Lactobacilli Downregulate Transcription Factors in Helicobacter pylori That Affect Motility, Acid Tolerance and Antimicrobial Peptide Survival
Abstract
1. Introduction
2. Results
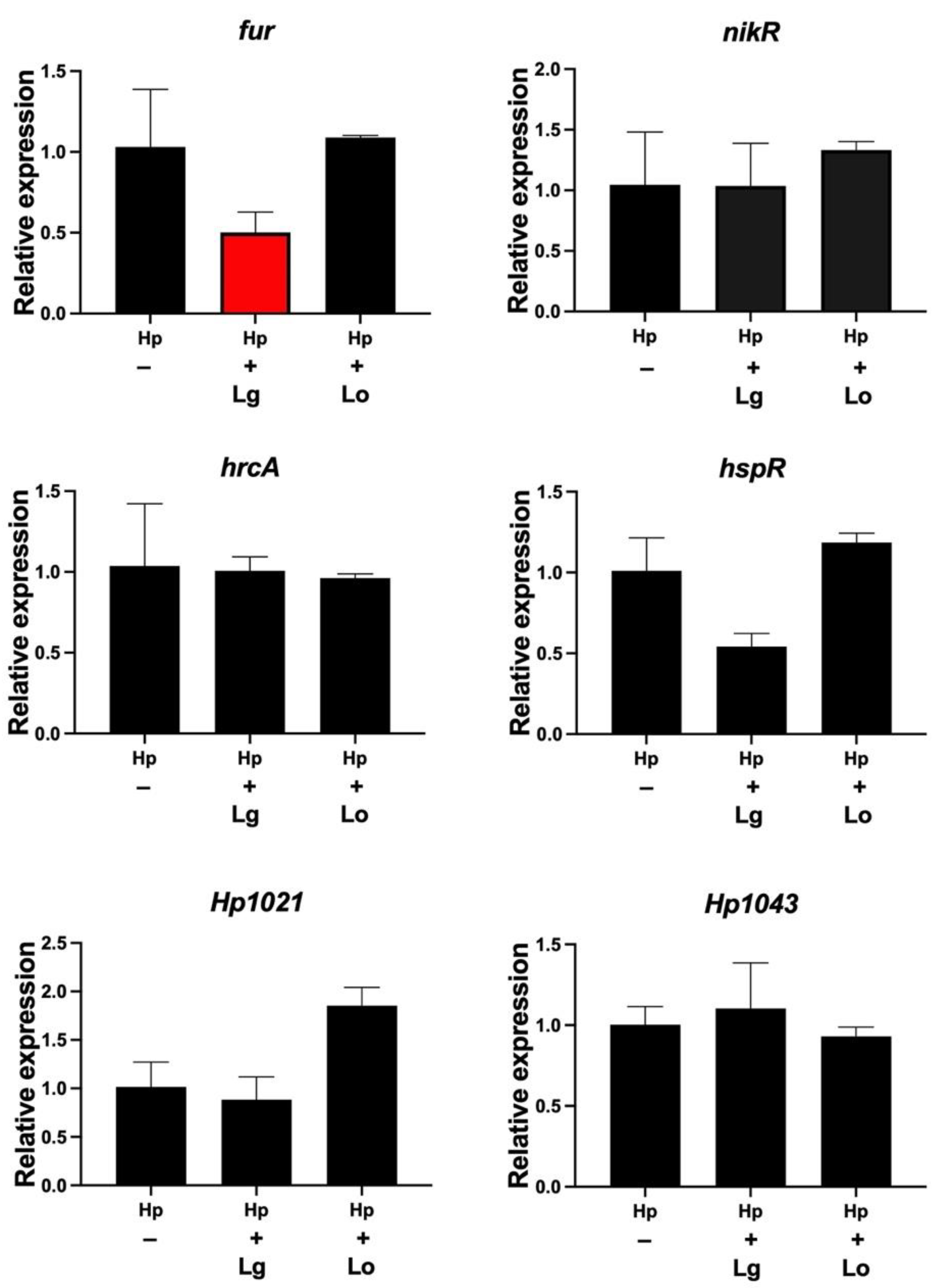
2.1. Lactobacilli Affect the Gene Expression of flgR and arsS in H. pylori
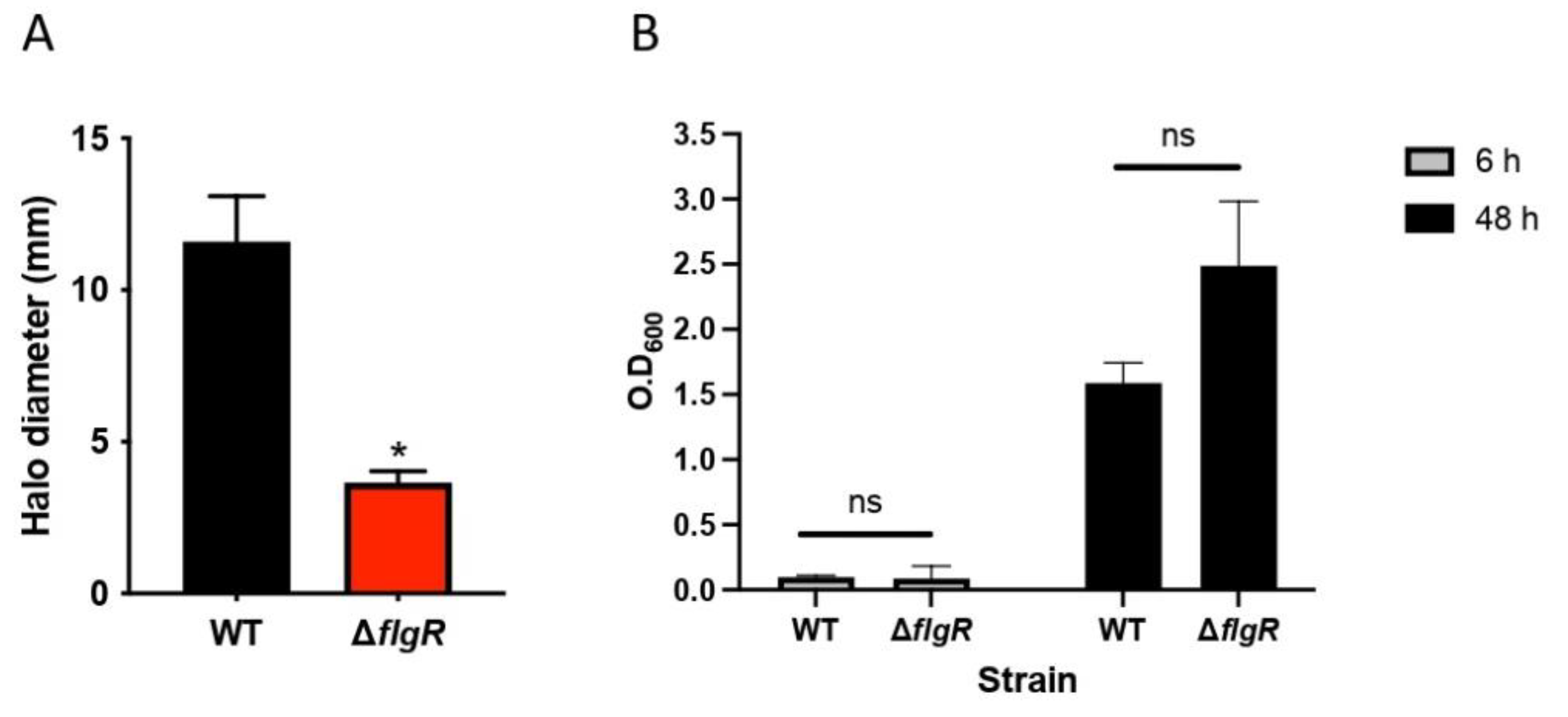
2.2. Lactobacilli Affect H. pylori Motility in a Strain-Specific Manner
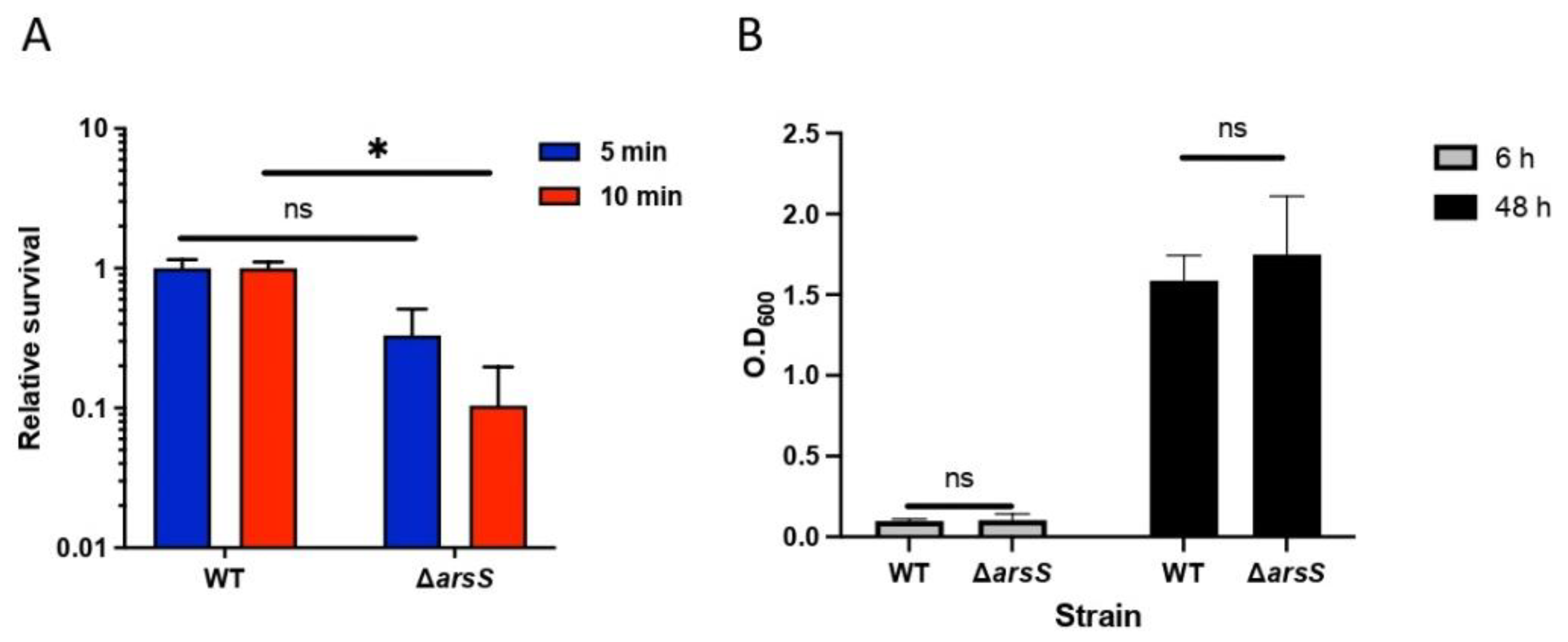
2.3. The ArsRS System Influences the Acid Sensitivity of H. pylori at pH 2
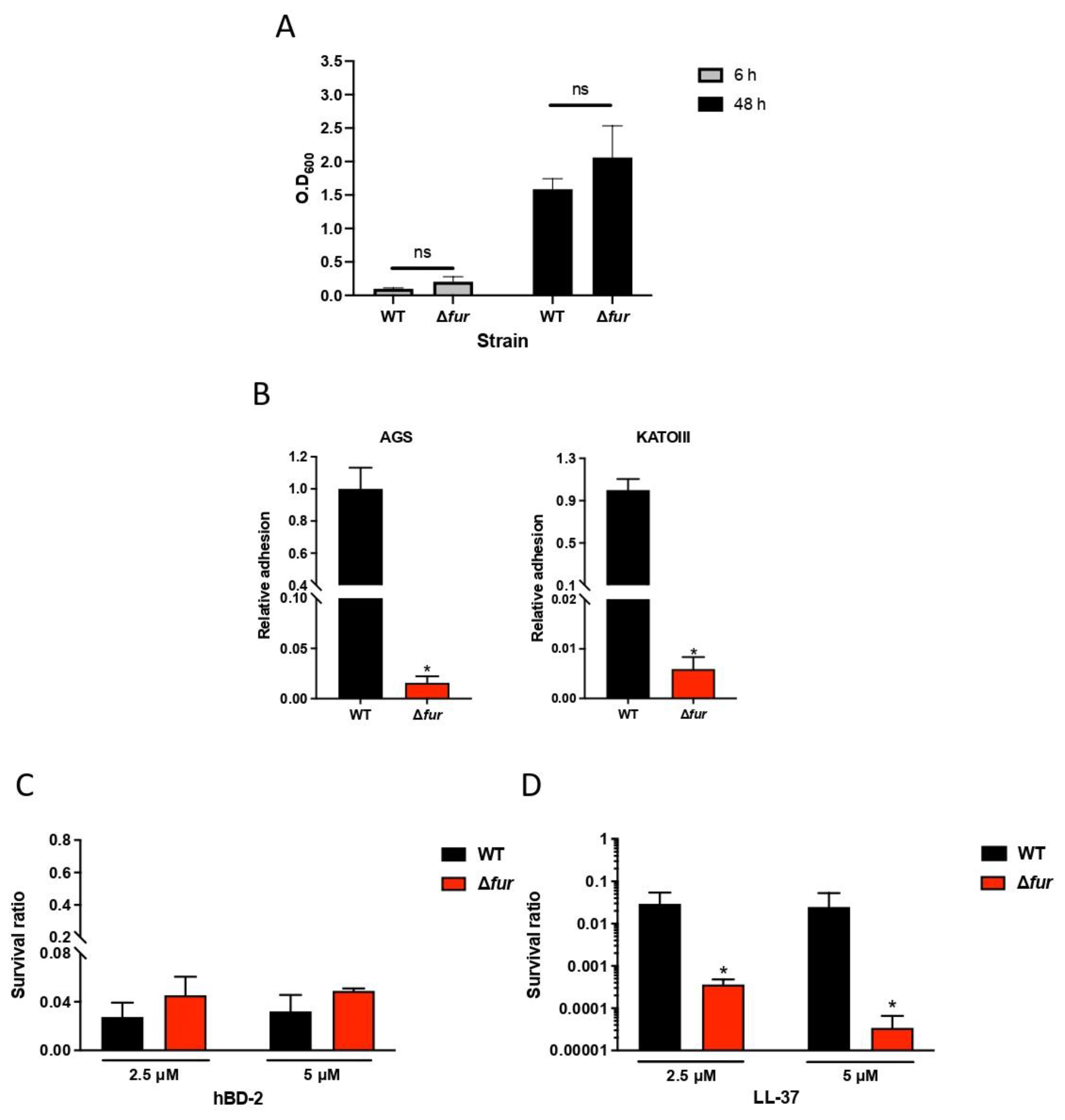
2.4. Lactobacillus Gasseri Downregulates the Ferric Uptake Regulator
2.5. Fur Is Involved in Antimicrobial Peptide LL-37 Resistance in H. pylori
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Media and Growth Conditions
4.2. Preparation of Conditioned Medium
4.3. qPCR Analysis
4.4. Construction of H. pylori Deletion Mutants
4.5. Motility Assay
4.6. Antimicrobial Peptide Susceptibility
4.7. Acid Survival Assay
4.8. Adhesion Assay
4.9. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yamaoka, Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 629. [Google Scholar] [CrossRef] [PubMed]
- Salama, N.R.; Hartung, M.L.; Müller, A. Life in the human stomach: Persistence strategies of the bacterial pathogen Helicobacter pylori. Nat. Rev. Microbiol. 2013, 11, 385–399. [Google Scholar] [CrossRef] [PubMed]
- Abadi, A.T.B. Strategies used by helicobacter pylori to establish persistent infection. World J. Gastroenterol. 2017, 23, 2870. [Google Scholar] [CrossRef] [PubMed]
- Danielli, A.; Amore, G.; Scarlato, V. Built shallow to maintain homeostasis and persistent infection: Insight into the transcriptional regulatory network of the gastric human pathogen Helicobacter pylori. PLoS Pathog. 2010, 6, e1000938. [Google Scholar] [CrossRef]
