Super-Resolution Microscopy to Study Interorganelle Contact Sites
Abstract
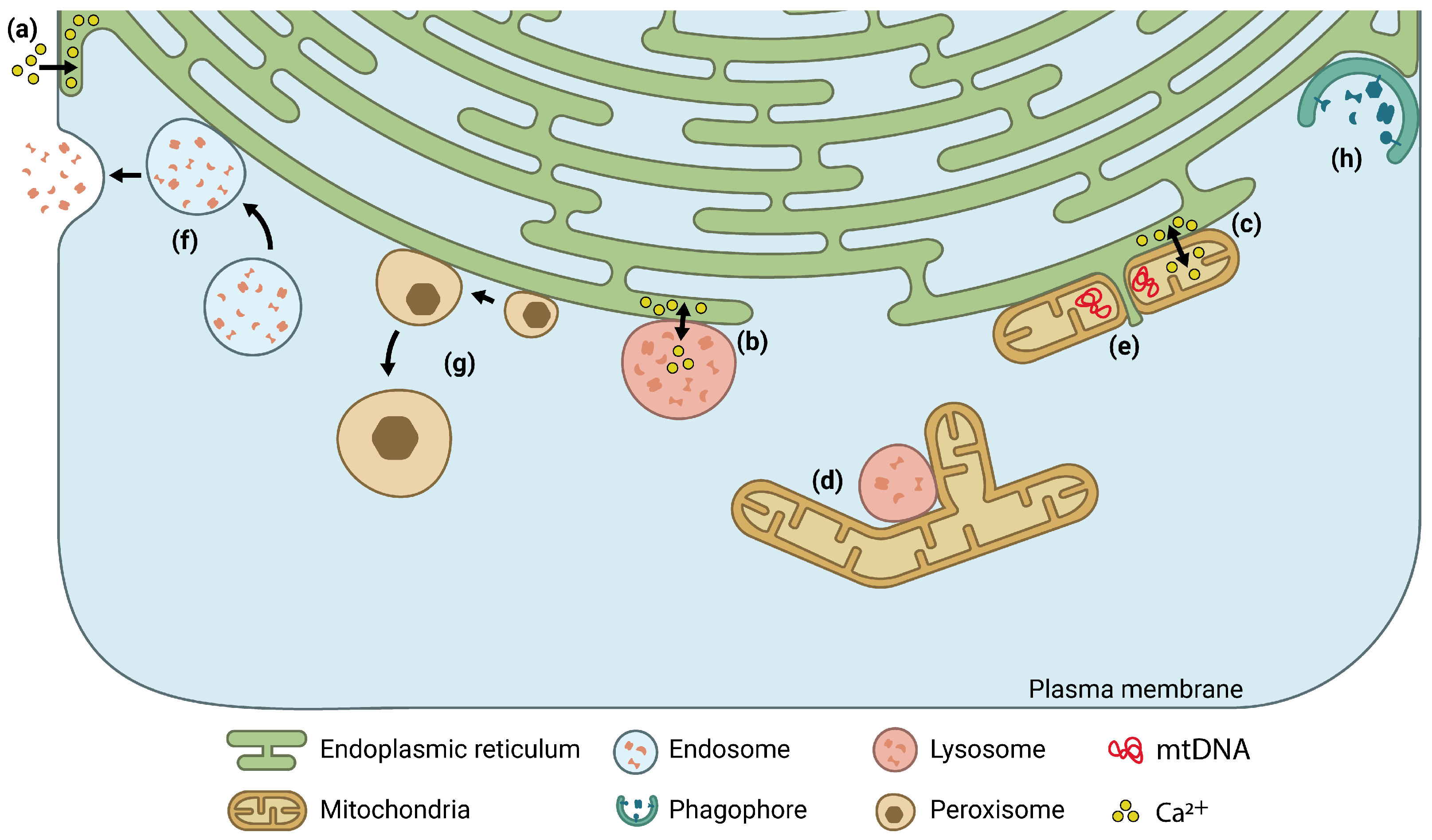
1. Introduction
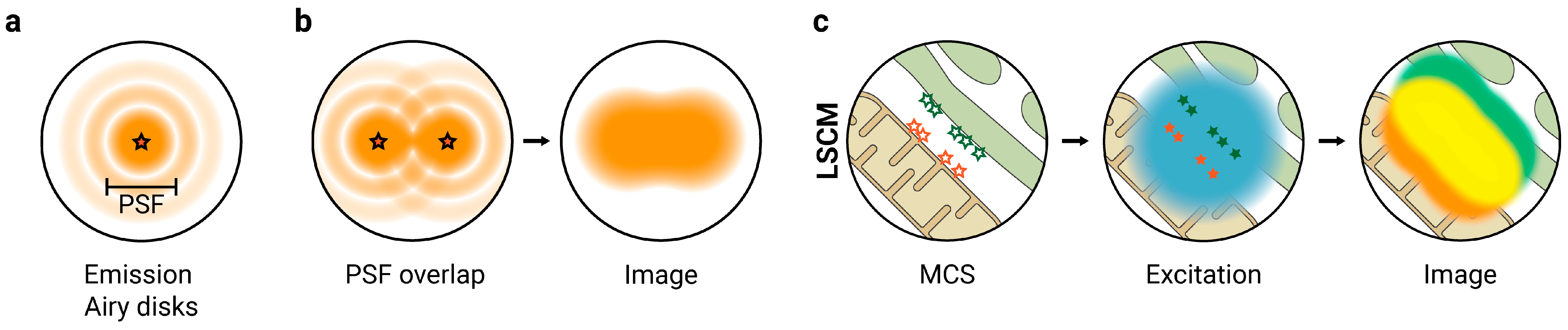
2. Super-Resolution Fluorescence Microscopy to Study MCS
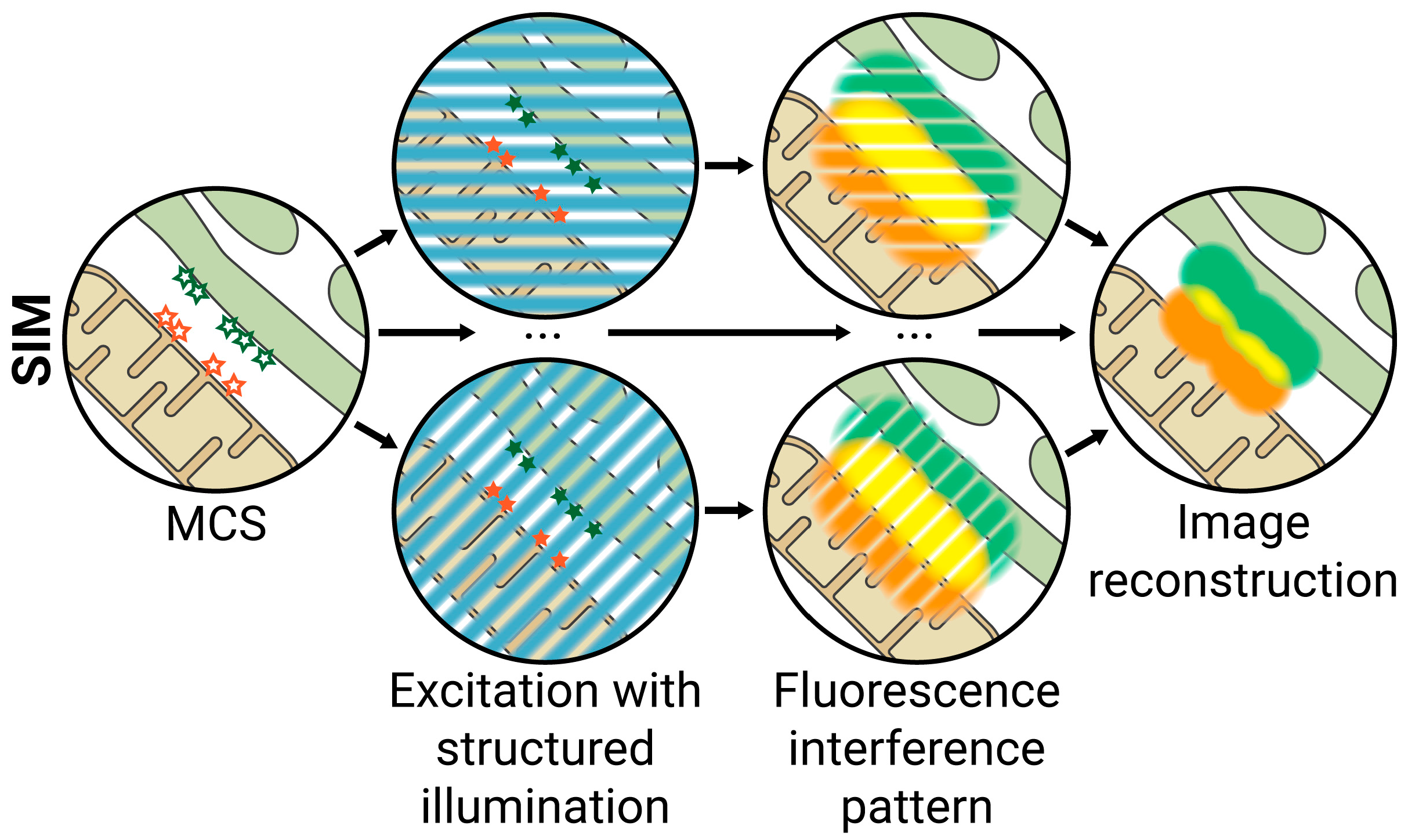
2.1. SIM
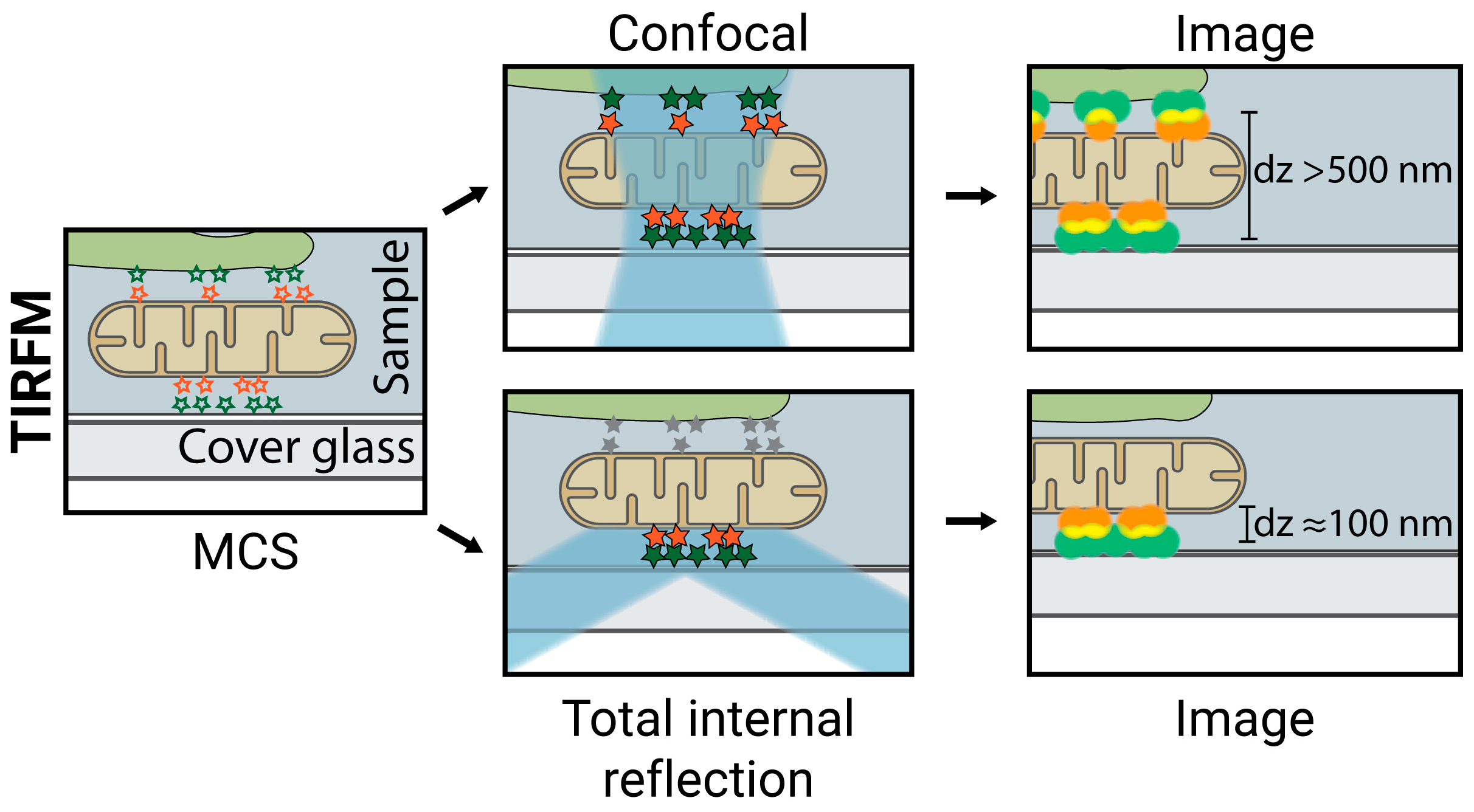
2.2. TIRFM
2.3. STED Microscopy
2.4. SMLM
3. Concluding Remarks and Outlook
Author Contributions
Funding
Conflicts of Interest
References
- Bernhard, W.; Rouiller, C. Close Topographical Relationship between Mitochondria and Ergastoplasm of Liver Cells in a Definite Phase of Cellular Activity. J. Biophys. Biochem. Cytol. 1956, 2, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Copeland, D.E.; Dalton, A.J. An Association between Mitochondria and the Endoplasmic Reticulum in Cells of the Pseudobranch Gland of a Teleost. J. Biophys. Biochem. Cytol. 1959, 5, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Vance, J.E. Phospholipid Synthesis in a Membrane Fraction Associated with Mitochondria. J. Biol. Chem. 1990, 265, 7248–7256. [Google Scholar] [CrossRef] [PubMed]
- Scorrano, L.; De Matteis, M.A.; Emr, S.; Giordano, F.; Hajnóczky, G.; Kornmann, B.; Lackner, L.L.; Levine, T.P.; Pellegrini, L.; Reinisch, K.; et al. Coming Together to Define Membrane Contact Sites. Nat. Commun. 2019, 10, 1287. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg-Bord, M.; Shai, N.; Schuldiner, M.; Bohnert, M. A Tether Is a Tether Is a Tether: Tethering at Membrane Contact Sites. Dev. Cell 2016, 39, 395–409. [Google Scholar] [CrossRef]
- Prinz, W.A.; Toulmay, A.; Balla, T. The Functional Universe of Membrane Contact Sites. Nat. Rev. Mol. Cell Biol. 2019, 21, 7–24. [Google Scholar] [CrossRef]
- Friedman, J.R.; Lackner, L.L.; West, M.; DiBenedetto, J.R.; Nunnari, J.; Voeltz, G.K. ER Tubules Mark Sites of Mitochondrial Division. Science 2011, 334, 358–362. [Google Scholar] [CrossRef]
- Manor, U.; Bartholomew, S.; Golani, G.; Christenson, E.; Kozlov, M.; Higgs, H.; Spudich, J.; Lippincott-Schwartz, J. A Mitochondria-Anchored Isoform of the Actin-Nucleating Spire Protein Regulates Mitochondrial Division. Elife 2015, 4, 828. [Google Scholar] [CrossRef]
- Korobova, F.; Ramabhadran, V.; Higgs, H.N. An Actin-Dependent Step in Mitochondrial Fission Mediated by the ER-Associated Formin INF2. Science 2013, 339, 464–467. [Google Scholar] [CrossRef]
- Korobova, F.; Gauvin, T.J.; Higgs, H.N. A Role for Myosin II in Mammalian Mitochondrial Fission. Curr. Biol. 2014, 24, 409–414. [Google Scholar] [CrossRef]
- Kwak, C.; Shin, S.; Park, J.-S.; Jung, M.; Nhung, T.T.M.; Kang, M.-G.; Lee, C.; Kwon, T.-H.; Park, S.K.; Mun, J.Y.; et al. Contact-ID, a Tool for Profiling Organelle Contact Sites, Reveals Regulatory Proteins of Mitochondrial-Associated Membrane Formation. Proc. Natl. Acad. Sci. USA 2020, 117, 12109–12120. [Google Scholar] [CrossRef] [PubMed]
- Kotschy, A.; Szlavik, Z.; Murray, J.; Davidson, J.; Maragno, A.L.; Le Toumelin-Braizat, G.; Chanrion, M.; Kelly, G.L.; Gong, J.-N.; Moujalled, D.M.; et al. The MCL1 Inhibitor S63845 Is Tolerable and Effective in Diverse Cancer Models. Nature 2016, 538, 477–482. [Google Scholar] [CrossRef] [PubMed]
