Li, D.; Javed, H.U.; Sabir, I.A.; Wang, L.; Jiu, S.; Song, S.; Ma, C.; Wang, D.; Zhang, C.; et al. Comparative Fungal Diversity and Dynamics in Plant Compartments at Different Developmental Stages under Root-Zone Restricted Grapevines. BMC Microbiol. 2021, 21, 317. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Jackson, R.B. The Diversity and Biogeography of Soil Bacterial Communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Griggs, R.G.; Steenwerth, K.L.; Mills, D.A.; Cantu, D.; Bokulich, N.A. Sources and Assembly of Microbial Communities in Vineyards as a Functional Component of Winegrowing. Front. Microbiol. 2021, 12, 836. [Google Scholar] [CrossRef]
- Fierer, N. Microbial Diversity Across Space and Time. In Accessing Uncultivated Microorganisms; Wiley: Hoboken, NJ, USA, 2008; pp. 95–116. ISBN 9781683671466. [Google Scholar] [CrossRef]
- Holland, T.C.; Bowen, P.A.; Bogdanoff, C.P.; Lowery, T.D.; Shaposhnikova, O.; Smith, S.; Hart, M.M. Evaluating the Diversity of Soil Microbial Communities in Vineyards Relative to Adjacent Native Ecosystems. Appl. Soil Ecol. 2016, 100, 91–103. [Google Scholar] [CrossRef]
- Li, J.G.; Shen, M.C.; Hou, J.F.; Li, L.; Wu, J.X.; Dong, Y.H. Effect of Different Levels of Nitrogen on Rhizosphere Bacterial Community Structure in Intensive Monoculture of Greenhouse Lettuce. Sci. Rep. 2016, 6, 25305. [Google Scholar] [CrossRef] [PubMed]
- de Melo, L.H.V.; Rocha, F.Y.O.; Vidal, M.S.; Gitahy, P.D.M.; Arruda, G.M.; Barreto, C.P.; Alves, P.B.; Ramos, E.T.D.A.; Rossi, C.N.; Schwab, S.; et al. Diversity and Biotechnological Potential of Endophytic Bacillus Species Originating from the Stem Apoplast Fluid of Sugarcane Plants. Appl. Soil Ecol. 2021, 166, 103985. [Google Scholar] [CrossRef]
- White, J.F.; Kingsley, K.L.; Zhang, Q.; Verma, R.; Obi, N.; Dvinskikh, S.; Elmore, M.T.; Verma, S.K.; Gond, S.K.; Kowalski, K.P. Review: Endophytic Microbes and Their Potential Applications in Crop Management. Pest Manag. Sci. 2019, 75, 2558–2565. [Google Scholar] [CrossRef]
- Johnstone, T.C.; Nolan, E.M. Beyond Iron: Non-Classical Biological Functions of Bacterial Siderophores. Dalt. Trans. 2015, 44, 6320–6339. [Google Scholar] [CrossRef]
- Xia, Q.; Rufty, T.; Shi, W. Soil Microbial Diversity and Composition: Links to Soil Texture and Associated Properties. Soil Biol. Biochem. 2020, 149, 107953. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhang, Y.; Zhang, F. Abundant and Rare Bacteria Possess Different Diversity and Function in Crop Monoculture and Rotation Systems across Regional Farmland. Soil Biol. Biochem. 2022, 171, 108742. [Google Scholar] [CrossRef]
- Qi, D.; Wieneke, X.; Tao, J.; Zhou, X.; Desilva, U. Soil PH Is the Primary Factor Correlating with Soil Microbiome in Karst Rocky Desertification Regions in the Wushan County, Chongqing, China. Front. Microbiol. 2018, 9, 1027. [Google Scholar] [CrossRef]
- Meena, V.S.; Maurya, B.R.; Verma, J.P.; Meena, R.S. Potassium Solubilizing Microorganisms for Sustainable Agriculture. In Potassium Solubilizing Microorganisms for Sustainable Agriculture; Springer: Cham, Switzerland, 2016; pp. 1–331. [Google Scholar] [CrossRef]
- Leaungvutiviroj, C.; Ruangphisarn, P.; Hansanimitkul, P.; Shinkawa, H.; Sasaki, K. Development of a New Biofertilizer with a High Capacity for N2 Fixation, Phosphate and Potassium Solubilization and Auxin Production. Biosci. Biotechnol. Biochem. 2010, 74, 1098–1101. [Google Scholar] [CrossRef]
