Transcriptome Analysis Reveals Immunomodulatory Effect of Spore-Displayed p75 on Human Intestinal Epithelial Caco-2 Cells
Abstract
:1. Introduction
2. Results
2.1. Summary of Transcriptome Sequencing Data
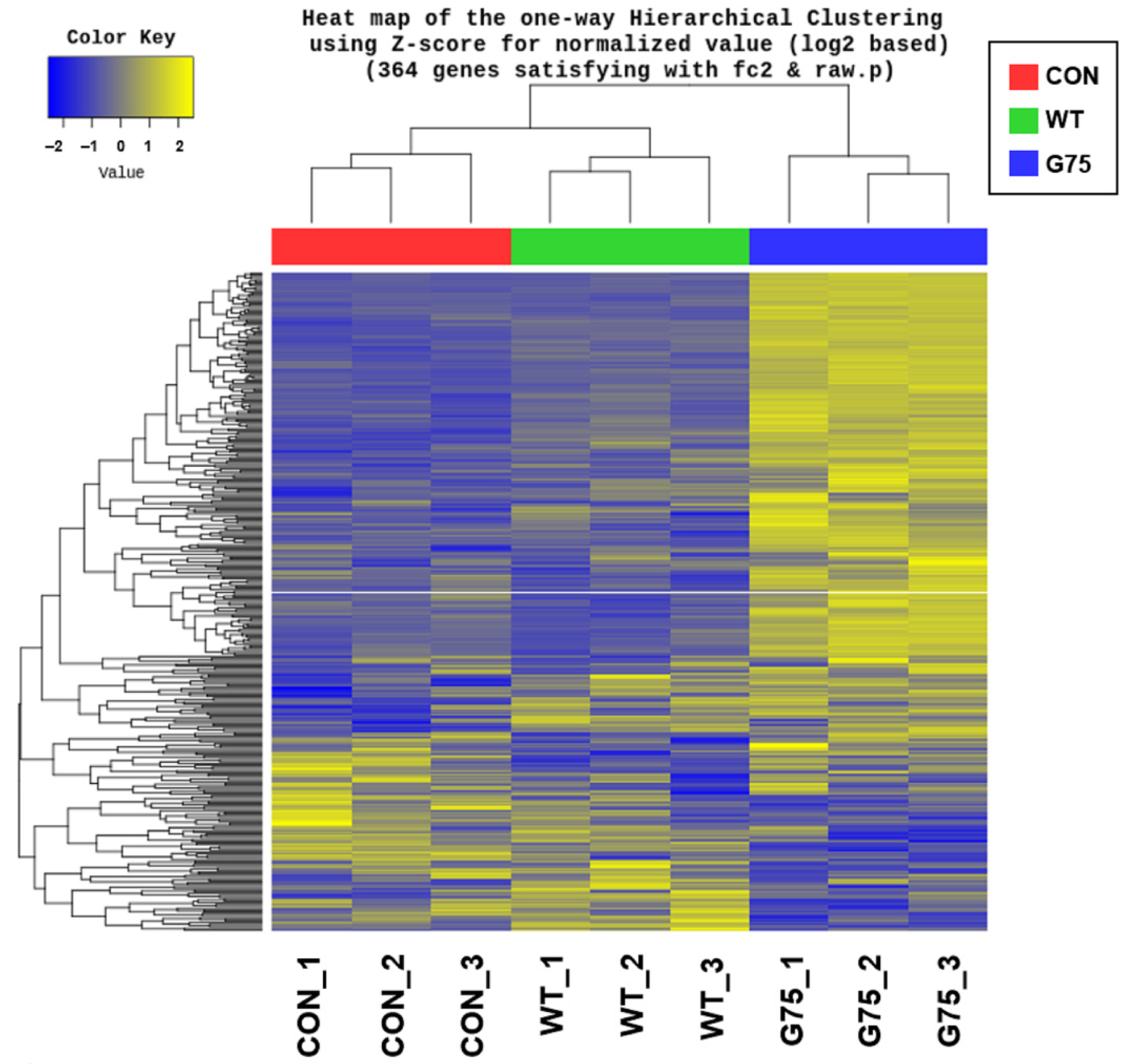
2.2. Analysis of Differentially Expressed Genes in Caco-2 Cells Stimulated with CotG-p75
2.3. Gene Ontology Enrichment Analysis
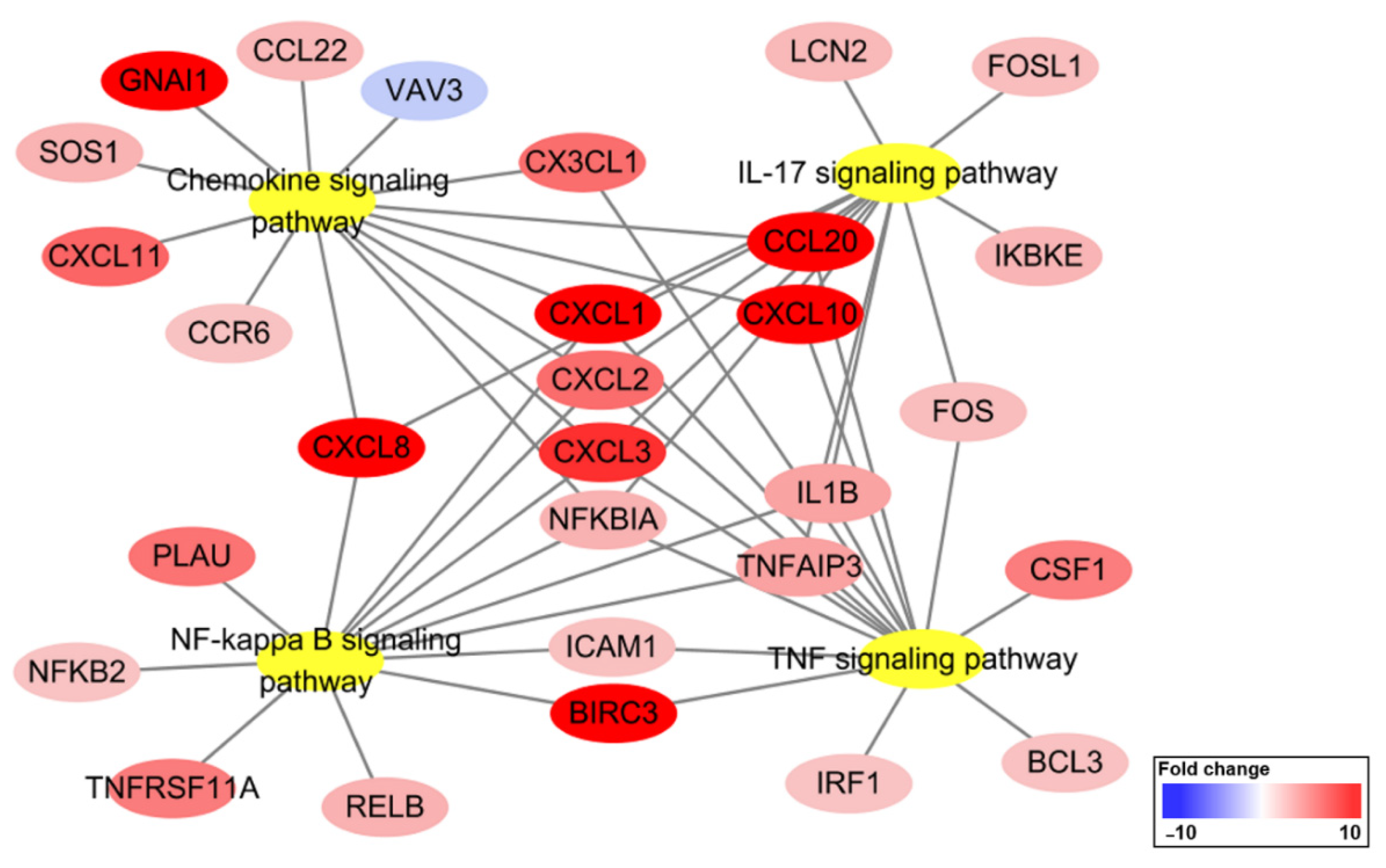
2.4. KEGG Pathway Enrichment Analysis
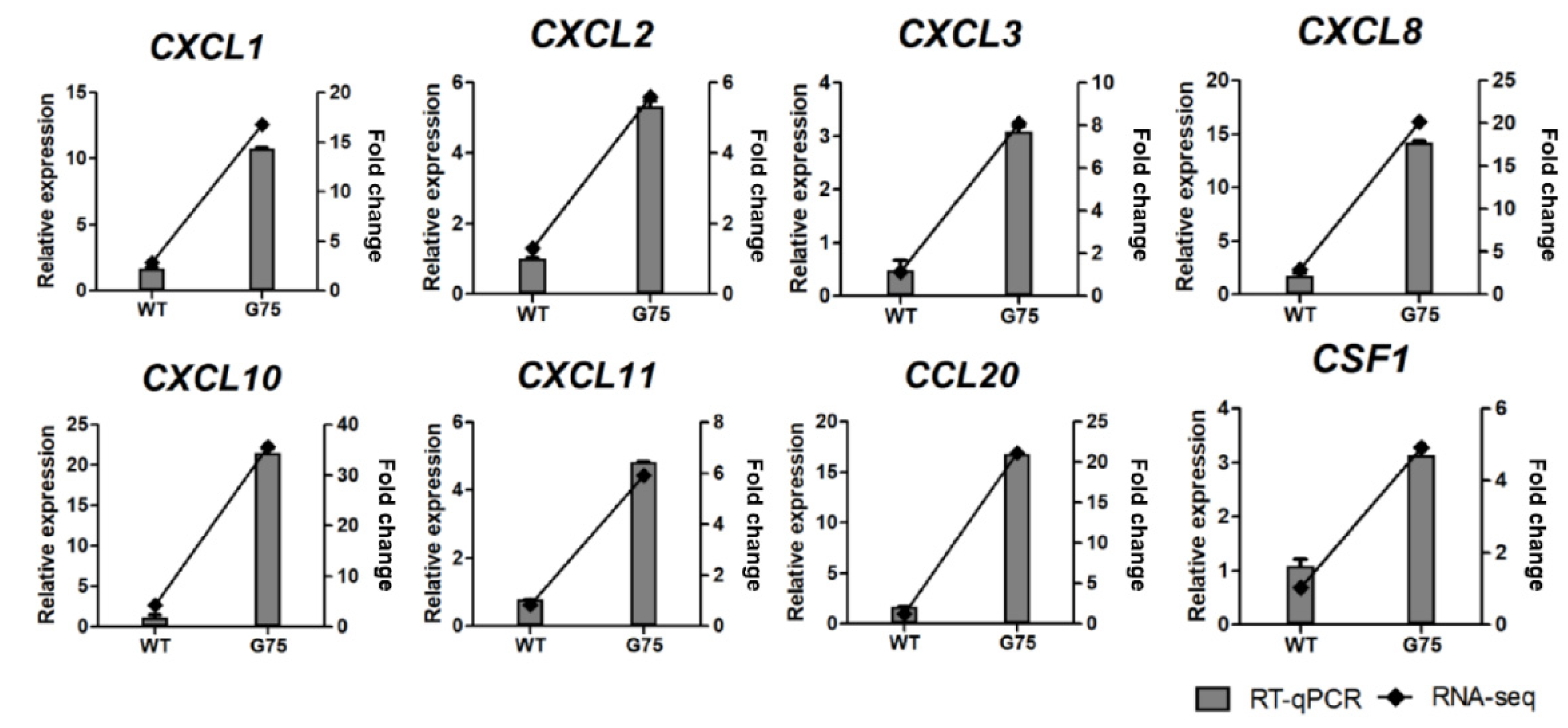
2.5. RNA-Seq Data Validation by RT-qPCR
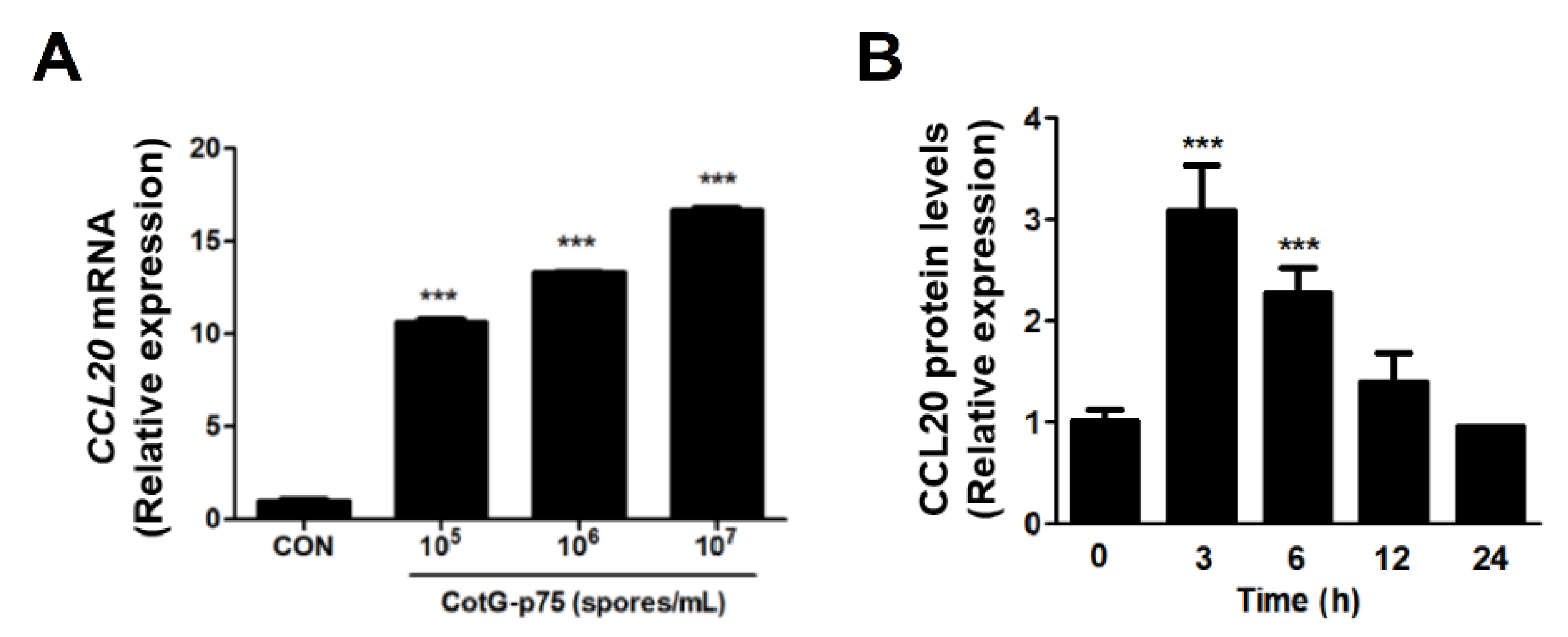
2.6. Effects of CotG-p75 on HBEGF and CCL20 Protein Expression
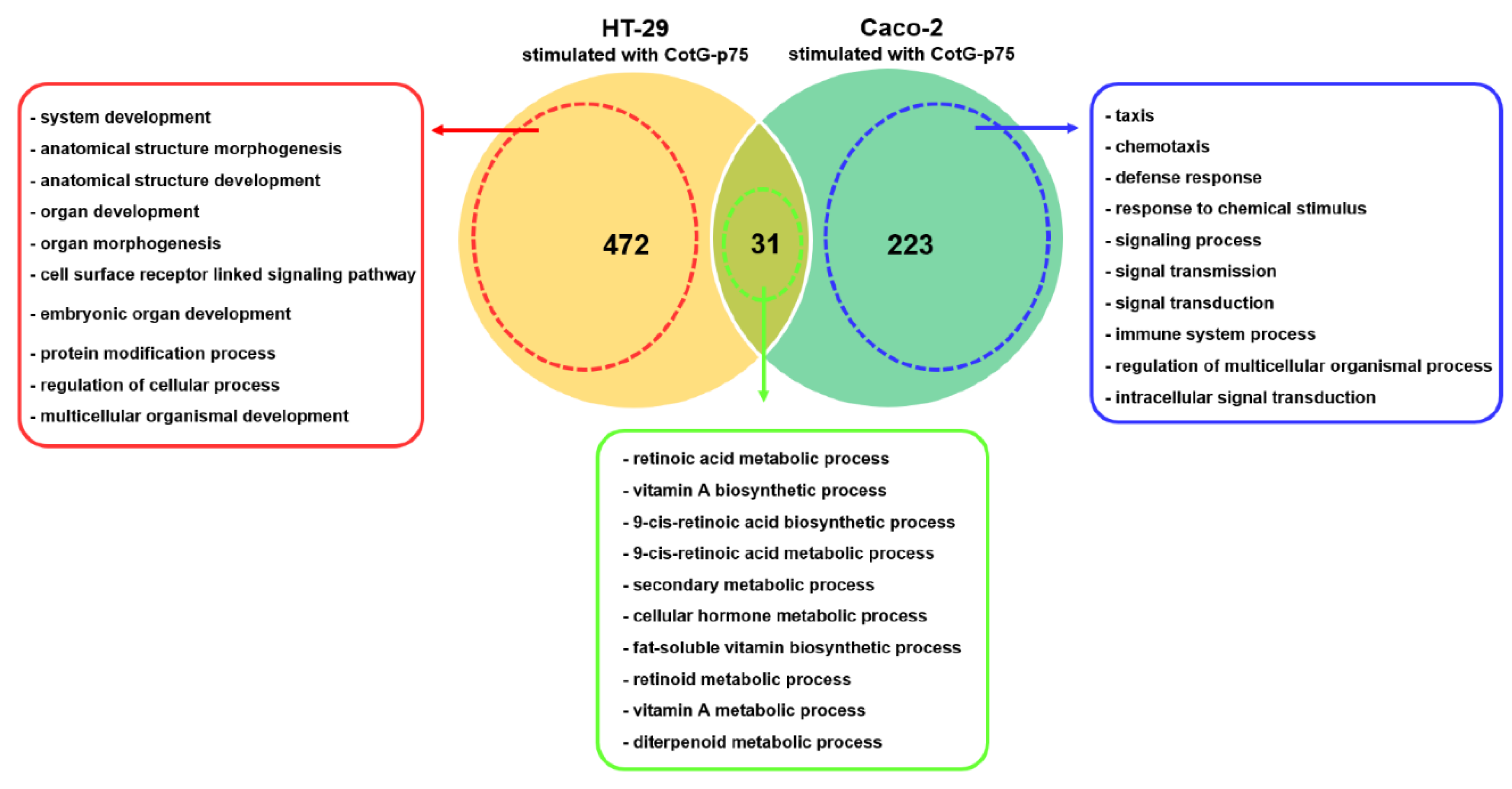
2.7. Comparison of Transcriptional Responses of HT-29 and Caco-2 Cells Stimulated by CotG-p75
3. Discussion
4. Materials and Methods
4.1. Spore Preparation
4.2. Cell Culture and Treatments
4.3. Cell Viability Assay
4.4. Total RNA Isolation, cDNA Library Construction, and RNA-Sequencing
4.5. Analysis of Differentially Expressed Genes (DEGs)
4.6. Functional Analyses of DEGs
4.7. Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR) Confirmation
4.8. Heparin Binding-Epidermal Growth Factor (HBEGF) and CC Motif Chemokine Ligand (CCL20) Protein Expression
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Erickson, K.L.; Hubbard, N.E. Probiotic immunomodulation in health and disease. J. Nutr. 2000, 130, 403S–409S. [Google Scholar] [CrossRef] [Green Version]
- Madsen, K.; Cornish, A.; Soper, P.; McKaigney, C.; Jijon, H.; Yachimec, C.; Doyle, J.; Jewell, L.; De Simone, C. Probiotic bacteria enhance murine and human intestinal epithelial barrier function. Gastroenterology 2001, 121, 580–591. [Google Scholar] [CrossRef] [Green Version]
- Yahfoufi, N.; Mallet, J.; Graham, E.; Matar, C. Role of probiotics and prebiotics in immunomodulation. Curr. Opin. Food Sci. 2018, 20, 82–91. [Google Scholar] [CrossRef]
- Gorbach, S.L.; Newton, I. The discovery of Lactobacillus GG. Nutr. Today 1996, 31, 5S. [Google Scholar] [CrossRef]
- Doron, S.; Snydman, D.R.; Gorbach, S.L. Lactobacillus GG: Bacteriology and clinical applications. Gastroenterol. Clin. 2005, 34, 483–498. [Google Scholar] [CrossRef]
- Capurso, L. Thirty years of Lactobacillus rhamnosus GG: A review. J. Clin. Gastroenterol. 2019, 53, S1–S41. [Google Scholar] [CrossRef]
- He, F.; Morita, H.; Kubota, A.; Ouwehand, A.C.; Hosoda, M.; Hiramatsu, M.; Kurisaki, J.I. Effect of orally administered non-viable Lactobacillus cells on murine humoral immune responses. Microbiol. Immunol. 2005, 49, 993–997. [Google Scholar] [CrossRef]
- Yan, F.; Liu, L.; Cao, H.; Moore, D.J.; Washington, M.K.; Wang, B.; Peek, R.; Acra, S.A.; Polk, D.B. Neonatal colonization of mice with LGG promotes intestinal development and decreases susceptibility to colitis in adulthood. Mucosal Immunol. 2017, 10, 117–127. [Google Scholar] [CrossRef] [Green Version]