- Hua, R.; Cheng, D.; Coyaud, É.; Freeman, S.; Di Pietro, E.; Wang, Y.; Vissa, A.; Yip, C.M.; Fairn, G.D.; Braverman, N.; et al. VAPs and ACBD5 Tether Peroxisomes to the ER for Peroxisome Maintenance and Lipid Homeostasis. J. Cell Biol. 2017, 216, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Hung, V.; Lam, S.S.; Udeshi, N.D.; Svinkina, T.; Guzman, G.; Mootha, V.K.; Carr, S.A.; Ting, A.Y. Proteomic Mapping of Cytosol-Facing Outer Mitochondrial and ER Membranes in Living Human Cells by Proximity Biotinylation. Elife 2017, 6, 463. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.F.; Branon, T.C.; Rajeev, S.; Svinkina, T.; Udeshi, N.D.; Thoudam, T.; Kwak, C.; Rhee, H.-W.; Lee, I.-K.; Carr, S.A.; et al. Split-TurboID Enables Contact-Dependent Proximity Labeling in Cells. Proc. Natl. Acad. Sci. USA 2020, 117, 12143–12154. [Google Scholar] [CrossRef]
- Area-Gomez, E.; Del Carmen Lara Castillo, M.; Tambini, M.D.; Guardia-Laguarta, C.; de Groof, A.J.C.; Madra, M.; Ikenouchi, J.; Umeda, M.; Bird, T.D.; Sturley, S.L.; et al. Upregulated Function of Mitochondria-Associated ER Membranes in Alzheimer Disease. EMBO J. 2012, 31, 4106–4123. [Google Scholar] [CrossRef]
- Hayashi, T.; Fujimoto, M. Detergent-Resistant Microdomains Determine the Localization of Sigma-1 Receptors to the Endoplasmic Reticulum-Mitochondria Junction. Mol. Pharmacol. 2010, 77, 517–528. [Google Scholar] [CrossRef]
- King, C.; Sengupta, P.; Seo, A.Y.; Lippincott-Schwartz, J. ER Membranes Exhibit Phase Behavior at Sites of Organelle Contact. Proc. Natl. Acad. Sci. USA 2020, 117, 7225–7235. [Google Scholar] [CrossRef]
- Lorizate, M.; Terrones, O.; Nieto-Garai, J.A.; Rojo-Bartolomé, I.; Ciceri, D.; Morana, O.; Olazar-Intxausti, J.; Arboleya, A.; Martin, A.; Szynkiewicz, M.; et al. Super-Resolution Microscopy Using a Bioorthogonal-Based Cholesterol Probe Provides Unprecedented Capabilities for Imaging Nanoscale Lipid Heterogeneity in Living Cells. Small Methods 2021, 5, 2100430. [Google Scholar] [CrossRef]
- Nieto-Garai, J.A.; Lorizate, M.; Contreras, F.-X. Shedding Light on Membrane Rafts Structure and Dynamics in Living Cells. Biochim. Biophys. Acta Biomembr. 2021, 1864, 183813. [Google Scholar] [CrossRef]
- Bianchetti, G.; Di Giacinto, F.; De Spirito, M.; Maulucci, G. Machine-Learning Assisted Confocal Imaging of Intracellular Sites of Triglycerides and Cholesteryl Esters Formation and Storage. Anal. Chim. Acta 2020, 1121, 57–66. [Google Scholar] [CrossRef]
- Rizzuto, R.; Pinton, P.; Carrington, W.; Fay, F.S.; Fogarty, K.E.; Lifshitz, L.M.; Tuft, R.A.; Pozzan, T. Close Contacts with the Endoplasmic Reticulum as Determinants of Mitochondrial Ca2+ Responses. Science 1998, 280, 1763–1766. [Google Scholar] [CrossRef] [PubMed]
- Csordás, G.; Weaver, D.; Hajnóczky, G. Endoplasmic Reticulum-Mitochondrial Contactology: Structure and Signaling Functions. Trends Cell Biol. 2018, 28, 523–540. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Carvalho, P.; Voeltz, G.K. Here, There, and Everywhere: The Importance of ER Membrane Contact Sites. Science 2018, 361, 835. [Google Scholar] [CrossRef] [PubMed]