- Sheng, X.F.; Zhao, F.; He, L.Y.; Qiu, G.; Chen, L. Isolation and Characterization of Silicate Mineral-Solubilizing Bacillus Globisporus Q12 from the Surfaces of Weathered Feldspar. Can. J. Microbiol. 2008, 54, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Hameeda, B.; Harini, G.; Rupela, O.P.; Wani, S.P.; Reddy, G. Growth Promotion of Maize by Phosphate-Solubilizing Bacteria Isolated from Composts and Macrofauna. Microbiol. Res. 2008, 163, 234–242. [Google Scholar] [CrossRef]
- Chung, H.; Park, M.; Madhaiyan, M.; Seshadri, S.; Song, J.; Cho, H.; Sa, T. Isolation and Characterization of Phosphate Solubilizing Bacteria from the Rhizosphere of Crop Plants of Korea. Soil Biol. Biochem. 2005, 37, 1970–1974. [Google Scholar] [CrossRef]
- Wan, W.; Qin, Y.; Wu, H.; Zuo, W.; He, H.; Tan, J.; Wang, Y.; He, D. Isolation and Characterization of Phosphorus Solubilizing Bacteria With Multiple Phosphorus Sources Utilizing Capability and Their Potential for Lead Immobilization in Soil. Front. Microbiol. 2020, 11, 752. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Tang, Y.; Hu, C.; Zhan, T.; Zhang, S.; Cai, M.; Zhao, X. Soil Applied Ca, Mg and B Altered Phyllosphere and Rhizosphere Bacterial Microbiome and Reduced Huanglongbing Incidence in Gannan Navel Orange. Sci. Total Environ. 2021, 791, 148046. [Google Scholar] [CrossRef]
- Mehetre, G.; Shah, M.; Dastager, S.G.; Dharne, M.S. Untapped Bacterial Diversity and Metabolic Potential within Unkeshwar Hot Springs, India. Arch. Microbiol. 2018, 200, 753–770. [Google Scholar] [CrossRef]
- Kochetkova, T.V.; Toshchakov, S.V.; Zayulina, K.S.; Elcheninov, A.G.; Zavarzina, D.G.; Lavrushin, V.Y.; Bonch-Osmolovskaya, E.A.; Kublanov, I. V Hot in Cold: Microbial Life in the Hottest Springs in Permafrost. Microorganisms 2020, 8, 1308. [Google Scholar] [CrossRef]
- Khan, A.G. Role of Soil Microbes in the Rhizospheres of Plants Growing on Trace Metal Contaminated Soils in Phytoremediation. J. Trace Elem. Med. Biol. 2005, 18, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Prasanna, R.; Bidyarani, N.; Babu, S.; Hossain, F.; Shivay, Y.S.; Nain, L. Cyanobacterial Inoculation Elicits Plant Defense Response and Enhanced Zn Mobilization in Maize Hybrids. Cogent Food Agric. 2015, 1, 998507. [Google Scholar] [CrossRef]
- Obreht, Z.; Kerby, N.W.; Gantar, M.; Rowell, P. Effects of Root-Associated N2-Fixing Cyanobacteria on the Growth and Nitrogen Content of Wheat (Triticum vulgare L.) Seedlings. Biol. Fertil. Soils 1993, 15, 68–72. [Google Scholar] [CrossRef]
- Edwards, J.; Johnson, C.; Santos-medellín, C.; Lurie, E.; Kumar, N. Structure, Variation, and Assembly of the Root-Associated Microbiomes of Rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, K.S.; Geisen, S.; Morriën, E.; Snoek, B.L.; van der Putten, W.H. Network Analyses Can Advance Above-Belowground Ecology. Trends Plant Sci. 2018, 23, 759–768. [Google Scholar] [CrossRef]
- Morriën, E.; Hannula, S.E.; Snoek, L.B.; Helmsing, N.R.; Zweers, H.; de Hollander, M.; Soto, R.L.; Bouffaud, M.-L.; Buée, M.; Dimmers, W.; et al. Soil Networks Become More Connected and Take up More Carbon as Nature Restoration Progresses. Nat. Commun. 2017, 8, 14349. [Google Scholar] [CrossRef]
- Banerjee, S.; Walder, F.; Büchi, L.; Meyer, M.; Held, A.Y.; Gattinger, A.; Keller, T.; Charles, R.; van der Heijden, M.G.A. Agricultural Intensification Reduces Microbial Network Complexity and the Abundance of Keystone Taxa in Roots. ISME J. 2019, 13, 1722–1736. [Google Scholar] [CrossRef]
