The Increased Amyloidogenicity of Spike RBD and pH-Dependent Binding to ACE2 May Contribute to the Transmissibility and Pathogenic Properties of SARS-CoV-2 Omicron as Suggested by In Silico Study
Abstract
1. Introduction
2. Results
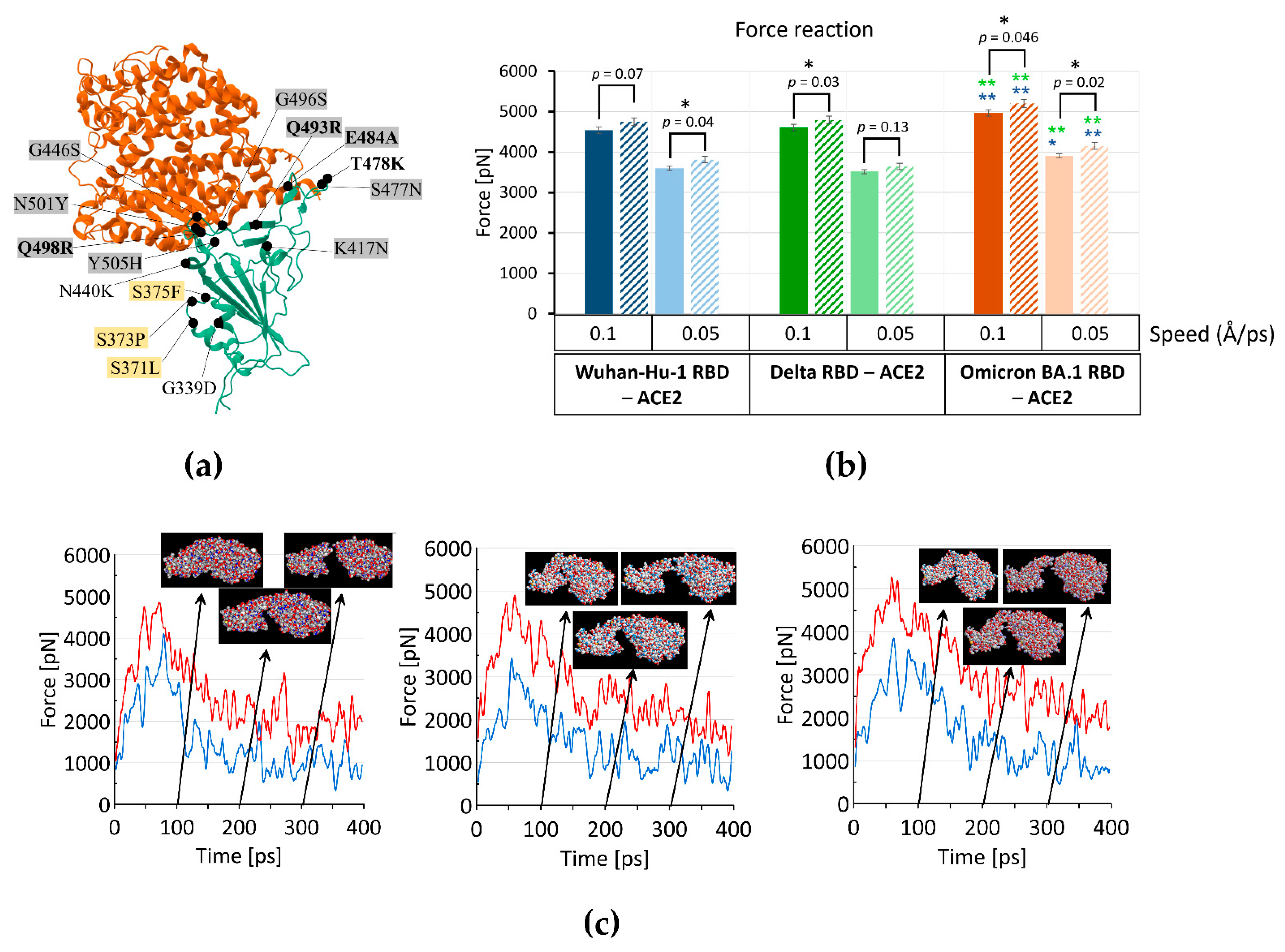
2.1. Omicron RBD Better Binds to ACE2 in Protonated form When Compared to Delta by MD Simulations
2.2. Amyloidogenic Properties of Spike RBDs of SARS-CoV-2 Variants

3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ruan, Y.; Hou, M.; Tang, X.; He, X.; Lu, X.; Lu, J.; Wu, C.I.; Wen, H. The Runaway Evolution of SARS-CoV-2 Leading to the Highly Evolved Delta Strain. Mol. Biol. Evol. 2022, 39, msac046. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Yi, S.V. On the Origin and Evolution of SARS-CoV-2. Exp. Mol. Med. 2021, 53, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Thakur, V.; Bhola, S.; Thakur, P.; Patel, S.K.S.; Kulshrestha, S.; Ratho, R.K.; Kumar, P. Waves and Variants of SARS-CoV-2: Understanding the Causes and Effect of the COVID-19 Catastrophe. Infection 2022, 50, 309–325. [Google Scholar] [CrossRef]
- Kumar, R.; Verma, H.; Singhvi, N.; Sood, U.; Gupta, V.; Singh, M.; Kumari, R.; Hira, P.; Nagar, S.; Talwar, C.; et al. Comparative Genomic Analysis of Rapidly Evolving SARS-CoV-2 Reveals Mosaic Pattern of Phylogeographical Distribution. mSystems 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Attwood, S.W.; Hill, S.C.; Aanensen, D.M.; Connor, T.R.; Pybus, O.G. Phylogenetic and Phylodynamic Approaches to Understanding and Combating the Early SARS-CoV-2 Pandemic. Nat. Rev. Genet. 2022, 23, 547–562. [Google Scholar] [CrossRef] [PubMed]
- Duarte, C.M.; Ketcheson, D.I.; Eguíluz, V.M.; Agustí, S.; Fernández-Gracia, J.; Jamil, T.; Laiolo, E.; Gojobori, T.; Alam, I. Rapid Evolution of SARS-CoV-2 Challenges Human Defenses. Sci. Rep. 2022, 12, 6457. [Google Scholar] [CrossRef]
- Nunes, D.R.; Braconi, C.T.; Ludwig-Begall, L.F.; Arns, C.W.; Durães-Carvalho, R. Deep Phylogenetic-Based Clustering Analysis Uncovers New and Shared Mutations in SARS-CoV-2 Variants as a Result of Directional and Convergent Evolution. PLoS ONE 2022, 17, e0268389. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Hofmann, H.; Pyrc, K.; Van Der Hoek, L.; Geier, M.; Berkhout, B.; Pöhlmann, S. Human Coronavirus NL63 Employs the Severe Acute Respiratory Syndrome Coronavirus Receptor for Cellular Entry. Proc. Natl. Acad. Sci. USA 2005, 102, 7988–7993. [Google Scholar] [CrossRef]
- Li, W.; Moore, M.J.; Vasllieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greeneugh, T.C.; et al. Angiotensin-Converting Enzyme 2 Is a Functional Receptor for the SARS Coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural Basis for the Recognition of SARS-CoV-2 by Full-Length Human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef] [PubMed]