- Nascimbeni, A.C.; Giordano, F.; Dupont, N.; Grasso, D.; Vaccaro, M.I.; Codogno, P.; Morel, E. ER –Plasma Membrane Contact Sites Contribute to Autophagosome Biogenesis by Regulation of Local PI 3P Synthesis. EMBO J. 2017, 36, 2018–2033. [Google Scholar] [CrossRef]
- Huang, X.; Jiang, C.; Yu, L.; Yang, A. Current and Emerging Approaches for Studying Inter-Organelle Membrane Contact Sites. Front. Cell Dev. Biol. 2020, 8, 195. [Google Scholar] [CrossRef]
- Madec, A.M.; Perrier, J.; Panthu, B.; Dingreville, F. Role of Mitochondria-Associated Endoplasmic Reticulum Membrane (MAMs) Interactions and Calcium Exchange in the Development of Type 2 Diabetes. Int. Rev. Cell Mol. Biol. 2021, 363, 169–202. [Google Scholar] [CrossRef]
- Rimessi, A.; Pedriali, G.; Vezzani, B.; Tarocco, A.; Marchi, S.; Wieckowski, M.R.; Giorgi, C.; Pinton, P. Interorganellar Calcium Signaling in the Regulation of Cell Metabolism: A Cancer Perspective. Semin. Cell Dev. Biol. 2020, 98, 167–180. [Google Scholar] [CrossRef]
- Petkovic, M.; O’Brien, C.E.; Jan, Y.N. Interorganelle Communication, Aging, and Neurodegeneration. Genes Dev. 2021, 35, 449–469. [Google Scholar] [CrossRef]
- Paillusson, S.; Stoica, R.; Gomez-Suaga, P.; Lau, D.H.W.; Mueller, S.; Miller, T.; Miller, C.C.J. There’s Something Wrong with My MAM; the ER-Mitochondria Axis and Neurodegenerative Diseases. Trends Neurosci. 2016, 39, 146–157. [Google Scholar] [CrossRef]
- Fernández-Busnadiego, R.; Saheki, Y.; De Camilli, P. Three-Dimensional Architecture of Extended Synaptotagmin-Mediated Endoplasmic Reticulum-Plasma Membrane Contact Sites. Proc. Natl. Acad. Sci. USA 2015, 112, E2004–E2013. [Google Scholar] [CrossRef] [PubMed]
- de Brito, O.M.; Scorrano, L. Mitofusin 2 Tethers Endoplasmic Reticulum to Mitochondria. Nature 2008, 456, 605–610. [Google Scholar] [CrossRef] [PubMed]
- Csordás, G.; Renken, C.; Várnai, P.; Walter, L.; Weaver, D.; Buttle, K.F.; Balla, T.; Mannella, C.A.; Hajnóczky, G. Structural and Functional Features and Significance of the Physical Linkage between ER and Mitochondria. J. Cell Biol. 2006, 2006, 16. [Google Scholar] [CrossRef] [PubMed]
- Wieckowski, M.R.M.R.; Giorgi, C.; Lebiedzinska, M.; Duszynski, J.; Pinton, P. Isolation of Mitochondria-Associated Membranes and Mitochondria from Animal Tissues and Cells. Nat. Protoc. 2009, 2009, 151. [Google Scholar] [CrossRef]
- Cook, K.C.; Tsopurashvili, E.; Needham, J.M.; Thompson, S.R.; Cristea, I.M. Restructured Membrane Contacts Rewire Organelles for Human Cytomegalovirus Infection. Nat. Commun. 2022, 13, 4720. [Google Scholar] [CrossRef]
- Sezgin, E. Super-Resolution Optical Microscopy for Studying Membrane Structure and Dynamics. J. Phys. Condens. Matter 2017, 29, 273001. [Google Scholar] [CrossRef]
- Schermelleh, L.; Ferrand, A.; Huser, T.; Eggeling, C.; Sauer, M.; Biehlmaier, O.; Drummen, G.P.C. Super-Resolution Microscopy Demystified. Nat. Cell Biol. 2019, 21, 72–84. [Google Scholar] [CrossRef]