- Dale, B.A.; Fredericks, L.P. Antimicrobial peptides in the oral environment: Expression and function in health and disease. Curr. Issues Mol. Biol. 2005, 7, 119–133. [Google Scholar] [PubMed]
- Kosciuczuk, E.M.; Lisowski, P.; Jarczak, J.; Strzalkowska, N.; Jozwik, A.; Horbanczuk, J.; Krzyzewski, J.; Zwierzchowski, L.; Bagnicka, E. Cathelicidins: Family of antimicrobial peptides. A review. Mol. Biol. Rep. 2012, 39, 10957–10970. [Google Scholar] [CrossRef]
- Ong, P.Y.; Ohtake, T.; Brandt, C.; Strickland, I.; Boguniewicz, M.; Ganz, T.; Gallo, R.L.; Leung, D.Y. Endogenous antimicrobial peptides and skin infections in atopic dermatitis. N. Engl. J. Med. 2002, 347, 1151–1160. [Google Scholar] [CrossRef]
- Schaller-Bals, S.; Schulze, A.; Bals, R. Increased levels of antimicrobial peptides in tracheal aspirates of newborn infants during infection. Am. J. Respir. Crit. Care Med. 2002, 165, 992–995. [Google Scholar] [CrossRef]
- Hase, K.; Murakami, M.; Iimura, M.; Cole, S.P.; Horibe, Y.; Ohtake, T.; Obonyo, M.; Gallo, R.L.; Eckmann, L.; Kagnoff, M.F. Expression of LL-37 by human gastric epithelial cells as a potential host defense mechanism against Helicobacter pylori. Gastroenterology 2003, 125, 1613–1625. [Google Scholar] [CrossRef]
- Hamanaka, Y.; Nakashima, M.; Wada, A.; Ito, M.; Kurazono, H.; Hojo, H.; Nakahara, Y.; Kohno, S.; Hirayama, T.; Sekine, I. Expression of human beta-defensin 2 (hBD-2) in Helicobacter pylori induced gastritis: Antibacterial effect of hBD-2 against Helicobacter pylori. Gut 2001, 49, 481–487. [Google Scholar] [CrossRef]