- Donato, K.A.; Gareau, M.G.; Wang, Y.J.J.; Sherman, P.M. Lactobacillus rhamnosus GG attenuates interferon-γ and tumour necrosis factor-α-induced barrier dysfunction and pro-inflammatory signalling. Microbiology 2010, 156, 3288–3297. [Google Scholar] [CrossRef] [Green Version]
- Fong, F.L.Y.; Kirjavainen, P.V.; El-Nezami, H. Immunomodulation of Lactobacillus rhamnosus GG (LGG)-derived soluble factors on antigen-presenting cells of healthy blood donors. Sci. Rep. 2016, 6, 22845. [Google Scholar] [CrossRef]
- Claes, I.J.; Segers, M.E.; Verhoeven, T.L.; Dusselier, M.; Sels, B.F.; De Keersmaecker, S.C.; Vanderleyden, J.; Lebeer, S. Lipoteichoic acid is an important microbe-associated molecular pattern of Lactobacillus rhamnosus GG. Microb. Cell Fact. 2012, 11, 161. [Google Scholar] [CrossRef] [Green Version]
- Tao, Y.; Drabik, K.A.; Waypa, T.S.; Musch, M.W.; Alverdy, J.C.; Schneewind, O.; Chang, E.B.; Petrof, E.O. Soluble factors from Lactobacillus GG activate MAPKs and induce cytoprotective heat shock proteins in intestinal epithelial cells. Am. J. Physiol. Cell Physiol. 2006, 290, C1018–C1030. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Liu, L.; Moore, D.J.; Shen, X.; Peek, R.; Acra, S.A.; Li, H.; Ren, X.; Polk, D.B.; Yan, F. An LGG-derived protein promotes IgA production through upregulation of APRIL expression in intestinal epithelial cells. Mucosal Immunol. 2017, 10, 373–384. [Google Scholar] [CrossRef] [Green Version]
- Yan, F.; Polk, D.B. Characterization of a probiotic-derived soluble protein which reveals a mechanism of preventive and treatment effects of probiotics on intestinal inflammatory diseases. Gut Microbes 2012, 3, 25–28. [Google Scholar] [CrossRef] [Green Version]
- Yan, F.; Polk, D.B. Probiotic bacterium prevents cytokine-induced apoptosis in intestinal epithelial cells. J. Biol. Chem. 2002, 277, 50959–50965. [Google Scholar] [CrossRef] [Green Version]
- Yan, F.; Cao, H.; Cover, T.L.; Whitehead, R.; Washington, M.K.; Polk, D.B. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology 2007, 132, 562–575. [Google Scholar] [CrossRef] [Green Version]
- Seth, A.; Yan, F.; Polk, D.B.; Rao, R. Probiotics ameliorate the hydrogen peroxide-induced epithelial barrier disruption by a PKC-and MAP kinase-dependent mechanism. Am. J. Physiol. Gastrointest. Liver Physiol. 2008, 294, G1060–G1069. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.J.; Jun, J.S.; Moon, J.A.; Hong, K.W. Surface display of p75, a Lactobacillus rhamnosus GG derived protein, on Bacillus subtilis spores and its antibacterial activity against Listeria monocytogenes. AMB Express 2020, 10, 139. [Google Scholar] [CrossRef]