- Nixon-Abell, J.; Obara, C.J.; Weigel, A.V.; Li, D.; Legant, W.R.; Xu, C.S.; Pasolli, H.A.; Harvey, K.; Hess, H.F.; Betzig, E.; et al. Increased Spatiotemporal Resolution Reveals Highly Dynamic Dense Tubular Matrices in the Peripheral ER. Science 2016, 354, 126. [Google Scholar] [CrossRef]
- Guo, Y.; Li, D.; Zhang, S.; Yang, Y.; Liu, J.J.; Wang, X.; Liu, C.; Milkie, D.E.; Moore, R.P.; Tulu, U.S.; et al. Visualizing Intracellular Organelle and Cytoskeletal Interactions at Nanoscale Resolution on Millisecond Timescales. Cell 2018, 175, 1430–1442.e17. [Google Scholar] [CrossRef]
- Qin, J.; Guo, Y.; Xue, B.; Shi, P.; Chen, Y.; Su, Q.P.; Hao, H.; Zhao, S.; Wu, C.; Yu, L.; et al. ER-Mitochondria Contacts Promote MtDNA Nucleoids Active Transportation via Mitochondrial Dynamic Tubulation. Nat. Commun. 2020, 11, 4471. [Google Scholar] [CrossRef]
- Van Alstyne, M.; Lotti, F.; Dal Mas, A.; Area-Gomez, E.; Pellizzoni, L. Stasimon/Tmem41b Localizes to Mitochondria-Associated ER Membranes and Is Essential for Mouse Embryonic Development. Biochem. Biophys. Res. Commun. 2018, 506, 463–470. [Google Scholar] [CrossRef]
- Swayne, T.C.; Zhou, C.; Boldogh, I.R.; Charalel, J.K.; McFaline-Figueroa, J.R.; Thoms, S.; Yang, C.; Leung, G.; McInnes, J.; Erdmann, R.; et al. Role for CER and Mmr1p in Anchorage of Mitochondria at Sites of Polarized Surface Growth in Budding Yeast. Curr. Biol. 2011, 21, 1994–1999. [Google Scholar] [CrossRef]
- Han, Y.; Li, M.; Qiu, F.; Zhang, M.; Zhang, Y.-H. Cell-Permeable Organic Fluorescent Probes for Live-Cell Long-Term Super-Resolution Imaging Reveal Lysosome-Mitochondrion Interactions. Nat. Commun. 2017, 8, 1307. [Google Scholar] [CrossRef] [PubMed]
- Raiborg, C.; Wenzel, E.M.; Pedersen, N.M.; Olsvik, H.; Schink, K.O.; Schultz, S.W.; Vietri, M.; Nisi, V.; Bucci, C.; Brech, A.; et al. Repeated ER-Endosome Contacts Promote Endosome Translocation and Neurite Outgrowth. Nature 2015, 520, 234–238. [Google Scholar] [CrossRef]
- Xu, D.; Li, Y.; Wu, L.; Li, Y.; Zhao, D.; Yu, J.; Huang, T.; Ferguson, C.; Parton, R.G.; Yang, H.; et al. Rab18 Promotes Lipid Droplet (LD) Growth by Tethering the ER to LDs through SNARE and NRZ Interactions. J. Cell Biol. 2018, 217, 975–995. [Google Scholar] [CrossRef] [PubMed]
- Dong, D.; Huang, X.; Li, L.; Mao, H.; Mo, Y.; Zhang, G.; Zhang, Z.; Shen, J.; Liu, W.; Wu, Z.; et al. Super-Resolution Fluorescence-Assisted Diffraction Computational Tomography Reveals the Three-Dimensional Landscape of the Cellular Organelle Interactome. Light Sci. Appl. 2020, 9, 494. [Google Scholar] [CrossRef] [PubMed]
- Besprozvannaya, M.; Dickson, E.; Li, H.; Ginburg, K.S.; Bers, D.M.; Auwerx, J.; Nunnari, J. GRAM Domain Proteins Specialize Functionally Distinct ER-PM Contact Sites in Human Cells. Elife 2018, 7, 19. [Google Scholar] [CrossRef]
- Guo, M.; Chandris, P.; Giannini, J.P.; Trexler, A.J.; Fischer, R.; Chen, J.; Vishwasrao, H.D.; Rey-Suarez, I.; Wu, Y.; Wu, X.; et al. Single-Shot Super-Resolution Total Internal Reflection Fluorescence Microscopy. Nat. Methods 2018, 15, 425–428. [Google Scholar] [CrossRef]