- Nuding, S.; Gersemann, M.; Hosaka, Y.; Konietzny, S.; Schaefer, C.; Beisner, J.; Schroeder, B.O.; Ostaff, M.J.; Saigenji, K.; Ott, G.; et al. Gastric antimicrobial peptides fail to eradicate Helicobacter pylori infection due to selective induction and resistance. PLoS ONE 2013, 8, e73867. [Google Scholar] [CrossRef] [PubMed]
- Buffie, C.G.; Pamer, E.G. Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 2013, 13, 790. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, N.; Li, P.; Gu, Q. Probiotic therapy in Helicobacter pylori infection: A potential strategy against a serious pathogen? Appl. Microbiol. Biotechnol. 2019, 103, 1573–1588. [Google Scholar] [CrossRef] [PubMed]
- Urrutia-Baca, V.H.; Escamilla-García, E.; de la Garza-Ramos, M.A.; Tamez-Guerra, P.; Gomez-Flores, R.; Urbina-Ríos, C.S. In vitro antimicrobial activity and downregulation of virulence gene expression on Helicobacter pylori by reuterin. Probiotics Antimicrob. Proteins 2018, 10, 168–175. [Google Scholar] [CrossRef] [PubMed]
- de Klerk, N.; Maudsdotter, L.; Gebreegziabher, H.; Saroj, S.D.; Eriksson, B.; Eriksson, O.S.; Roos, S.; Linden, S.; Sjolinder, H.; Jonsson, A.B. Lactobacilli Reduce Helicobacter pylori Attachment to Host Gastric Epithelial Cells by Inhibiting Adhesion Gene Expression. Infect. Immun. 2016, 84, 1526–1535. [Google Scholar] [CrossRef]
- Danielli, A.; Scarlato, V. Regulatory circuits in Helicobacter pylori: Network motifs and regulators involved in metal-dependent responses. FEMS Microbiol. Rev. 2010, 34, 738–752. [Google Scholar] [CrossRef]
- Pflock, M.; Dietz, P.; Schar, J.; Beier, D. Genetic evidence for histidine kinase HP165 being an acid sensor of Helicobacter pylori. FEMS Microbiol. Lett. 2004, 234, 51–61. [Google Scholar] [CrossRef][Green Version]