- Abdul-Rasool, S.; Fielding, B.C. Understanding Human Coronavirus HCoV-NL63. Open Virol. J. 2010, 4, 76. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural Basis of Receptor Recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Pöhlmann, S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell 2020, 78, 779–784.e5. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.M.; Yang, W.L.; Yang, F.Y.; Zhang, L.; Huang, W.J.; Hou, W.; Fan, C.F.; Jin, R.H.; Feng, Y.M.; Wang, Y.C.; et al. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Signal Transduct. Target. Ther. 2021, 6, 134. [Google Scholar] [CrossRef] [PubMed]
- Ou, T.; Mou, H.; Zhang, L.; Ojha, A.; Choe, H.; Farzan, M. Hydroxychloroquine-Mediated Inhibition of SARS-CoV-2 Entry Is Attenuated by TMPRSS2. PLoS Pathog. 2021, 17, e1009212. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 Entry into Cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Pacheco, M.; Silva, P.L.; Cruz, F.F.; Battaglini, D.; Robba, C.; Pelosi, P.; Morales, M.M.; Caruso Neves, C.; Rocco, P.R.M. Pathogenesis of Multiple Organ Injury in COVID-19 and Potential Therapeutic Strategies. Front. Physiol. 2021, 12, 593223. [Google Scholar] [CrossRef]
- Teuwen, L.A.; Geldhof, V.; Pasut, A.; Carmeliet, P. COVID-19: The Vasculature Unleashed. Nat. Rev. Immunol. 2020, 20, 389–391. [Google Scholar] [CrossRef]
- Aksenova, A.Y. Von Willebrand Factor and Endothelial Damage: A Possible Association with COVID-19. Ecol. Genet. 2020, 18, 135–138. [Google Scholar] [CrossRef]
- Solomon, I.H.; Normandin, E.; Bhattacharyya, S.; Mukerji, S.S.; Keller, K.; Ali, A.S.; Adams, G.; Hornick, J.L.; Padera, R.F.; Sabeti, P. Neuropathological Features of COVID-19. N. Engl. J. Med. 2020, 383, 989–992. [Google Scholar] [CrossRef] [PubMed]
- Lippi, A.; Domingues, R.; Setz, C.; Outeiro, T.F.; Krisko, A. SARS-CoV-2: At the Crossroad Between Aging and Neurodegeneration. Mov. Disord. 2020, 35, 716–720. [Google Scholar] [CrossRef] [PubMed]
- Tavassoly, O.; Safavi, F.; Tavassoly, I. Seeding Brain Protein Aggregation by SARS-CoV-2 as a Possible Long-Term Complication of COVID-19 Infection. ACS Chem. Neurosci. 2020, 11, 3704–3706. [Google Scholar] [CrossRef] [PubMed]
- Dolatshahi, M.; Sabahi, M.; Aarabi, M.H. Pathophysiological Clues to How the Emergent SARS-CoV-2 Can Potentially Increase the Susceptibility to Neurodegeneration. Mol. Neurobiol. 2021, 58, 2379–2394. [Google Scholar] [CrossRef]
- Shen, W.-B.; Logue, J.; Yang, P.; Baracco, L.; Elahi, M.; Reece, E.A.; Wang, B.; Li, L.; Blanchard, T.G.; Han, Z.; et al. SARS-CoV-2 Invades Cognitive Centers of the Brain and Induces Alzheimer’s-like Neuropathology. bioRxiv 2022. [Google Scholar] [CrossRef]