- Cencič, A.; Langerholc, T. Functional cell models of the gut and their applications in food microbiology—A review. Int. J. Food Microbiol. 2010, 141, S4–S14. [Google Scholar] [CrossRef]
- Zweibaum, A.; Laburthe, M.; Grasset, E.; Louvard, D. Use of cultured cell lines in studies of intestinal cell differentiation and function. In Comprehensive Physiology; Wiley: Hoboken, NJ, USA, 2010; pp. 223–255. [Google Scholar]
- Liévin-Le Moal, V.; Servin, A.L. The front line of enteric host defense against unwelcome intrusion of harmful microorganisms: Mucins, antimicrobial peptides, and microbiota. Clin. Microbiol. Rev. 2006, 19, 315–337. [Google Scholar] [CrossRef]
- Lea, T. Caco-2 cell line. In The Impact of Food Bioactives on Health; Springer: Berlin/Heidelberg, Germany, 2015; pp. 103–111. [Google Scholar]
- Engle, M.; Goetz, G.; Alpers, D. Caco-2 cells express a combination of colonocyte and enterocyte phenotypes. J. Cell. Physiol. 1998, 174, 362–369. [Google Scholar] [CrossRef]
- Shimizu, M. Interaction between food substances and the intestinal epithelium. Biosci. Biotechnol. Biochem. 2010, 74, 232–241. [Google Scholar] [CrossRef] [Green Version]
- Knipp, G.T.; Ho, N.F.; Barsuhn, C.L.; Borchardt, R.T. Paracellular diffusion in Caco-2 cell monolayers: Effect of perturbation on the transport of hydrophilic compounds that vary in charge and size. J. Pharm. Sci. 1997, 86, 1105–1110. [Google Scholar] [CrossRef]
- Bourgine, J.; Billaut-Laden, I.; Happillon, M.; Lo-Guidice, J.-M.; Maunoury, V.; Imbenotte, M.; Broly, F. Gene expression profiling of systems involved in the metabolism and the disposition of xenobiotics: Comparison between human intestinal biopsy samples and colon cell lines. Drug Metab. Dispos. 2012, 40, 694–705. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.-J.; Kim, M.-J.; Son, D.-Y.; Kang, S.-S.; Hong, K.-W. Effects of Spore-Displayed p75 Protein from Lacticaseibacillus rhamnosus GG on the Transcriptional Response of HT-29 Cells. Microorganisms 2022, 10, 1276. [Google Scholar] [CrossRef]
- Warabi, E.; Wada, Y.; Kajiwara, H.; Kobayashi, M.; Koshiba, N.; Hisada, T.; Shibata, M.; Ando, J.; Tsuchiya, M.; Kodama, T. Effect on endothelial cell gene expression of shear stress, oxygen concentration, and low-density lipoprotein as studied by a novel flow cell culture system. Free Radical Biol. Med. 2004, 37, 682–694. [Google Scholar] [CrossRef]
- Brunet De La Grange, P.; Barthe, C.; Lippert, E.; Hermitte, F.; Belloc, F.; Lacombe, F.; Ivanovic, Z.; Praloran, V. Oxygen concentration influences mRNA processing and expression of the cd34 gene. J. Cell. Biochem. 2006, 97, 135–144. [Google Scholar] [CrossRef]