- Bury-Mone, S.; Thiberge, J.M.; Contreras, M.; Maitournam, A.; Labigne, A.; De Reuse, H. Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Mol. Microbiol. 2004, 53, 623–638. [Google Scholar] [CrossRef]
- Wen, Y.; Feng, J.; Scott, D.R.; Marcus, E.A.; Sachs, G. The HP0165-HP0166 two-component system (ArsRS) regulates acid-induced expression of HP1186 alpha-carbonic anhydrase in Helicobacter pylori by activating the pH-dependent promoter. J. Bacteriol. 2007, 189, 2426–2434. [Google Scholar] [CrossRef]
- Pich, O.Q.; Merrell, D.S. The ferric uptake regulator of Helicobacter pylori: A critical player in the battle for iron and colonization of the stomach. Future Microbiol. 2013, 8, 725–738. [Google Scholar] [CrossRef]
- Grubman, A.; Kaparakis, M.; Viala, J.; Allison, C.; Badea, L.; Karrar, A.; Boneca, I.G.; Le Bourhis, L.; Reeve, S.; Smith, I.A. The innate immune molecule, NOD1, regulates direct killing of Helicobacter pylori by antimicrobial peptides. Cell. Microbiol. 2010, 12, 626–639. [Google Scholar] [CrossRef] [PubMed]
- Miles, S.; Piazuelo, M.B.; Semino-Mora, C.; Washington, M.K.; Dubois, A.; Peek, R.M., Jr.; Correa, P.; Merrell, D.S. Detailed in vivo analysis of the role of Helicobacter pylori Fur in colonization and disease. Infect. Immun. 2010, 78, 3073–3082. [Google Scholar] [CrossRef] [PubMed]
- Ottemann, K.M.; Lowenthal, A.C. Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infect. Immun. 2002, 70, 1984–1990. [Google Scholar] [CrossRef] [PubMed]
- Iriarte, M.; Stainier, I.; Mikulskis, A.V.; Cornelis, G.R. The fliA gene encoding sigma 28 in Yersinia enterocolitica. J. Bacteriol. 1995, 177, 2299–2304. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Haiko, J.; Westerlund-Wikström, B. The role of the bacterial flagellum in adhesion and virulence. Biology 2013, 2, 1242–1267. [Google Scholar] [CrossRef]