- Merello, M.; Bhatia, K.P.; Obeso, J.A. SARS-CoV-2 and the Risk of Parkinson’s Disease: Facts and Fantasy. Lancet Neurol. 2021, 20, 94–95. [Google Scholar] [CrossRef]
- Sinha, N.; Thakur, A.K. Likelihood of Amyloid Formation in COVID-19-Induced ARDS. Trends Microbiol. 2021, 29, 967–969. [Google Scholar] [CrossRef]
- Pretorius, E.; Venter, C.; Laubscher, G.J.; Lourens, P.J.; Steenkamp, J.; Kell, D.B. Prevalence of Readily Detected Amyloid Blood Clots in ‘Unclotted’ Type 2 Diabetes Mellitus and COVID-19 Plasma: A Preliminary Report. Cardiovasc. Diabetol. 2020, 19, 193. [Google Scholar] [CrossRef]
- Semerdzhiev, S.A.; Fakhree, M.A.A.; Segers-Nolten, I.; Blum, C.; Claessens, M.M.A.E. Interactions between SARS-CoV-2 N-Protein and α-Synuclein Accelerate Amyloid Formation. ACS Chem. Neurosci. 2022, 13, 143–150. [Google Scholar] [CrossRef]
- Song, E.; Zhang, C.; Israelow, B.; Lu-Culligan, A.; Prado, A.V.; Skriabine, S.; Lu, P.; El Weizman, O.; Liu, F.; Dai, Y.; et al. Neuroinvasion of SARS-CoV-2 in Human and Mouse Brain. J. Exp. Med. 2021, 218, e20202135. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, I.N.; Hartshorn, K.L. The Role of Antimicrobial Peptides in Influenza Virus Infection and Their Potential as Antiviral and Immunomodulatory Therapy. Pharmaceuticals 2016, 9, 53. [Google Scholar] [CrossRef] [PubMed]
- Kurpe, S.R.; Grishin, S.Y.; Surin, A.K.; Panfilov, A.V.; Slizen, M.V.; Chowdhury, S.D.; Galzitskaya, O.V. Antimicrobial and Amyloidogenic Activity of Peptides. Can Antimicrobial Peptides Be Used against SARS-CoV-2? Int. J. Mol. Sci. 2020, 21, 9552. [Google Scholar] [CrossRef] [PubMed]
- Bourgade, K.; Garneau, H.; Giroux, G.; Le Page, A.Y.; Bocti, C.; Dupuis, G.; Frost, E.H.; Fülöp, T. β-Amyloid Peptides Display Protective Activity against the Human Alzheimer’s Disease-Associated Herpes Simplex Virus-1. Biogerontology 2015, 16, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Ezzat, K.; Pernemalm, M.; Pålsson, S.; Roberts, T.C.; Järver, P.; Dondalska, A.; Bestas, B.; Sobkowiak, M.J.; Levänen, B.; Sköld, M.; et al. The Viral Protein Corona Directs Viral Pathogenesis and Amyloid Aggregation. Nat. Commun. 2019, 10, 2331. [Google Scholar] [CrossRef]
- WuDunn, D.; Spear, P.G. Initial Interaction of Herpes Simplex Virus with Cells Is Binding to Heparan Sulfate. J. Virol. 1989, 63, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Iannuzzi, C.; Irace, G.; Sirangelo, I. The Effect of Glycosaminoglycans (GAGs) on Amyloid Aggregation and Toxicity. Molecules 2015, 20, 2510–2528. [Google Scholar] [CrossRef] [PubMed]
- Mycroft-West, C.J.; Su, D.; Pagani, I.; Rudd, T.R.; Elli, S.; Gandhi, N.S.; Guimond, S.E.; Miller, G.J.; Meneghetti, M.C.Z.; Nader, H.B.; et al. Heparin Inhibits Cellular Invasion by SARS-CoV-2: Structural Dependence of the Interaction of the Spike S1 Receptor-Binding Domain with Heparin. Thromb. Haemost. 2020, 120, 1700. [Google Scholar] [CrossRef]