- Banerjee, S.; Schlaeppi, K.; van der Heijden, M.G.A. Keystone Taxa as Drivers of Microbiome Structure and Functioning. Nat. Rev. Microbiol. 2018, 16, 567–576. [Google Scholar] [CrossRef]
- Ma, L.; Zhang, J.; Li, Z.; Xin, X.; Guo, Z.; Wang, D.; Li, D. Long-Term Phosphorus Deficiency Decreased Bacterial-Fungal Network Complexity and Efficiency across Three Soil Types in China as Revealed by Network Analysis. Appl. Soil Ecol. 2020, 148, 103506. [Google Scholar] [CrossRef]
- Zhao, D.; Shen, F.; Zeng, J.; Huang, R.; Yu, Z.; Wu, Q.L. Science of the Total Environment Network Analysis Reveals Seasonal Variation of Co-Occurrence Correlations between Cyanobacteria and Other Bacterioplankton. Sci. Total Environ. 2016, 573, 817–825. [Google Scholar] [CrossRef]
- Fierer, N. Embracing the Unknown: Disentangling the Complexities of the Soil Microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Meng, D.; Qin, C.; Liu, X.; Liang, Y.; Xiao, Y.; Liu, Z. Integrated Network Analysis Reveals the Importance of Microbial Interactions for Maize Growth. Appl. Microbiol. Biotechnol. 2018, 102, 3805–3818. [Google Scholar] [CrossRef] [PubMed]
- Lurie, S.; Ovadia, R.; Nissim-Levi, A.; Oren-Shamir, M.; Kaplunov, T.; Zutahy, Y.; Weksler, H.; Lichter, A. Abscisic Acid Improves Colour Development in “Crimson Seedless” Grapes in the Vineyard and on Detached Berries. J. Hortic. Sci. Biotechnol. 2009, 84, 639–644. [Google Scholar] [CrossRef]
- Ruck, J.A. Chemical Methods for Analysis of Fruit and Vegetable Products. In Chemical Methods for Analysis of Fruit and Vegetable Products; Canada Department of Agriculture: Ottawa, ON, Canada, 1969; p. 1154. [Google Scholar] [CrossRef]
- Chapman, H.D. Cation-exchange Capacity. Methods Soil Anal. Part 2 Chem. Microbiol. Prop. 1965, 9, 891–901. [Google Scholar]
- Estefan, G.; Sommer, R.; Ryan, J. Methods of Soil, Plant, and Water Analysis. A Man. West Asia North Africa Reg. 2013, 3, 65–119. [Google Scholar]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet 2011, 17, 10. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Kazutaka, K.; Misakwa, K.; Kei-ichi, K.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing Taxonomic Classification of Marker-Gene Amplicon Sequences with QIIME 2’s Q2-Feature-Classifier Plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; Desantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An Improved Greengenes Taxonomy with Explicit Ranks for Ecological and Evolutionary Analyses of Bacteria and Archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Bray, J.R.; Curtis, J.T. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 1957, 27, 326–349. [Google Scholar] [CrossRef]
- Legendre, P.; Legendre, L. Numerical Ecology, 3rd ed.; Elsevier: Amsterdam, the Netherlands, 2012; p. 1006. ISBN 0444538690. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community Ecology Package. 2019. R Package Vegan, Vers. 2.2-1. World Agrocentre 2015, 3, 7–81. [Google Scholar]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic Biomarker Discovery and Explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for Prediction of Metagenome Functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Deng, Y.; Jiang, Y.H.; Yang, Y.; He, Z.; Luo, F.; Zhou, J. Molecular Ecological Network Analyses. BMC Bioinform. 2012, 13, 113. [Google Scholar] [CrossRef]
- Guimerà, R.; Nunes Amaral, L.A. Functional Cartography of Complex Metabolic Networks—Supplementary Material. Nature 2005, 433, 895–900. [Google Scholar] [CrossRef]