- Tremblay, E.; Auclair, J.; Delvin, E.; Levy, E.; Ménard, D.; Pshezhetsky, A.V.; Rivard, N.; Seidman, E.G.; Sinnett, D.; Vachon, P.H. Gene expression profiles of normal proliferating and differentiating human intestinal epithelial cells: A comparison with the Caco-2 cell model. J. Cell. Biochem. 2006, 99, 1175–1186. [Google Scholar] [CrossRef]
- Yarden, Y. The EGFR family and its ligands in human cancer: Signalling mechanisms and therapeutic opportunities. Eur. J. Cancer 2001, 37, 3–8. [Google Scholar] [CrossRef]
- Yarden, Y.; Sliwkowski, M.X. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2001, 2, 127–137. [Google Scholar] [CrossRef]
- Van Baarlen, P.; Troost, F.J.; van Hemert, S.; van der Meer, C.; de Vos, W.M.; de Groot, P.J.; Hooiveld, G.J.; Brummer, R.-J.M.; Kleerebezem, M. Differential NF-κB pathways induction by Lactobacillus plantarum in the duodenum of healthy humans correlating with immune tolerance. Proc. Natl. Acad. Sci. USA 2009, 106, 2371–2376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, F.; Cao, H.; Cover, T.L.; Washington, M.K.; Shi, Y.; Liu, L.; Chaturvedi, R.; Peek, R.M.; Wilson, K.T.; Polk, D.B. Colon-specific delivery of a probiotic-derived soluble protein ameliorates intestinal inflammation in mice through an EGFR-dependent mechanism. J. Clin. Investig. 2011, 121, 2242–2253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, F.; Liu, L.; Dempsey, P.J.; Tsai, Y.-H.; Raines, E.W.; Wilson, C.L.; Cao, H.; Cao, Z.; Liu, L.; Polk, D.B. A Lactobacillus rhamnosus GG-derived soluble protein, p40, stimulates ligand release from intestinal epithelial cells to transactivate epidermal growth factor receptor. J. Biol. Chem. 2013, 288, 30742–30751. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Cao, H.; Liu, L.; Wang, B.; Walker, W.A.; Acra, S.A.; Yan, F. Activation of epidermal growth factor receptor mediates mucin production stimulated by p40, a Lactobacillus rhamnosus GG-derived protein. J. Biol. Chem. 2014, 289, 20234–20244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boone, D.L.; Turer, E.E.; Lee, E.G.; Ahmad, R.-C.; Wheeler, M.T.; Tsui, C.; Hurley, P.; Chien, M.; Chai, S.; Hitotsumatsu, O. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat. Immunol. 2004, 5, 1052–1060. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.-C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef] [Green Version]
- Nenci, A.; Becker, C.; Wullaert, A.; Gareus, R.; Van Loo, G.; Danese, S.; Huth, M.; Nikolaev, A.; Neufert, C.; Madison, B. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature 2007, 446, 557–561. [Google Scholar] [CrossRef]
- Zaph, C.; Troy, A.E.; Taylor, B.C.; Berman-Booty, L.D.; Guild, K.J.; Du, Y.; Yost, E.A.; Gruber, A.D.; May, M.J.; Greten, F.R. Epithelial-cell-intrinsic IKK-β expression regulates intestinal immune homeostasis. Nature 2007, 446, 552–556. [Google Scholar] [CrossRef]