- Idrees, D.; Kumar, V. SARS-CoV-2 Spike Protein Interactions with Amyloidogenic Proteins: Potential Clues to Neurodegeneration. Biochem. Biophys. Res. Commun. 2021, 554, 94. [Google Scholar] [CrossRef]
- Tavassoly, O.; Safavi, F.; Tavassoly, I. Heparin-Binding Peptides as Novel Therapies to Stop SARS-CoV-2 Cellular Entry and Infection. Mol. Pharmacol. 2020, 98, 612–619. [Google Scholar] [CrossRef]
- Hsu, J.T.A.; Tien, C.F.; Yu, G.Y.; Shen, S.; Lee, Y.H.; Hsu, P.C.; Wang, Y.; Chao, P.K.; Tsay, H.J.; Shie, F.S. The Effects of Aβ 1-42 Binding to the SARS-CoV-2 Spike Protein S1 Subunit and Angiotensin-Converting Enzyme 2. Int. J. Mol. Sci. 2021, 22, 8226. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Hossinger, A.; Heumüller, S.E.; Hornberger, A.; Buravlova, O.; Konstantoulea, K.; Müller, S.A.; Paulsen, L.; Rousseau, F.; Schymkowitz, J.; et al. Highly Efficient Intercellular Spreading of Protein Misfolding Mediated by Viral Ligand-Receptor Interactions. Nat. Commun. 2021, 12, 5739. [Google Scholar] [CrossRef] [PubMed]
- Nyström, S.; Hammarström, P. Amyloidogenesis of SARS-CoV-2 Spike Protein. bioRxiv 2021. [Google Scholar] [CrossRef] [PubMed]
- Castelletto, V.; Hamley, I.W. Amyloid and Hydrogel Formation of a Peptide Sequence from a Coronavirus Spike Protein. ACS Nano 2022, 16, 1857–1867. [Google Scholar] [CrossRef]
- Petrlova, J.; Samsudin, F.; Bond, P.J.; Schmidtchen, A. SARS-CoV-2 Spike Protein Aggregation Is Triggered by Bacterial Lipopolysaccharide. FEBS Lett. 2022, 596, 2566–2575. [Google Scholar] [CrossRef]
- Tetz, G.; Tetz, V. Prion-like Domains in Spike Protein of SARS-CoV-2 Differ across Its Variants and Enable Changes in Affinity to ACE2. Microorganisms 2022, 10, 280. [Google Scholar] [CrossRef]
- Loganathan, S.K.; Schleicher, K.; Malik, A.; Quevedo, R.; Langille, E.; Teng, K.; Oh, R.H.; Rathod, B.; Tsai, R.; Samavarchi-Tehrani, P.; et al. Rare Driver Mutations in Head and Neck Squamous Cell Carcinomas Converge on NOTCH Signaling. Science 2020, 367, 1264–1269. [Google Scholar] [CrossRef]
- Mohabatkar, H.; Behbahani, M.; Moradi, M. A Concise in silico Prediction Report of a Potential Prion-Like Domain in SARS-CoV-2 Polyprotein. J. Microbiol. Biotechnol. Food Sci. 2021, 11, e4813. [Google Scholar] [CrossRef]
- Shahzad, S.; Willcox, M. The Possible Role of Prion-Like Viral Protein Domains on the Emergence of Novel Viruses as SARS-CoV-2. J. Mol. Evol. 2022, 90, 227–230. [Google Scholar] [CrossRef]
- Oz, M.; Lorke, D.E. Multifunctional Angiotensin Converting Enzyme 2, the SARS-CoV-2 Entry Receptor, and Critical Appraisal of Its Role in Acute Lung Injury. Biomed. Pharmacother. 2021, 136, 111193. [Google Scholar] [CrossRef]