- Kao, C.-Y.; Sheu, B.-S.; Wu, J.-J. Helicobacter pylori infection: An overview of bacterial virulence factors and pathogenesis. Biomed. J. 2016, 39, 14–23. [Google Scholar] [CrossRef]
- Danielli, A.; Roncarati, D.; Delany, I.; Chiarini, V.; Rappuoli, R.; Scarlato, V. In vivo dissection of the Helicobacter pylori Fur regulatory circuit by genome-wide location analysis. J. Bacteriol. 2006, 188, 4654–4662. [Google Scholar] [CrossRef]
- Jeng, L.; Yamshchikov, A.V.; Judd, S.E.; Blumberg, H.M.; Martin, G.S.; Ziegler, T.R.; Tangpricha, V. Alterations in vitamin D status and anti-microbial peptide levels in patients in the intensive care unit with sepsis. J. Transl. Med. 2009, 7, 28. [Google Scholar] [CrossRef]
- Graham, D.Y.; Fischbach, L. Helicobacter pylori treatment in the era of increasing antibiotic resistance. Gut 2010, 59, 1143–1153. [Google Scholar] [CrossRef]
- Savoldi, A.; Carrara, E.; Graham, D.Y.; Conti, M.; Tacconelli, E. Prevalence of antibiotic resistance in Helicobacter pylori: A systematic review and meta-analysis in World Health Organization regions. Gastroenterology 2018, 155, 1372–1382.e17. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Sang, J.; He, H.; Wan, X.; Lin, Y.; Li, L.; Li, Y.; Yu, C. Probiotic supplementation does not improve eradication rate of Helicobacter pylori infection compared to placebo based on standard therapy: A meta-analysis. Sci. Rep. 2016, 6, 23522. [Google Scholar] [CrossRef] [PubMed]
- Losurdo, G.; Cubisino, R.; Barone, M.; Principi, M.; Leandro, G.; Ierardi, E.; Di Leo, A. Probiotic monotherapy and Helicobacter pylori eradication: A systematic review with pooled-data analysis. World J. Gastroenterol. 2018, 24, 139. [Google Scholar] [CrossRef] [PubMed]
