Genetic Variation of SAMM50 Is Not an Independent Risk Factor for Alcoholic Hepatocellular Carcinoma in Caucasian Patients
Abstract
:1. Introduction
2. Results
2.1. Study Population
2.2. Genotype Distribution
2.3. Frequency of the SAMM50 Minor Variants
2.4. Uni- and Multivariate Analysis for Various Risk Factors
2.5. Analysis of Linkage Disequilibrium between SAMM50 Variants and the PNPLA3 148M Variant
3. Patients and Methods
3.1. Patients
3.2. Methods
Determination of SAMM50 and PNPLA3 Genotypes
3.3. Statistical Analysis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ANOVA | Analysis of Variance |
| ALT | alanine aminotransferase |
| AST | aspartate aminotransferase |
| HCC | hepatocellular carcinoma |
| HCV | hepatitis C virus |
| HSD17B13 | 17β-Hydroxysteroid dehydrogenase type 13 |
| GGT | Gamma-glutamyl transferase |
| MAF | minor allele frequency |
| MELD | Model for End-Stage Liver Disease |
| NAFLD | non-alcoholic fatty liver disease |
| NASH | non-alcoholic steatohepatitis |
| NF-kB | nuclear factor ‘kappa-light-chain-enhancer’ of activated B-cells |
| PNPLA3 | patatin-like phospholipase domain-containing protein 3 |
| OR | odds ratio |
| SAMM50 | Sorting and assembly machinery component 50 homolog |
| TLR | toll-like receptor |
| TM6SF2 | Transmembrane 6 superfamily 2 |
References
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular Carcinoma. Nat. Rev. Dis. Primer 2021, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Galle, P.R.; Forner, A.; Llovet, J.M.; Mazzaferro, V.; Piscaglia, F.; Raoul, J.-L.; Schirmacher, P.; Vilgrain, V. EASL Clinical Practice Guidelines: Management of Hepatocellular Carcinoma. J. Hepatol. 2018, 69, 182–236. [Google Scholar] [CrossRef] [Green Version]
- Heimbach, J.K.; Kulik, L.M.; Finn, R.S.; Sirlin, C.B.; Abecassis, M.M.; Roberts, L.R.; Zhu, A.X.; Murad, M.H.; Marrero, J.A. AASLD Guidelines for the Treatment of Hepatocellular Carcinoma. Hepatology 2018, 67, 358–380. [Google Scholar] [CrossRef] [Green Version]
- Frenette, C.T.; Isaacson, A.J.; Bargellini, I.; Saab, S.; Singal, A.G. A Practical Guideline for Hepatocellular Carcinoma Screening in Patients at Risk. Mayo Clin. Proc. Innov. Qual. Outcomes 2019, 3, 302–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singal, A.G.; Lampertico, P.; Nahon, P. Epidemiology and Surveillance for Hepatocellular Carcinoma: New Trends. J. Hepatol. 2020, 72, 250–261. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Liu, L. Changes in the Epidemiology of Hepatocellular Carcinoma in Asia. Cancers 2022, 14, 4473. [Google Scholar] [CrossRef]