- Peacock, T.P.; Brown, J.C.; Zhou, J.; Thakur, N.; Newman, J.; Kugathasan, R.; Sukhova, K.; Kaforou, M.; Bailey, D.; Barclay, W.S. The SARS-CoV-2 Variant, Omicron, Shows Rapid Replication in Human Primary Nasal Epithelial Cultures and Efficiently Uses the Endosomal Route of Entry. bioRxiv 2022. [Google Scholar] [CrossRef]
- Meng, B.; Abdullahi, A.; Ferreira, I.A.T.M.; Goonawardane, N.; Saito, A.; Kimura, I.; Yamasoba, D.; Gerber, P.P.; Fatihi, S.; Rathore, S.; et al. Altered TMPRSS2 Usage by SARS-CoV-2 Omicron Impacts Infectivity and Fusogenicity. Nature 2022, 603, 706–714. [Google Scholar] [CrossRef] [PubMed]
- Willett, B.J.; Grove, J.; MacLean, O.A.; Wilkie, C.; Logan, N.; De Lorenzo, G.; Furnon, W.; Scott, S.; Manali, M.; Szemiel, A.; et al. The Hyper-Transmissible SARS-CoV-2 Omicron Variant Exhibits Significant Antigenic Change, Vaccine Escape and a Switch in Cell Entry Mechanism. medRxiv 2022. [Google Scholar] [CrossRef]
- Abdelnabi, R.; Foo, C.S.; Zhang, X.; Lemmens, V.; Maes, P.; Slechten, B.; Raymenants, J.; André, E.; Weynand, B.; Dallmeier, K.; et al. The Omicron (B.1.1.529) SARS-CoV-2 Variant of Concern Does Not Readily Infect Syrian Hamsters. Antivir. Res. 2022, 198, 105253. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Cai, Y.; Lavine, C.L.; Peng, H.; Zhu, H.; Anand, K.; Tong, P.; Gautam, A.; Mayer, M.L.; Rits-Volloch, S.; et al. Structural and Functional Impact by SARS-CoV-2 Omicron Spike Mutations. Cell Rep. 2022, 39, 110729. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Liu, P.; Wang, N.; Wang, L.; Fan, K.; Zhu, Q.; Wang, K.; Chen, R.; Feng, R.; Jia, Z.; et al. Structural and Functional Characterizations of Infectivity and Immune Evasion of SARS-CoV-2 Omicron. Cell 2022, 185, 860–871.e13. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Li, L.; Liu, S.; Wang, Q.; Zhang, D.; Xu, Z.; Han, P.; Li, X.; Peng, Q.; Su, C.; et al. Receptor Binding and Complex Structures of Human ACE2 to Spike RBD from Omicron and Delta SARS-CoV-2. Cell 2022, 185, 630–640.e10. [Google Scholar] [CrossRef]
- Pipitò, L.; Rujan, R.M.; Reynolds, C.A.; Deganutti, G. Molecular Dynamics Studies Reveal Structural and Functional Features of the SARS-CoV-2 Spike Protein. BioEssays 2022, 44, 2200060. [Google Scholar] [CrossRef]
- Starr, T.N.; Greaney, A.J.; Hilton, S.K.; Ellis, D.; Crawford, K.H.D.; Dingens, A.S.; Navarro, M.J.; Bowen, J.E.; Tortorici, M.A.; Walls, A.C.; et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 2020, 182, 1295–1310.e20. [Google Scholar] [CrossRef]
- Zou, J.; Kurhade, C.; Xia, H.; Liu, M.; Xie, X.; Ren, P.; Shi, P.Y. Cross-Neutralization of Omicron BA.1 against BA.2 and BA.3 SARS-CoV-2. Nat. Commun. 2022, 13, 2956. [Google Scholar] [CrossRef]