| Pre-Veraison | Veraison | Maturity | ||||
|---|---|---|---|---|---|---|
| Parameters | Control | RR | Control | RR | Control | RR |
| TA | 4.81 ± 0.4 b | 5.63 ± 0.3 b | 2.73 ± 0.2 a | 1.95 ± 0.09 b | 1.17 ± 0.05 a | 0.79 ± 0.01 b |
| Anthocyanin | 0.09 ± 0.08 a | 0.12 ± 0.01 a | 3.43 ± 0.2 a | 3.51 ± 0.1 a | 99.31 ± 8.2 b | 117.21 ± 11.7 a |
| pH | 2.73 ± 0.1 a | 2.48 ± 0.2 b | 3.13 ± 0.01 b | 3.37 ± 0.05 a | 3.81 ± 0.07 b | 4.16 ± 0.03 a |
| Vitamin C | 1.42 ± 0.41 a | 1.47 ± 0.23 a | 1.77 ± 0.30 a | 1.80 ± 0.32 a | 2.02 ± 0.1 b | 2.21 ± 0.39 ab |
| Full Bloom | Veraison | Maturity | ||||
|---|---|---|---|---|---|---|
| Control | Treatment | Control | Treatment | Control | Treatment | |
| Available N | 859.18 ± 55.89 b | 187.28 ± 9.78 d | 1153.49 ± 46.02 a | 171.29 ± 1.44 de | 257.84 ± 2.12 c | 130.40 ± 1.44 e |
| Total N | 3000 ± 0.01 ab | 2800 ± 0.02 b | 3000 ± 0.01 ab | 3110 ± 0.01 a | 2200 ± 0.01 c | 2100 ± 0.02 c |
| Available P | 402.16 ± 2.25 a | 220 ± 1.00 e | 397.06 ± 1.45 b | 352.5 ± 3.77 c | 214.16 ± 2.25 f | 271.33 ± 1.53 d |
| Total P | 611.2 ± 1.08 e | 1660.38 ± 142.99 a | 1369.8 ± 153.45 b | 1124.87 ± 23.07 c | 938.9 ± 7.98 d | 952.63 ± 7.46 d |
| Available K | 1063.38 ± 44.25 b | 433.23 ± 7.04 d | 1129.39 ± 17.28 a | 563.15 ± 9.17 c | 304.24 ± 2.12 e | 334.37 ± 2.03 e |
| Total K | 8290.13 ± 400.4 c | 17,308.16 ± 1371.81 a | 17,047.66 ± 1520.11 a | 14,850.33 ± 903.31 b | 16,221.16 ± 1511.78 ab | 15,971.5 ± 584.52 ab |
| Fe | 55.91 ± 2.16 b | 54.35 ± 1.57 bc | 68.70 ± 3.01 a | 51.37 ± 1.05 c | 55.58 ± 3.86 b | 50.62 ± 0.75 c |
| Mn | 18.08 ± 0.76 d | 18.57 ± 0.62 d | 34.06 ± 0.51 a | 22.06 ± 1.70 c | 25.96 ± 1.42 b | 27.83 ± 1.05 b |
| Zn | 18.83 ± 0.86 a | 13.26 ± 0.50 b | 8.40 ± 0.44 c | 12.77 ± 1.13 b | 8.67 ± 1.21 c | 8.71 ± 0.99 c |
| Mg | 384.79 ± 7.52 b | 152.03 ± 3.76 e | 451.32 ± 4.10 a | 214.26 ± 7.59 c | 194.86 ± 4.33 d | 198.89 ± 3.41 d |
| CEC | 14.00 ± 0.50 c | 15.73 ± 0.56 ab | 11.9 ± 0.2 d | 16.47 ± 0.54 a | 10.70 ± 0.37 e | 15.43 ± 0.56 b |
| OM | 3.93 ± 0.15 a | 4 ± 0.1 a | 3.53 ± 0.15 bc | 3.80 ± 0.26 ab | 3.23 ± 0.15 c | 3.58 ± 0.19 b |
| pH | 6.36 ± 0.45 a | 6.10 ± 0.06 a | 6.45 ± 0.45 a | 6.38 ± 0.30 a | 6.45 ± 0.23 a | 6.67 ± 0.25 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zahid, M.S.; Hussain, M.; Song, Y.; Li, J.; Guo, D.; Li, X.; Song, S.; Wang, L.; Xu, W.; Wang, S. Root-Zone Restriction Regulates Soil Factors and Bacterial Community Assembly of Grapevine. Int. J. Mol. Sci. 2022, 23, 15628. https://doi.org/10.3390/ijms232415628
Zahid MS, Hussain M, Song Y, Li J, Guo D, Li X, Song S, Wang L, Xu W, Wang S. Root-Zone Restriction Regulates Soil Factors and Bacterial Community Assembly of Grapevine. International Journal of Molecular Sciences. 2022; 23(24):15628. https://doi.org/10.3390/ijms232415628
Chicago/Turabian StyleZahid, Muhammad Salman, Muzammil Hussain, Yue Song, Jiajia Li, Dinghan Guo, Xiangyi Li, Shiren Song, Lei Wang, Wenping Xu, and Shiping Wang. 2022. "Root-Zone Restriction Regulates Soil Factors and Bacterial Community Assembly of Grapevine" International Journal of Molecular Sciences 23, no. 24: 15628. https://doi.org/10.3390/ijms232415628
APA StyleZahid, M. S., Hussain, M., Song, Y., Li, J., Guo, D., Li, X., Song, S., Wang, L., Xu, W., & Wang, S. (2022). Root-Zone Restriction Regulates Soil Factors and Bacterial Community Assembly of Grapevine. International Journal of Molecular Sciences, 23(24), 15628. https://doi.org/10.3390/ijms232415628