- Steinbrecher, K.A.; Harmel-Laws, E.; Sitcheran, R.; Baldwin, A.S. Loss of epithelial RelA results in deregulated intestinal proliferative/apoptotic homeostasis and susceptibility to inflammation. J. Immunol. 2008, 180, 2588–2599. [Google Scholar] [CrossRef] [Green Version]
- Lee, E.G.; Boone, D.L.; Chai, S.; Libby, S.L.; Chien, M.; Lodolce, J.P.; Ma, A. Failure to regulate TNF-induced NF-κB and cell death responses in A20-deficient mice. Science 2000, 289, 2350–2354. [Google Scholar] [CrossRef]
- Arsenescu, R.; Bruno, M.; Rogier, E.; Stefka, A.; McMahan, A.; Wright, T.; Nasser, M.; De Villiers, W.; Kaetzel, C. Signature biomarkers in Crohn’s disease: Toward a molecular classification. Mucosal Immunol. 2008, 1, 399–411. [Google Scholar] [CrossRef] [Green Version]
- Kolodziej, L.E.; Lodolce, J.P.; Chang, J.E.; Schneider, J.R.; Grimm, W.A.; Bartulis, S.J.; Zhu, X.; Messer, J.S.; Murphy, S.F.; Reddy, N. TNFAIP3 maintains intestinal barrier function and supports epithelial cell tight junctions. PLoS ONE 2011, 6, e26352. [Google Scholar] [CrossRef] [Green Version]
- Jacobs, M.D.; Harrison, S.C. Structure of an IκBα/NF-κB complex. Cell 1998, 95, 749–758. [Google Scholar] [CrossRef] [Green Version]
- Chu, Z.-L.; McKinsey, T.A.; Liu, L.; Gentry, J.J.; Malim, M.H.; Ballard, D.W. Suppression of tumor necrosis factor-induced cell death by inhibitor of apoptosis c-IAP2 is under NF-κB control. Proc. Natl. Acad. Sci. USA 1997, 94, 10057–10062. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.; Zhou, Y.; Shen, P. NF-κB and its regulation on the immune system. Cell Mol. Immunol. 2004, 1, 343–350. [Google Scholar]
- Luster, A.D. Chemokines—Chemotactic cytokines that mediate inflammation. N. Engl. J. Med. 1998, 338, 436–445. [Google Scholar] [CrossRef]
- Vongsa, R.A.; Zimmerman, N.P.; Dwinell, M.B. CCR6 regulation of the actin cytoskeleton orchestrates human beta defensin-2-and CCL20-mediated restitution of colonic epithelial cells. J. Biol. Chem. 2009, 284, 10034–10045. [Google Scholar] [CrossRef] [Green Version]
- Cai, S.; Batra, S.; Lira, S.A.; Kolls, J.K.; Jeyaseelan, S. CXCL1 regulates pulmonary host defense to Klebsiella infection via CXCL2, CXCL5, NF-κB, and MAPKs. J. Immunol. 2010, 185, 6214–6225. [Google Scholar] [CrossRef] [Green Version]
- Wolf, M.; Moser, B. Antimicrobial activities of chemokines: Not just a side-effect? Front. Immunol. 2012, 3, 213. [Google Scholar] [CrossRef] [Green Version]
- Van Baarlen, P.; Troost, F.; van der Meer, C.; Hooiveld, G.; Boekschoten, M.; Brummer, R.J.; Kleerebezem, M. Human mucosal in vivo transcriptome responses to three lactobacilli indicate how probiotics may modulate human cellular pathways. Proc. Natl. Acad. Sci. USA 2011, 108, 4562–4569. [Google Scholar] [CrossRef] [Green Version]