- Stickel, F.; Buch, S.; Nischalke, H.D.; Weiss, K.H.; Gotthardt, D.; Fischer, J.; Rosendahl, J.; Marot, A.; Elamly, M.; Casper, M.; et al. Genetic Variants in PNPLA3 and TM6SF2 Predispose to the Development of Hepatocellular Carcinoma in Individuals with Alcohol-Related Cirrhosis. Am. J. Gastroenterol. 2018, 113, 1475–1483. [Google Scholar] [CrossRef]
- Buch, S.; Innes, H.; Lutz, P.L.; Nischalke, H.D.; Marquardt, J.U.; Fischer, J.; Weiss, K.H.; Rosendahl, J.; Marot, A.; Krawczyk, M.; et al. Genetic Variation in TERT Modifies the Risk of Hepatocellular Carcinoma in Alcohol-Related Cirrhosis: Results from a Genome-Wide Case-Control Study. Gut 2022. [Google Scholar] [CrossRef]
- Abul-Husn, N.S.; Cheng, X.; Li, A.H.; Xin, Y.; Schurmann, C.; Stevis, P.; Liu, Y.; Kozlitina, J.; Stender, S.; Wood, G.C.; et al. A Protein-Truncating HSD17B13 Variant and Protection from Chronic Liver Disease. N. Engl. J. Med. 2018, 378, 1096–1106. [Google Scholar] [CrossRef]
- Stickel, F.; Lutz, P.; Buch, S.; Nischalke, H.D.; Silva, I.; Rausch, V.; Fischer, J.; Weiss, K.H.; Gotthardt, D.; Rosendahl, J.; et al. Genetic Variation in HSD17B13 Reduces the Risk of Developing Cirrhosis and Hepatocellular Carcinoma in Alcohol Misusers. Hepatology 2020, 72, 88–102. [Google Scholar] [CrossRef]
- Fujiwara, N.; Hoshida, Y. Hepatocellular Carcinoma Risk Stratification by Genetic Profiling in Patients with Cirrhosis. Semin. Liver Dis. 2019, 39, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Bianco, C.; Jamialahmadi, O.; Pelusi, S.; Baselli, G.; Dongiovanni, P.; Zanoni, I.; Santoro, L.; Maier, S.; Liguori, A.; Meroni, M.; et al. Non-Invasive Stratification of Hepatocellular Carcinoma Risk in Non-Alcoholic Fatty Liver Using Polygenic Risk Scores. J. Hepatol. 2021, 74, 775–782. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.E.; Diergaarde, B.; Kuipers, A.L.; Adibi, J.J.; Luu, H.N.; Chang, X.; Dorajoo, R.; Heng, C.; Khor, C.; Wang, R.; et al. NAFLD Polygenic Risk Score and Risk of Hepatocellular Carcinoma in an East Asian Population. Hepatol. Commun. 2022, 6, 2310–2321. [Google Scholar] [CrossRef]
- Degasperi, E.; Galmozzi, E.; Pelusi, S.; D’Ambrosio, R.; Soffredini, R.; Borghi, M.; Perbellini, R.; Facchetti, F.; Iavarone, M.; Sangiovanni, A.; et al. Hepatic Fat-Genetic Risk Score Predicts Hepatocellular Carcinoma in Patients with Cirrhotic HCV Treated with DAAs. Hepatology 2020, 72, 1912–1923. [Google Scholar] [CrossRef]