- Liou, J.; Kim, M.L.; Heo, W.D.; Jones, J.T.; Myers, J.W.; Ferrell, J.E.; Meyer, T. STIM Is a Ca2+ Sensor Essential for Ca2+-Store-Depletion-Triggered Ca2+ Influx. Curr. Biol. 2005, 15, 1235–1241. [Google Scholar] [CrossRef]
- Wu, M.M.; Buchanan, J.A.; Luik, R.M.; Lewis, R.S. Ca2+ Store Depletion Causes STIM1 to Accumulate in ER Regions Closely Associated with the Plasma Membrane. J. Cell Biol. 2006, 174, 803–813. [Google Scholar] [CrossRef]
- Kang, F.; Zhou, M.; Huang, X.; Fan, J.; Wei, L.; Boulanger, J.; Liu, Z.; Salamero, J.; Liu, Y.; Chen, L. E-Syt1 Re-Arranges STIM1 Clusters to Stabilize Ring-Shaped ER-PM Contact Sites and Accelerate Ca2+ Store Replenishment. Sci. Rep. 2019, 9, 3975. [Google Scholar] [CrossRef] [PubMed]
- Atakpa, P.; Thillaiappan, N.B.; Mataragka, S.; Prole, D.L.; Taylor, C.W. IP3 Receptors Preferentially Associate with ER-Lysosome Contact Sites and Selectively Deliver Ca2+ to Lysosomes. Cell Rep. 2018, 25, 3180–3193.e7. [Google Scholar] [CrossRef]
- Kim, Y.J.; Guzman-Hernandez, M.L.; Wisniewski, E.; Balla, T. Phosphatidylinositol-Phosphatidic Acid Exchange by Nir2 at ER-PM Contact Sites Maintains Phosphoinositide Signaling Competence. Dev. Cell 2015, 33, 549–561. [Google Scholar] [CrossRef]
- Bottanelli, F.; Kromann, E.B.; Allgeyer, E.S.; Erdmann, R.S.; Wood Baguley, S.; Sirinakis, G.; Schepartz, A.; Baddeley, D.; Toomre, D.K.; Rothman, J.E.; et al. Two-Colour Live-Cell Nanoscale Imaging of Intracellular Targets. Nat. Commun. 2016, 7, 10778. [Google Scholar] [CrossRef] [PubMed]
- Damenti, M.; Coceano, G.; Pennacchietti, F.; Bodén, A.; Testa, I. STED and Parallelized RESOLFT Optical Nanoscopy of the Tubular Endoplasmic Reticulum and Its Mitochondrial Contacts in Neuronal Cells. Neurobiol. Dis. 2021, 155, 105361. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Stephan, T.; Chen, P.; Chen, J.; Riedel, D.; Yang, Z.; Jakobs, S.; Chen, Z. Multi-Color Live-Cell STED Nanoscopy of Mitochondria with a Gentle Inner Membrane Stain. bioRxiv 2022. [Google Scholar] [CrossRef]
- Gemmink, A.; Daemen, S.; Kuijpers, H.J.H.H.; Schaart, G.; Duimel, H.; López-Iglesias, C.; van Zandvoort, M.A.M.J.M.J.; Knoops, K.; Hesselink, M.K.C.C. Super-Resolution Microscopy Localizes Perilipin 5 at Lipid Droplet-Mitochondria Interaction Sites and at Lipid Droplets Juxtaposing to Perilipin 2. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 1423–1432. [Google Scholar] [CrossRef]
- Shim, S.H.; Xia, C.; Zhong, G.; Babcock, H.P.; Vaughan, J.C.; Huang, B.; Wang, X.; Xu, C.; Bi, G.Q.; Zhuang, X. Super-Resolution Fluorescence Imaging of Organelles in Live Cells with Photoswitchable Membrane Probes. Proc. Natl. Acad. Sci. USA 2012, 109, 13978–13983. [Google Scholar] [CrossRef]
- Modi, S.; López-Doménech, G.; Halff, E.F.; Covill-Cooke, C.; Ivankovic, D.; Melandri, D.; Arancibia-Cárcamo, I.L.; Burden, J.J.; Lowe, A.R.; Kittler, J.T. Miro Clusters Regulate ER-Mitochondria Contact Sites and Link Cristae Organization to the Mitochondrial Transport Machinery. Nat. Commun. 2019, 10, 4399. [Google Scholar] [CrossRef]