- O’Callaghan, J.; Buttó, L.F.; MacSharry, J.; Nally, K.; O’Toole, P.W. Influence of adhesion and bacteriocin production by Lactobacillus salivarius on the intestinal epithelial cell transcriptional response. Appl. Environ. Microbiol. 2012, 78, 5196–5203. [Google Scholar] [CrossRef]
- O’Flaherty, S.; Klaenhammer, T.R. Influence of exposure time on gene expression by human intestinal epithelial cells exposed to Lactobacillus acidophilus. Appl. Environ. Microbiol. 2012, 78, 5028–5032. [Google Scholar] [CrossRef] [Green Version]
- Baldwin, A.S. Series introduction: The transcription factor NF-κB and human disease. J. Clin. Investig. 2001, 107, 3–6. [Google Scholar] [CrossRef] [Green Version]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 19 November 2022).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef] [Green Version]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]








| GO ID | GO Description | Adjusted p-Value a | Cluster Frequency b | Total Frequency c |
|---|---|---|---|---|
| Biological process | ||||
| 0042221 | response to chemical stimulus | 3.70 × 10−15 | 54/202 (26.7%) | 1466/17,784 (8.2%) |
| 0042330 | taxis | 5.20 × 10−15 | 20/202 (9.9%) | 169/17,784 (0.9%) |
| 0006935 | chemotaxis | 5.20 × 10−15 | 20/202 (9.9%) | 169/17,784 (0.9%) |
| 0006952 | defense response | 6.97 × 10−13 | 32/202 (15.8%) | 621/17,784 (3.4%) |
| 0009605 | response to external stimulus | 1.03 × 10−12 | 30/202 (14.8%) | 552/17,784 (3.1%) |
| 0048518 | positive regulation of biological process | 1.43 × 10−12 | 63/202 (31.1%) | 2210/17,784 (12.4%) |
| 0009611 | response to wounding | 3.92 × 10−12 | 29/202 (14.3%) | 543/17,784 (3.0%) |
| 0048522 | positive regulation of cellular process | 8.84 × 10−12 | 58/202 (28.7%) | 2006/17,784 (11.2%) |
| 0002376 | immune system process | 3.68 × 10−11 | 37/202 (18.3%) | 949/17,784 (5.3%) |
| 0007626 | locomotory behavior | 4.31 × 10−11 | 20/202 (9.9%) | 273/17,784 (1.5%) |
| 0048513 | organ development | 1.33 × 10−10 | 52/202 (25.7%) | 1792/17,784 (10.0%) |
| 0023046 | signaling process | 1.63 × 10−10 | 58/202 (28.7%) | 2157/17,784 (12.1%) |
| 0023060 | signal transmission | 1.63 × 10−10 | 58/202 (28.7%) | 2157/17,784 (12.1%) |
| 0048731 | system development | 2.26 × 10−10 | 62/202 (30.6%) | 2422/17,784 (13.6%) |
| 0051239 | regulation of multicellular organismal process | 2.71 × 10−10 | 38/202 (18.8%) | 1068/17,784 (6.0%) |
| 0032502 | developmental process | 3.55 × 10−10 | 74/202 (36.6%) | 3234/17,784 (18.1%) |
| 0007275 | multicellular organismal development | 3.84 × 10−10 | 70/202 (34.6%) | 2971/17,784 (16.7%) |
| 0048856 | anatomical structure development | 4.62 × 10−10 | 65/202 (32.1%) | 2656/17,784 (14.9%) |