- Liévin-Le Moal, V.; Servin, A.L. Anti-infective activities of lactobacillus strains in the human intestinal microbiota: From probiotics to gastrointestinal anti-infectious biotherapeutic agents. Clin. Microbiol. Rev. 2014, 27, 167–199. [Google Scholar] [CrossRef]
- Lebeer, S.; Bron, P.A.; Marco, M.L.; Van Pijkeren, J.-P.; Motherway, M.O.C.; Hill, C.; Pot, B.; Roos, S.; Klaenhammer, T. Identification of probiotic effector molecules: Present state and future perspectives. Curr. Opin. Biotechnol. 2018, 49, 217–223. [Google Scholar] [CrossRef]
- Carey, C.M.; Kostrzynska, M.; Ojha, S.; Thompson, S. The effect of probiotics and organic acids on Shiga-toxin 2 gene expression in enterohemorrhagic Escherichia coli O157:H7. J. Microbiol. Methods 2008, 73, 125–132. [Google Scholar] [CrossRef]
- Bjorkholm, B.; Lundin, A.; Sillén, A.; Guillemin, K.; Salama, N.; Rubio, C.; Gordon, J.I.; Falk, P.; Engstrand, L. Comparison of genetic divergence and fitness between two subclones of Helicobacter pylori. Infect. Immun. 2001, 69, 7832–7838. [Google Scholar] [CrossRef]
- Ge, Z.; Taylor, D.E. H. pylori DNA Transformation by Natural Competence and Electroporation. Methods Mol. Med. 1997, 8, 145–152. [Google Scholar]
- Rotcheewaphan, S.; Belisle, J.T.; Webb, K.J.; Kim, H.J.; Spencer, J.S.; Borlee, B.R. Diguanylate cyclase activity of the Mycobacterium leprae T cell antigen ML1419c. Microbiology 2016, 162, 1651–1661. [Google Scholar] [CrossRef]


| Strain or Plasmid | Characteristics a | Source or Reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | Invitrogen | |
| H. pylori 67:21 | Isolated from a patient with gastric ulcer | [37] |
| H. pylori 67:21ΔarsR | Cmr; H. pylori 67:21 arsR mutant | This study |