- Hsieh, T.-S.; Chen, Y.-J.; Chang, C.-L.; Lee, W.-R.; Liou, J. Cortical Actin Contributes to Spatial Organization of ER-PM Junctions. Mol. Biol. Cell 2017, 28, 3171–3180. [Google Scholar] [CrossRef]
- Yan, R.; Chen, K.; Xu, K. Probing Nanoscale Diffusional Heterogeneities in Cellular Membranes through Multidimensional Single-Molecule and Super-Resolution Microscopy. J. Am. Chem. Soc. 2020, 142, 18866. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, M.G.L. Surpassing the Lateral Resolution Limit by a Factor of Two Using Structured Illumination Microscopy. J. Microsc. 2000, 198, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, M.G.L.; Shao, L.; Carlton, P.M.; Wang, C.J.R.; Golubovskaya, I.N.; Cande, W.Z.; Agard, D.A.; Sedat, J.W. Three-Dimensional Resolution Doubling in Wide-Field Fluorescence Microscopy by Structured Illumination. Biophys. J. 2008, 94, 4957–4970. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, M.G.L. Nonlinear Structured-Illumination Microscopy: Wide-Field Fluorescence Imaging with Theoretically Unlimited Resolution. Proc. Natl. Acad. Sci. USA 2005, 102, 13081–13086. [Google Scholar] [CrossRef]
- Gottschalk, B.; Koshenov, Z.; Bachkoenig, O.A.; Rost, R.; Malli, R.; Graier, W.F. MFN2 Mediates ER-Mitochondrial Coupling during ER Stress through Specialized Stable Contact Sites. Front. Cell Dev. Biol. 2022, 10. [Google Scholar] [CrossRef]
- Fang, G.; Chen, H.; Shao, X.; Wang, H.; Zhan, D.; Wang, R.; Meng, P.; Fang, H.; Liu, F.; Ling, P.; et al. Single Image Capture of Bioactive Ion Crosstalk within Inter-Organelle Membrane Contacts at Nanometer Resolution. Small Methods 2022, 6, 2200321. [Google Scholar] [CrossRef]
- Wong, Y.C.; Kim, S.; Cisneros, J.; Molakal, C.G.; Song, P.; Lubbe, S.J.; Krainc, D. Mid51/Fis1 Mitochondrial Oligomerization Complex Drives Lysosomal Untethering and Network Dynamics. J. Cell Biol. 2022, 221, 140. [Google Scholar] [CrossRef]
- Axelrod, D. Cell-Substrate Contacts Illuminated by Total Internal Reflection Fluorescence. J. Cell Biol. 1981, 89, 141–145. [Google Scholar] [CrossRef]
- Poulter, N.S.; Pitkeathly, W.T.E.; Smith, P.J.; Rappoport, J.Z. The Physical Basis of Total Internal Reflection Fluorescence (TIRF) Microscopy and Its Cellular Applications. Methods Mol. Biol. 2015, 1251, 1–23. [Google Scholar] [CrossRef]
- Hell, S.W.; Wichmann, J. Breaking the Diffraction Resolution Limit by Stimulated Emission: Stimulated-Emission-Depletion Fluorescence Microscopy. Opt. Lett. 1994, 19, 780. [Google Scholar] [CrossRef]
- Klar, T.A.; Hell, S.W. Subdiffraction Resolution in Far-Field Fluorescence Microscopy. Opt. Lett. 1999, 24, 954. [Google Scholar] [CrossRef] [PubMed]
- Vicidomini, G.; Bianchini, P.; Diaspro, A. STED Super-Resolved Microscopy. Nat. Methods 2018, 15, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Masullo, L.A.; Bodén, A.; Pennacchietti, F.; Coceano, G.; Ratz, M.; Testa, I. Enhanced Photon Collection Enables Four Dimensional Fluorescence Nanoscopy of Living Systems. Nat. Commun. 2018, 9, 3281. [Google Scholar] [CrossRef] [PubMed]