| 0065007 | biological regulation | 5.60 × 10−10 | 122/202 (60.3%) | 6941/17,784 (39.0%) |
| 0006954 | inflammatory response | 5.80 × 10−10 | 20/202 (9.9%) | 316/17,784 (1.7%) |
| Molecular function | ||||
| 0008009 | chemokine activity | 2.47 × 10−9 | 9/202 (4.4%) | 47/17,784 (0.2%) |
| 0042379 | chemokine receptor binding | 6.35 × 10−9 | 9/202 (4.4%) | 52/17,784 (0.2%) |
| 0001664 | G-protein-coupled receptor binding | 4.92 × 10−8 | 12/202 (5.9%) | 136/17,784 (0.7%) |
| 0005102 | receptor binding | 5.53 × 10−8 | 31/202 (15.3%) | 922/17,784 (5.1%) |
| 0005126 | cytokine receptor binding | 2.13 × 10−7 | 13/202 (6.4%) | 186/17,784 (1.0%) |
| 0005125 | cytokine activity | 4.12 × 10−7 | 13/202 (6.4%) | 197/17,784 (1.1%) |
| Cellular component | ||||
| 0005615 | extracellular space | 1.91 × 10−11 | 33/202 (16.3%) | 747/17,784 (4.2%) |
| 0044421 | extracellular region part | 1.00 × 10−10 | 37/202 (18.3%) | 983/17,784 (5.5%) |
| 0005576 | extracellular region | 1.91 × 10−7 | 49/202 (24.2%) | 2025/17,784 11.3% |
| Pathway ID | KEGG Pathway | Counts | Adjusted p-Val. a | Genes |
|---|---|---|---|---|
| has04668 | TNF signaling pathway | 15 | 7.13 × 10−13 | CSF1, FOS, CXCL1, CXCL2, CXCL3, BIRC3, ICAM1, IL1B, CXCL10, IRF1, NFKBIA, BCL3, CCL20, CX3CL1, TNFAIP3 |
| has04657 | IL-17 signaling pathway | 13 | 2.95 × 10−11 | FOS, CXCL1, CXCL2, CXCL3, IL1B, CXCL8, CXCL10, LCN2, NFKBIA, CCL20, TNFAIP3, FOSL1, IKBKE |
| has04064 | NF-κB signaling pathway | 13 | 5.90 × 10−11 | CXCL1, CXCL2, CXCL3, BIRC3, ICAM1, IL1B, CXCL8, NFKB2, NFKBIA, PLAU, RELB, TNFAIP3, TNFRSF11A |
| has04062 | Chemokine signaling pathway | 14 | 2.50 × 10−9 | VAV3, CCR6, GNAI1, CXCL1, CXCL2, CXCL3, CXCL8, CXCL10, NFKBIA, CCL20, CCL22, CXCL11, CX3CL1, SOS1 |
| has05323 | Rheumatoid arthritis | 11 | 5.90 × 10−9 | CSF1, FOS, 245972, CXCL1, CXCL2, CXCL3, ICAM1, IL1B, CXCL8, CCL20, TNFRSF11A |
| has04061 | Viral protein interaction with cytokine and cytokine receptor | 11 | 9.49 × 10−9 | CCR6, CSF1, CXCL1, CXCL2, CXCL3, CXCL8, CXCL10, CCL20, CCL22, CXCL11, CX3CL1 |
| has04625 | C-type lectin receptor signaling pathway | 11 | 1.16 × 10−8 | EGR2, MRAS, IL1B, IRF1, ITPR1, NFKB2, NFKBIA, RELB, BCL3, CX3CL1, IKBKE |
| has05417 | Lipid and atherosclerosis | 13 | 5.84 × 10−8 | 10451, CYP1A1, FOS, CXCL1, CXCL2, CXCL3, ICAM1, IL1B, CXCL8, LAMA4, NFKBIA, OLR1, IKBKE |
| has05200 | Pathways in cancer | 18 | 7.05 × 10−8 | ETS1, FGF9, FOS, RASGRP3, GNAI1, BIRC3, CXCL8, LAMA4, NFKB2, NFKBIA, NOS2, WNT4, SOS1, STAT5A, TGFA, WNT5A, WNT6, WNT7B |
| has04010 | MAPK signaling pathway | 14 | 1.33 × 10−7 | CSF1, DUSP4, DUSP6, FGF9, MRAS, FOS, RASGRP3, IL1B, AREG, MEF2C, NFKB2, RELB, SOS1, TGFA |
| Gene Symbol | Gene Name | Fold Change | Raw p-Value |
|---|---|---|---|
| CXCL10 | C-X-C motif chemokine ligand 10 | 35.58 | 5.23 × 10−34 |