| H. pylori 67:21ΔflgR | Cmr; H. pylori 67:21 flgR mutant | This study |
| H. pylori 67:21Δfur | Cmr; H. pylori 67:21 fur mutant | This study |
| L. gasseri Kx 110 A1 | Isolated from human gastric biopsy specimen | [15] |
| L. oris Kx 112 A1 | Isolated from human gastric biopsy specimen | [15] |
| Plasmids | ||
| pACYC184 | Cmr, Tetr; E. coli cloning vector | ATCC |
| pJET1.2./blunt | Ampr; E. coli cloning vector | ThermoFisher Scientific |
| pJET-arsR-cmr | pJET1.2./blunt derivative containing arsR-Cmr fusion PCR product | This study |
| pJET-flgR-cmr | pJET1.2./blunt derivative containing flgR-Cmr fusion PCR product | This study |
| pJET-fur-cmr | pJET1.2./blunt derivative containing fur-Cmr fusion PCR product | This study |
| Primers | Sequence (5′-3′) a | Purpose | Source |
|---|---|---|---|
| arsS_up_F | GGCATTAGTGCGGCTAACACACAAAAT | Cloning | This study |
| arsS_up_R | ACTGATTTAGTGTATGATGGAACCCCTTAACTCCTTATTAGAAT | Cloning | This study |
| arsS_down_F | ATAATAAGCGGATGAATGGCAGAAAAACAAAAAGAGAGAACATG | Cloning | This study |
| arsS_down_R | TTAGTGGAATAACTCATGATGGGCGTGT | Cloning | This study |
| arsS_Cmr_F | GGAGTTAAGGGGTTCCATCATACACTAAATCAGTAAGT | Cloning | This study |
| arsS_Cmr_R | CTTTTTGTTTTTCTGCCATTCATCCGCTTATTATCACT | Cloning | This study |
| flgR_up_F | TAGAAGATCAAGAATTTTTAATTTCGT | Cloning | This study |
| flgR_up_R | TTAGTGTATGATGGTCTTCTTCCTTTCTAAAAATATCT | Cloning | This study |
| flgR_down_F | ATAAGCGGATGAATGGCAATAAAGGCACGATCTTTTTAGAT | Cloning | This study |
| flgR_down_R | CCACGACGCCTAAAAGCTCTCGC | Cloning | This study |
| flgR_Cmr_F | AAAGGAAGAAGACCATCATACACTAAATCAGTAAGT | Cloning | This study |
| flgR_Cmr_R | CTTTATTGCCATTCATCCGCTTATTATCACTTAT | Cloning | This study |