- Whitfield, J.B.; Schwantes-An, T.-H.; Darlay, R.; Aithal, G.P.; Atkinson, S.R.; Bataller, R.; Botwin, G.; Chalasani, N.P.; Cordell, H.J.; Daly, A.K.; et al. A Genetic Risk Score and Diabetes Predict Development of Alcohol-Related Cirrhosis in Drinkers. J. Hepatol. 2022, 76, 275–282. [Google Scholar] [CrossRef]
- Wang, Z.; Budhu, A.S.; Shen, Y.; Wong, L.L.; Hernandez, B.Y.; Tiirikainen, M.; Ma, X.; Irwin, M.L.; Lu, L.; Zhao, H.; et al. Genetic Susceptibility to Hepatocellular Carcinoma in Chromosome 22q13.31, Findings of a Genome-wide Association Study. JGH Open 2021, 5, 1363–1372. [Google Scholar] [CrossRef] [PubMed]
- Kitamoto, T.; Kitamoto, A.; Yoneda, M.; Hyogo, H.; Ochi, H.; Nakamura, T.; Teranishi, H.; Mizusawa, S.; Ueno, T.; Chayama, K.; et al. Genome-Wide Scan Revealed That Polymorphisms in the PNPLA3, SAMM50, and PARVB Genes Are Associated with Development and Progression of Nonalcoholic Fatty Liver Disease in Japan. Hum. Genet. 2013, 132, 783–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Shen, W.; Wu, G.; Qin, C.; Zhang, Y.; Wang, Y.; Song, G.; Xiao, C.; Zhang, X.; Deng, G.; et al. The Role of SAMM50 in Non-alcoholic Fatty Liver Disease: From Genetics to Mechanisms. FEBS Open Bio 2021, 11, 1893–1906. [Google Scholar] [CrossRef]
- Yuan, X.; Waterworth, D.; Perry, J.R.B.; Lim, N.; Song, K.; Chambers, J.C.; Zhang, W.; Vollenweider, P.; Stirnadel, H.; Johnson, T.; et al. Population-Based Genome-Wide Association Studies Reveal Six Loci Influencing Plasma Levels of Liver Enzymes. Am. J. Hum. Genet. 2008, 83, 520–528. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Conti, D.V.; Bogumil, D.; Sheng, X.; Noureddin, M.; Wilkens, L.R.; Le Marchand, L.; Rosen, H.R.; Haiman, C.A.; Setiawan, V.W. Association of Genetic Risk Score with NAFLD in An Ethnically Diverse Cohort. Hepatol. Commun. 2021, 5, 1689–1703. [Google Scholar] [CrossRef]
- Ott, C.; Ross, K.; Straub, S.; Thiede, B.; Götz, M.; Goosmann, C.; Krischke, M.; Mueller, M.J.; Krohne, G.; Rudel, T.; et al. Sam50 Functions in Mitochondrial Intermembrane Space Bridging and Biogenesis of Respiratory Complexes. Mol. Cell. Biol. 2012, 32, 1173–1188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.-J.; Shon, D.-H.; Kim, J.-H.; Ryu, Y.-H.; Ko, Y. SAMM50 Regulates Thermogenesis of Beige Adipocytes Differentiated from Human Adipose-Derived Stem Cells by Balancing Mitochondrial Dynamics. Int. J. Mol. Sci. 2022, 23, 6764. [Google Scholar] [CrossRef] [PubMed]