- Rust, M.J.; Bates, M.; Zhuang, X. Sub-Diffraction-Limit Imaging by Stochastic Optical Reconstruction Microscopy (STORM). Nat. Methods 2006, 3, 793–795. [Google Scholar] [CrossRef]
- Hess, S.T.; Girirajan, T.P.K.; Mason, M.D. Ultra-High Resolution Imaging by Fluorescence Photoactivation Localization Microscopy. Biophys. J. 2006, 91, 4258–4272. [Google Scholar] [CrossRef]
- Betzig, E.; Patterson, G.H.; Sougrat, R.; Lindwasser, O.W.; Olenych, S.; Bonifacino, J.S.; Davidson, M.W.; Lippincott-Schwartz, J.; Hess, H.F. Imaging Intracellular Fluorescent Proteins at Nanometer Resolution. Science 2006, 313, 1642–1645. [Google Scholar] [CrossRef]
- Sharonov, A.; Hochstrasser, R.M. Wide-Field Subdiffraction Imaging by Accumulated Binding of Diffusing Probes. Proc. Natl. Acad. Sci. USA 2006, 103, 18911–18916. [Google Scholar] [CrossRef]
- Mehlitz, A.; Karunakaran, K.; Herweg, J.A.; Krohne, G.; van de Linde, S.; Rieck, E.; Sauer, M.; Rudel, T. The Chlamydial Organism Simkania Negevensis Forms ER Vacuole Contact Sites and Inhibits ER-Stress. Cell. Microbiol. 2014, 16, 1224–1243. [Google Scholar] [CrossRef]
- Balzarotti, F.; Eilers, Y.; Gwosch, K.C.; Gynnå, A.H.; Westphal, V.; Stefani, F.D.; Elf, J.; Hell, S.W. Nanometer Resolution Imaging and Tracking of Fluorescent Molecules with Minimal Photon Fluxes. Science 2017, 355, 606–612. [Google Scholar] [CrossRef]
- Eilers, Y.; Ta, H.; Gwosch, K.C.; Balzarotti, F.; Hell, S.W. MINFLUX Monitors Rapid Molecular Jumps with Superior Spatiotemporal Resolution. Proc. Natl. Acad. Sci. USA 2018, 115, 6117–6122. [Google Scholar] [CrossRef]
- Gwosch, K.C.; Pape, J.K.; Balzarotti, F.; Hoess, P.; Ellenberg, J.; Ries, J.; Hell, S.W. MINFLUX Nanoscopy Delivers 3D Multicolor Nanometer Resolution in Cells. Nat. Methods 2020, 17, 880. [Google Scholar] [CrossRef] [PubMed]
- Pape, J.K.; Stephan, T.; Balzarotti, F.; Büchner, R.; Lange, F.; Riedel, D.; Jakobs, S.; Hell, S.W. Multicolor 3D MINFLUX Nanoscopy of Mitochondrial MICOS Proteins. Proc. Natl. Acad. Sci. USA 2020, 117, 20607–20614. [Google Scholar] [CrossRef] [PubMed]
- Corradi, V.; Sejdiu, B.I.; Mesa-Galloso, H.; Abdizadeh, H.; Noskov, S.Y.; Marrink, S.J.; Tieleman, D.P. Emerging Diversity in Lipid-Protein Interactions. Chem. Rev. 2019, 119, 5775–5848. [Google Scholar] [CrossRef]
- Lingwood, D.; Simons, K. Lipid Rafts As a Membrane-Organizing Principle. Science 2010, 327, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Cebecauer, M.; Amaro, M.; Jurkiewicz, P.; Sarmento, M.J.; Šachl, R.; Cwiklik, L.; Hof, M. Membrane Lipid Nanodomains. Chem. Rev. 2018, 118, 11259–11297. [Google Scholar] [CrossRef] [PubMed]
- Blouin, C.M.; Hamon, Y.; Gonnord, P.; Boularan, C.; Kagan, J.; Viaris de Lesegno, C.; Ruez, R.; Mailfert, S.; Bertaux, N.; Loew, D.; et al. Glycosylation-Dependent IFN-ΓR Partitioning in Lipid and Actin Nanodomains Is Critical for JAK Activation. Cell 2016, 166, 920–934. [Google Scholar] [CrossRef]
- Morana, O.; Nieto-Garai, J.A.; Björkholm, P.; Bernardino de la Serna, J.; Terrones, O.; Arboleya, A.; Ciceri, D.; Rojo-Bartolomé, I.; Blouin, C.M.; Lamaze, C.; et al. Identification of a New Cholesterol-Binding Site within the IFN-γ Receptor That Is Required for Signal Transduction. Adv. Sci. 2022, 2022, 170. [Google Scholar] [CrossRef] [PubMed]