| fur_up_F | CTACCCTGAAGCGCGCATCAT | Cloning | This study |
| fur_up_R | AGTGTATGATGGCCTTATCCGTAAAATGATTTTTATAACT | Cloning | This study |
| fur_down_F | ATGAATGGCAGAGTGAATGTTAAAAGATTTTAAAAAAG | Cloning | This study |
| fur_down_R | GAAAAGCTCTTTTGTGGAGTTTTTTG | Cloning | This study |
| fur_Cmr_F | ATTTTACGGATAAGGCCATCATACACTAAATCAGTAAGTTG | Cloning | This study |
| fur_Cmr_R | ATCTTTTAACATTCACTCTGCCATTCATCCGCTTATTAT | Cloning | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zuo, F.; Somiah, T.; Gebremariam, H.G.; Jonsson, A.-B. Lactobacilli Downregulate Transcription Factors in Helicobacter pylori That Affect Motility, Acid Tolerance and Antimicrobial Peptide Survival. Int. J. Mol. Sci. 2022, 23, 15451. https://doi.org/10.3390/ijms232415451
Zuo F, Somiah T, Gebremariam HG, Jonsson A-B. Lactobacilli Downregulate Transcription Factors in Helicobacter pylori That Affect Motility, Acid Tolerance and Antimicrobial Peptide Survival. International Journal of Molecular Sciences. 2022; 23(24):15451. https://doi.org/10.3390/ijms232415451
Chicago/Turabian StyleZuo, Fanglei, Tanvi Somiah, Hanna G. Gebremariam, and Ann-Beth Jonsson. 2022. "Lactobacilli Downregulate Transcription Factors in Helicobacter pylori That Affect Motility, Acid Tolerance and Antimicrobial Peptide Survival" International Journal of Molecular Sciences 23, no. 24: 15451. https://doi.org/10.3390/ijms232415451
APA StyleZuo, F., Somiah, T., Gebremariam, H. G., & Jonsson, A.-B. (2022). Lactobacilli Downregulate Transcription Factors in Helicobacter pylori That Affect Motility, Acid Tolerance and Antimicrobial Peptide Survival. International Journal of Molecular Sciences, 23(24), 15451. https://doi.org/10.3390/ijms232415451