| CCL20 | C-C motif chemokine ligand 20 | 21.17 | 0.00 |
| CXCL8 | C-X-C motif chemokine ligand 8 | 20.17 | 4.15 × 10−38 |
| CXCL1 | C-X-C motif chemokine ligand 1 | 16.81 | 9.56 × 10−41 |
| BIRC3 | baculoviral IAP repeat containing 3 | 11.90 | 3.76 × 10−218 |
| CXCL3 | C-X-C motif chemokine ligand 3 | 8.10 | 3.93 × 10−52 |
| CXCL11 | C-X-C motif chemokine ligand 11 | 5.91 | 2.38 × 10−3 |
| CXCL2 | C-X-C motif chemokine ligand 2 | 5.59 | 6.66 × 10−41 |
| CX3CL1 | C-X3-C motif chemokine ligand 1 | 5.54 | 9.45 × 10−9 |
| PLAU | plasminogen activator, urokinase | 5.36 | 8.88 × 10−22 |
| TNFRSF11A | TNF receptor superfamily member 11a | 4.98 | 3.77 × 10−7 |
| CSF1 | colony stimulating factor 1 | 4.92 | 1.23 × 10−25 |
| TNFAIP3 | TNF α induced protein 3 | 3.44 | 7.71 × 10−119 |
| IL1B | interleukin 1 β | 3.38 | 2.07 × 10−4 |
| RELB | RELB proto-oncogene, NF-κB subunit | 2.83 | 1.63 × 10−35 |
| NFKBIA | NF-κB inhibitor α | 2.80 | 1.67 × 10−75 |
| SOS1 | SOS Ras/Rac guanine nucleotide exchange factor 1 | 2.75 | 7.76 × 10−57 |
| IKBKE | inhibitor of nuclear factor κB kinase subunit epsilon | 2.67 | 5.34 × 10−33 |
| CCL22 | C-C motif chemokine ligand 22 | 2.57 | 4.78 × 10−2 |
| LCN2 | lipocalin 2 | 2.50 | 2.14 × 10−2 |
| FOS | Fos proto-oncogene, AP-1 transcription factor subunit | 2.36 | 6.24 × 10−23 |
| FOSL1 | FOS like 1, AP-1 transcription factor subunit | 2.34 | 4.92 × 10−11 |
| ICAM1 | intercellular adhesion molecule 1 | 2.24 | 9.38 × 10−43 |
| BCL3 | B-cell CLL/lymphoma 3 | 2.19 | 6.45 × 10−35 |
| CCR6 | C-C motif chemokine receptor 6 | 2.13 | 3.81 × 10−4 |
| IRF1 | interferon regulatory factor 1 | 2.11 | 7.13 × 10−26 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, S.-J.; Jun, J.-S.; Hong, K.-W. Transcriptome Analysis Reveals Immunomodulatory Effect of Spore-Displayed p75 on Human Intestinal Epithelial Caco-2 Cells. Int. J. Mol. Sci. 2022, 23, 14519. https://doi.org/10.3390/ijms232314519
Kang S-J, Jun J-S, Hong K-W. Transcriptome Analysis Reveals Immunomodulatory Effect of Spore-Displayed p75 on Human Intestinal Epithelial Caco-2 Cells. International Journal of Molecular Sciences. 2022; 23(23):14519. https://doi.org/10.3390/ijms232314519
Chicago/Turabian StyleKang, Soo-Ji, Ji-Su Jun, and Kwang-Won Hong. 2022. "Transcriptome Analysis Reveals Immunomodulatory Effect of Spore-Displayed p75 on Human Intestinal Epithelial Caco-2 Cells" International Journal of Molecular Sciences 23, no. 23: 14519. https://doi.org/10.3390/ijms232314519
APA StyleKang, S.-J., Jun, J.-S., & Hong, K.-W. (2022). Transcriptome Analysis Reveals Immunomodulatory Effect of Spore-Displayed p75 on Human Intestinal Epithelial Caco-2 Cells. International Journal of Molecular Sciences, 23(23), 14519. https://doi.org/10.3390/ijms232314519