- Karkucinska-Wieckowska, A.; Simoes, I.C.M.; Kalinowski, P.; Lebiedzinska-Arciszewska, M.; Zieniewicz, K.; Milkiewicz, P.; Górska-Ponikowska, M.; Pinton, P.; Malik, A.N.; Krawczyk, M.; et al. Mitochondria, Oxidative Stress and Nonalcoholic Fatty Liver Disease: A Complex Relationship. Eur. J. Clin. Investig. 2022, 52, e13622. [Google Scholar] [CrossRef] [PubMed]
- 1000 Genomes Project Consortium; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A Global Reference for Human Genetic Variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef] [Green Version]
- Seitz, H.K.; Bataller, R.; Cortez-Pinto, H.; Gao, B.; Gual, A.; Lackner, C.; Mathurin, P.; Mueller, S.; Szabo, G.; Tsukamoto, H. Alcoholic Liver Disease. Nat. Rev. Dis. Primer 2018, 4, 16. [Google Scholar] [CrossRef]
- Trépo, E.; Caruso, S.; Yang, J.; Imbeaud, S.; Couchy, G.; Bayard, Q.; Letouzé, E.; Ganne-Carrié, N.; Moreno, C.; Oussalah, A.; et al. Common Genetic Variation in Alcohol-Related Hepatocellular Carcinoma: A Case-Control Genome-Wide Association Study. Lancet Oncol. 2022, 23, 161–171. [Google Scholar] [CrossRef]
- Innes, H.; Morling, J.R.; Buch, S.; Hamill, V.; Stickel, F.; Guha, I.N. Performance of Routine Risk Scores for Predicting Cirrhosis-Related Morbidity in the Community. J. Hepatol. 2022, 77, 365–376. [Google Scholar] [CrossRef]
- Nischalke, H.D.; Fischer, J.; Klüners, A.; Matz-Soja, M.; Krämer, B.; Langhans, B.; Goeser, F.; Soyka, M.; Stickel, F.; Spengler, U.; et al. A Genetic Variant in Toll-like Receptor 5 Is Linked to Chemokine Levels and Hepatocellular Carcinoma in Steatohepatitis. Liver Int. Off. J. Int. Assoc. Study Liver 2021, 41, 2139–2148. [Google Scholar] [CrossRef]
- Nischalke, H.D.; Klüners, A.; Nattermann, J.; Berg, T.; Strassburg, C.P.; Lutz, P. Hepatocellular Carcinoma Prevention by Aspirin: Are Platelets the Link? Hepatol. Commun. 2021, 5, 2151–2152. [Google Scholar] [CrossRef]
- Liu, S.; Gao, Y.; Zhang, C.; Li, H.; Pan, S.; Wang, X.; Du, S.; Deng, Z.; Wang, L.; Song, Z.; et al. SAMM50 Affects Mitochondrial Morphology through the Association of Drp1 in Mammalian Cells. FEBS Lett. 2016, 590, 1313–1323. [Google Scholar] [CrossRef]