- Nieto-Garai, J.A.; Arboleya, A.; Otaegi, S.; Chojnacki, J.; Casas, J.; Fabriàs, G.; Contreras, F.; Kräusslich, H.; Lorizate, M. Cholesterol in the Viral Membrane Is a Molecular Switch Governing HIV-1 Env Clustering. Adv. Sci. 2021, 8, 2003468. [Google Scholar] [CrossRef]
- Katan, M.; Cockcroft, S. Phosphatidylinositol(4,5)Bisphosphate: Diverse Functions at the Plasma Membrane. Essays Biochem. 2020, 64, 513–531. [Google Scholar] [CrossRef]
- Klymchenko, A.S.; Kreder, R. Fluorescent Probes for Lipid Rafts: From Model Membranes to Living Cells. Chem. Biol. 2014, 21, 97–113. [Google Scholar] [CrossRef]
- Bianchetti, G.; Azoulay-Ginsburg, S.; Keshet-Levy, N.Y.; Malka, A.; Zilber, S.; Korshin, E.E.; Sasson, S.; De Spirito, M.; Gruzman, A.; Maulucci, G. Investigation of the Membrane Fluidity Regulation of Fatty Acid Intracellular Distribution by Fluorescence Lifetime Imaging of Novel Polarity Sensitive Fluorescent Derivatives. Int. J. Mol. Sci. 2021, 22, 3106. [Google Scholar] [CrossRef] [PubMed]


| SRFM Technique | Maximum Lateral (d) and Axial (dz) Resolutions (nm) | Advantages | Limitations | References |
|---|---|---|---|---|
| SIM | d ≈ 100, dz ≈ 300 | High sensitivity, common fluorophores, allows 3D imaging | Limited spatial resolution | [13,38,39,40,41,42,43,44,45,46] |
| TIRFM | d ≈ 200, dz ≈ 100 | High axial resolution, common fluorophores | Only images close to PM, limited lateral resolution | [47,48,49,50,51,52,53] |
| STED | d ≈ 30, dz ≈ 100 | High lateral and axial resolutions, allows 3D imaging | Limited multi-color imaging, photostable fluorophores | [25,54,55,56,57] |
| SMLM | d ≈ 20, dz ≈ 50 | Very high lateral and axial resolutions | Specialized fluorophores and buffers, limited temporal resolution, limited 3D imaging | [58,59,60,61] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nieto-Garai, J.A.; Olazar-Intxausti, J.; Anso, I.; Lorizate, M.; Terrones, O.; Contreras, F.-X. Super-Resolution Microscopy to Study Interorganelle Contact Sites. Int. J. Mol. Sci. 2022, 23, 15354. https://doi.org/10.3390/ijms232315354
Nieto-Garai JA, Olazar-Intxausti J, Anso I, Lorizate M, Terrones O, Contreras F-X. Super-Resolution Microscopy to Study Interorganelle Contact Sites. International Journal of Molecular Sciences. 2022; 23(23):15354. https://doi.org/10.3390/ijms232315354
Chicago/Turabian StyleNieto-Garai, Jon Ander, June Olazar-Intxausti, Itxaso Anso, Maier Lorizate, Oihana Terrones, and Francesc-Xabier Contreras. 2022. "Super-Resolution Microscopy to Study Interorganelle Contact Sites" International Journal of Molecular Sciences 23, no. 23: 15354. https://doi.org/10.3390/ijms232315354
APA StyleNieto-Garai, J. A., Olazar-Intxausti, J., Anso, I., Lorizate, M., Terrones, O., & Contreras, F.-X. (2022). Super-Resolution Microscopy to Study Interorganelle Contact Sites. International Journal of Molecular Sciences, 23(23), 15354. https://doi.org/10.3390/ijms232315354