- Chiusolo, V.; Jacquemin, G.; Yonca Bassoy, E.; Vinet, L.; Liguori, L.; Walch, M.; Kozjak-Pavlovic, V.; Martinvalet, D. Granzyme B Enters the Mitochondria in a Sam50-, Tim22- and MtHsp70-Dependent Manner to Induce Apoptosis. Cell Death Differ. 2017, 24, 747–758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lionello, S.; Marzaro, G.; Martinvalet, D. SAM50, a Side Door to the Mitochondria: The Case of Cytotoxic Proteases. Pharmacol. Res. 2020, 160, 105196. [Google Scholar] [CrossRef] [PubMed]
- Abudu, Y.P.; Shrestha, B.K.; Zhang, W.; Palara, A.; Brenne, H.B.; Larsen, K.B.; Wolfson, D.L.; Dumitriu, G.; Øie, C.I.; Ahluwalia, B.S.; et al. SAMM50 Acts with P62 in Piecemeal Basal- and OXPHOS-Induced Mitophagy of SAM and MICOS Components. J. Cell Biol. 2021, 220, e202009092. [Google Scholar] [CrossRef]
- Abudu, Y.P.; Mouilleron, S.; Tooze, S.A.; Lamark, T.; Johansen, T. SAMM50 Is a Receptor for Basal Piecemeal Mitophagy and Acts with SQSTM1/P62 in OXPHOS-Induced Mitophagy. Autophagy 2021, 17, 2656–2658. [Google Scholar] [CrossRef]
- BasuRay, S.; Smagris, E.; Cohen, J.C.; Hobbs, H.H. The PNPLA3 Variant Associated with Fatty Liver Disease (I148M) Accumulates on Lipid Droplets by Evading Ubiquitylation: Basuray et al. Hepatology 2017, 66, 1111–1124. [Google Scholar] [CrossRef] [Green Version]
- BasuRay, S.; Wang, Y.; Smagris, E.; Cohen, J.C.; Hobbs, H.H. Accumulation of PNPLA3 on Lipid Droplets Is the Basis of Associated Hepatic Steatosis. Proc. Natl. Acad. Sci. USA 2019, 116, 9521–9526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.; Zhao, Y.; Zhang, F.; Zhang, S.; Kwong, A.C.; Zhang, Y.; Hoffmann, H.-H.; Bushweller, L.; Wu, X.; Ashbrook, A.W.; et al. IL6/STAT3 Axis Dictates the PNPLA3-Mediated Susceptibility to Non-Alcoholic Fatty Liver Disease. J. Hepatol. 2022, in press. [Google Scholar] [CrossRef]
- Nischalke, H.D.; Lutz, P.; Bartok, E.; Krämer, B.; Langhans, B.; Frizler, R.; Berg, T.; Hampe, J.; Buch, S.; Datz, C.; et al. The PNPLA3 I148M Variant Promotes Lipid-Induced Hepatocyte Secretion of CXC Chemokines Establishing a Tumorigenic Milieu. J. Mol. Med. 2019, 97, 1589–1600. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, B.; Lindén, D.; Brolén, G.; Liljeblad, M.; Bjursell, M.; Romeo, S.; Loomba, R. Review Article: The Emerging Role of Genetics in Precision Medicine for Patients with Non-Alcoholic Steatohepatitis. Aliment. Pharmacol. Ther. 2020, 51, 1305–1320. [Google Scholar] [CrossRef]
- Xu, K.; Zheng, K.I.; Zhu, P.-W.; Liu, W.-Y.; Ma, H.-L.; Li, G.; Tang, L.-J.; Rios, R.S.; Targher, G.; Byrne, C.D.; et al. Interaction of SAMM50-Rs738491, PARVB-Rs5764455 and PNPLA3-Rs738409 Increases Susceptibility to Nonalcoholic Steatohepatitis. J. Clin. Transl. Hepatol. 2022, 10, 219–229. [Google Scholar] [CrossRef]
- Qiao, M.; Yang, J.; Zhu, Y.; Hu, J. Association of Sorting and Assembly Machinery Component 50 Homolog Gene Polymorphisms with Nonalcoholic Fatty Liver Disease Susceptibility. Medicine 2022, 101, e29958. [Google Scholar] [CrossRef] [PubMed]
- Edelman, D.; Kalia, H.; Delio, M.; Alani, M.; Krishnamurthy, K.; Abd, M.; Auton, A.; Wang, T.; Wolkoff, A.W.; Morrow, B.E. Genetic Analysis of Nonalcoholic Fatty Liver Disease within a Caribbean-Hispanic Population. Mol. Genet. Genom. Med. 2015, 3, 558–569. [Google Scholar] [CrossRef] [PubMed]

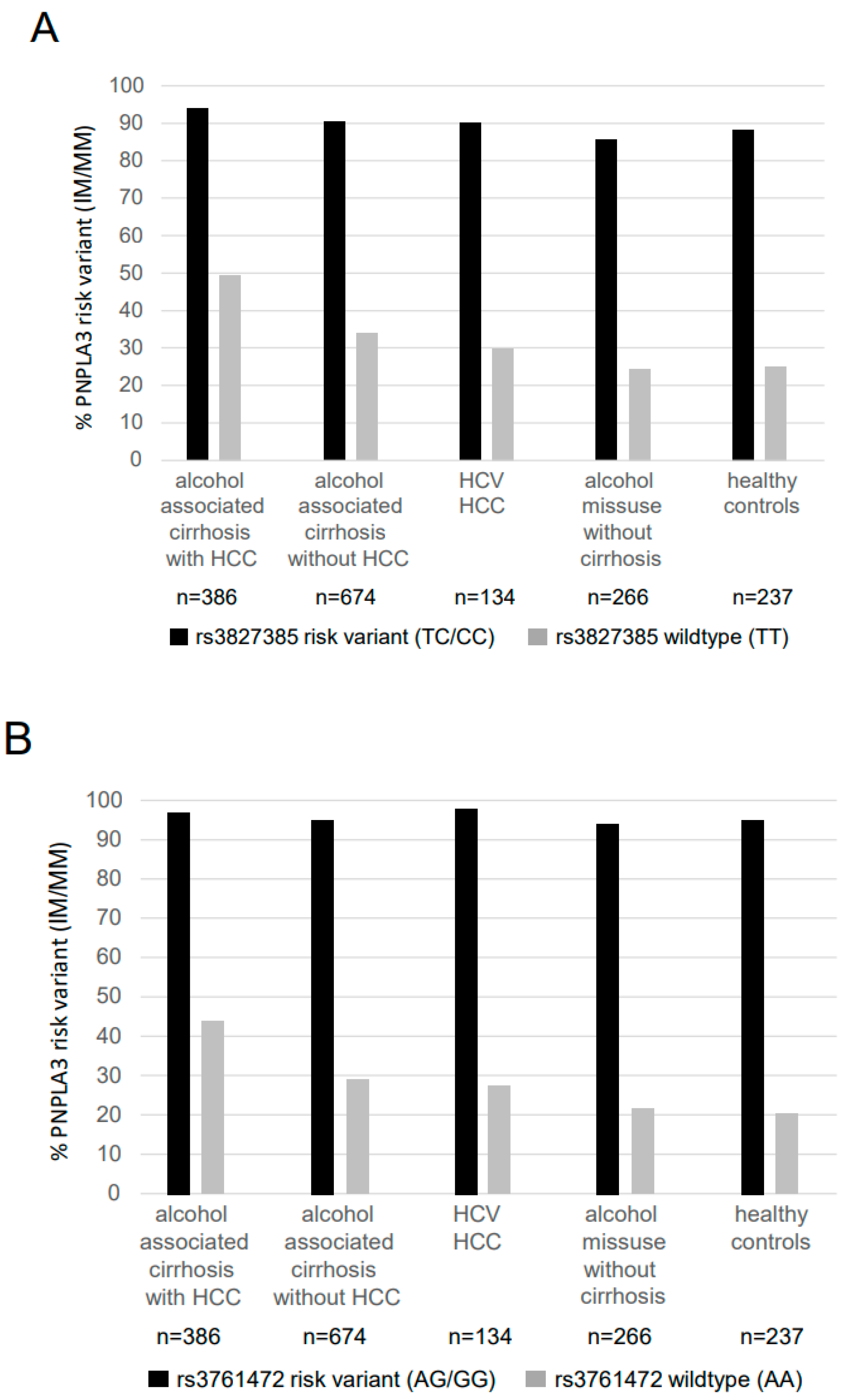
| Alcohol-Associated Cirrhosis | HCV HCC | Alcohol Misuse without Cirrhosis | Healthy Controls | ||
|---|---|---|---|---|---|
| with HCC | without HCC | ||||
| Total number | 386 | 674 | 134 | 266 | 237 |
| Age, mean (range) | 63.5 (36–87) a,b | 56.4 (27–92) c,d | 59.6 (38–82) d | 42.5 (18–81) e | 39.6 (20–75) |
| Sex (% male/female) | 88.3/11.7 f | 70.6/29.4 g,h | 62.1/37.9 g | 85.4/14.6 i | 58.6/41.4 |
| Bilirubin [mg/dL], (Mean ± SD) | 2.78 ± 4.16 j | 4.00 ± 6.71 g | 2.63 ± 3.49 | 0.80 ± 1.22 | n.d. |
| ALT [IU/L], (Mean ± SD) | 49.2 ± 56.7 | 43.1 ± 110.0 | 71.9 ± 56.2 | 44.5 ± 49.9 | n.d. |
| AST [IU/L], (Mean ± SD) | 88.8 ± 109.8 k | 72.5 ± 168.5 | 85.9 ± 81.1 | 53.4 ± 65.4 | n.d. |
| GGT [IU/L], (Mean ± SD) | 273.5 ± 335.2 k,l | 197.1 ± 227.1 | 118.6 ± 127.0 | 203.4 ± 381.6 | n.d. |
| Platelet count [* 103/µL], (Mean ± SD) | 150.9 ± 90.8 g | 152.3 ± 118.0 g | 111.4 ± 71.2 g | 229.3 ± 85.2 | n.d. |
| MELD (Mean ± SD) | 13.7 ± 6.8 n | 16.8 ± 7.2 | 15.2 ± 5.0 | n.d. | n.d. |
| Diabetes (%) | 46.4 m | 22.9 | 17.9 | n.d. | n.d. |
| Genotype | Alcohol-Associated Cirrhosis (n = 1060) | HCV HCC (n = 134) | Alcohol Misuse without Cirrhosis (n = 266) | Healthy Controls (n = 237) | |
|---|---|---|---|---|---|
| with HCC n = 386 | without HCC n = 674 | ||||
| SAMM50 rs3827385 | |||||
| TT | 162 (42.0%) | 381 (56.5%) | 84 (62.7%) | 176 (66.2%) | 160 (67.5%) |
| TC | 178 (46.1%) a,d,g | 246 (36.5%) b | 40 (29.9%) | 84 (31.6%) | 65 (27.4%) |
| CC | 46 (11.9%) a,d,f | 47 (7.0%) e | 10 (7.5%) | 6 (2.3%) | 12 (5.1%) |
| allele frequency T/C | 65.0%/35.0% a,d,g | 74.8%/25.2% b | 81.2%/18.8% | 82.0%/18.0% | 81.2%/18.8% |
| SAMM50 rs3761472 | |||||
| AA | 158 (40.9%) | 374 (55.5%) | 87 (64.9%) | 180 (67.7%) | 157 (66.2%) |
| AG | 179 (46.4%) a,d,g | 248 (36.8%) c,f | 38 (28.4%) | 77 (28.9%) | 67 (28.3%) |
| GG | 49 (12.7%) a,d,g | 52 (7.7%) | 9 (6.7%) | 9 (3.4%) | 13 (5.5%) |
| allele frequency A/G | 64.1%/35.9% a,d,g | 73.9%/26.1% c | 79.1%/20.9% | 82.1%/17.9% | 80.4%/19.6% |
| PNPLA3 rs738409 | |||||
| CC | 96 (24.9%) | 280 (41.5%) | 64 (47.8%) | 146 (54.9%) | 129 (54.4%) |
| GC | 199 (51.6%) a,d,g | 306 (45.4%) c | 54 (40.3%) | 104 (39.1%) | 89 (37.6%) |
| GG | 91 (23.6%) a,d,g | 88 (13.1%) c | 16 (11.9%) | 16 (6.0%) | 19 (8.0%) |
| allele frequency C/G | 50.6%/49.4% a,d,g | 64.2%/35.8% d | 67.9%/32.1% | 74.4%/25.6% | 73.2%/26.8% |
| Univariate Analysis | ||||
|---|---|---|---|---|
| 95% CI | ||||
| Parameter | P | OR | Lower | Upper |
| Age | 4.0 × 10−24 | 1.086 | 1.069 | 1.103 |
| Sex (male) | 1.1 × 10−9 | 3.151 | 2.178 | 4.559 |
| Diabetes | 2.4 × 10−13 | 2.909 | 2.186 | 3.871 |
| PNPLA3 148 (IM/MM) | 6.4 × 10−8 | 2.147 | 1.627 | 2.832 |
| rs3827385 (TC/CC) | 6.0 × 10−6 | 1.798 | 1.396 | 2.316 |
| rs3761472 (AG/GG) | 6.0 × 10−6 | 1.799 | 1.396 | 2.318 |
| Multivariate Analysis * | ||||
| 95% CI | ||||
| Parameter | P | OR | Lower | Upper |
| Age | 1.2 × 10−16 | 1.078 | 1.059 | 1.097 |
| Sex (male) | 8.1 × 10−8 | 3.174 | 2.081 | 4.840 |
| Diabetes | 6.3 × 10−5 | 1.930 | 1.399 | 2.662 |
| PNPLA3 148M | 1.6 × 10−5 | 2.083 | 1.492 | 2.909 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nischalke, H.D.; Schmalz, F.; Buch, S.; Fischer, J.; Möller, C.; Matz-Soja, M.; Krämer, B.; Langhans, B.; Klüners, A.; Soyka, M.; et al. Genetic Variation of SAMM50 Is Not an Independent Risk Factor for Alcoholic Hepatocellular Carcinoma in Caucasian Patients. Int. J. Mol. Sci. 2022, 23, 15353. https://doi.org/10.3390/ijms232315353
Nischalke HD, Schmalz F, Buch S, Fischer J, Möller C, Matz-Soja M, Krämer B, Langhans B, Klüners A, Soyka M, et al. Genetic Variation of SAMM50 Is Not an Independent Risk Factor for Alcoholic Hepatocellular Carcinoma in Caucasian Patients. International Journal of Molecular Sciences. 2022; 23(23):15353. https://doi.org/10.3390/ijms232315353
Chicago/Turabian StyleNischalke, Hans Dieter, Franziska Schmalz, Stephan Buch, Janett Fischer, Christine Möller, Madlen Matz-Soja, Benjamin Krämer, Bettina Langhans, Alexandra Klüners, Michael Soyka, and et al. 2022. "Genetic Variation of SAMM50 Is Not an Independent Risk Factor for Alcoholic Hepatocellular Carcinoma in Caucasian Patients" International Journal of Molecular Sciences 23, no. 23: 15353. https://doi.org/10.3390/ijms232315353