- Wu, L.; Zhou, L.; Mo, M.; Liu, T.; Wu, C.; Gong, C.; Lu, K.; Gong, L.; Zhu, W.; Xu, Z. SARS-CoV-2 Omicron RBD Shows Weaker Binding Affinity than the Currently Dominant Delta Variant to Human ACE2. Signal Transduct. Target. Ther. 2022, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Zahradník, J.; Marciano, S.; Shemesh, M.; Zoler, E.; Harari, D.; Chiaravalli, J.; Meyer, B.; Rudich, Y.; Li, C.; Marton, I.; et al. SARS-CoV-2 Variant Prediction and Antiviral Drug Design Are Enabled by RBD in Vitro Evolution. Nat. Microbiol. 2021, 6, 1188–1198. [Google Scholar] [CrossRef] [PubMed]
- Cameroni, E.; Bowen, J.E.; Rosen, L.E.; Saliba, C.; Zepeda, S.K.; Culap, K.; Pinto, D.; VanBlargan, L.A.; De Marco, A.; di Iulio, J.; et al. Broadly Neutralizing Antibodies Overcome SARS-CoV-2 Omicron Antigenic Shift. Nature 2022, 602, 664–670. [Google Scholar] [CrossRef] [PubMed]
- Dejnirattisai, W.; Huo, J.; Zhou, D.; Zahradník, J.; Supasa, P.; Liu, C.; Duyvesteyn, H.M.E.; Ginn, H.M.; Mentzer, A.J.; Tuekprakhon, A.; et al. SARS-CoV-2 Omicron-B.1.1.529 Leads to Widespread Escape from Neutralizing Antibody Responses. Cell 2022, 185, 467–484.e15. [Google Scholar] [CrossRef] [PubMed]
- Mannar, D.; Saville, J.W.; Zhu, X.; Srivastava, S.S.; Berezuk, A.M.; Tuttle, K.S.; Marquez, A.C.; Sekirov, I.; Subramaniam, S. SARS-CoV-2 Omicron Variant: Antibody Evasion and Cryo-EM Structure of Spike Protein–ACE2 Complex. Science 2022, 375, 760–764. [Google Scholar] [CrossRef]
- Han, P.; Su, C.; Zhang, Y.; Bai, C.; Zheng, A.; Qiao, C.; Wang, Q.; Niu, S.; Chen, Q.; Zhang, Y.; et al. Molecular Insights into Receptor Binding of Recent Emerging SARS-CoV-2 Variants. Nat. Commun. 2021, 12, 6103. [Google Scholar] [CrossRef]
- Toelzer, C.; Gupta, K.; Yadav, S.K.N.; Borucu, U.; Davidson, A.D.; Williamson, M.K.; Shoemark, D.K.; Garzoni, F.; Staufer, O.; Milligan, R.; et al. Free Fatty Acid Binding Pocket in the Locked Structure of SARS-CoV-2 Spike Protein. Science 2020, 370, 725–730. [Google Scholar] [CrossRef]
- Carrique, L.; Duyvesteyn, H.M.; Malinauskas, T.; Zhao, Y.; Ren, J.; Zhou, D.; Walter, T.S.; Radecke, J.; Huo, J.; Ruza, R.R.; et al. The SARS-CoV-2 Spike Harbours a Lipid Binding Pocket Which Modulates Stability of the Prefusion Trimer. bioRxiv 2020. [Google Scholar] [CrossRef]
- Campbell, F.; Archer, B.; Laurenson-Schafer, H.; Jinnai, Y.; Konings, F.; Batra, N.; Pavlin, B.; Vandemaele, K.; Van Kerkhove, M.D.; Jombart, T.; et al. Increased Transmissibility and Global Spread of SARSCoV-2 Variants of Concern as at June 2021. Eurosurveillance 2021, 26, 2100509. [Google Scholar] [CrossRef]
- Deng, X.; Garcia-Knight, M.A.; Khalid, M.M.; Servellita, V.; Wang, C.; Morris, M.K.; Sotomayor-González, A.; Glasner, D.R.; Reyes, K.R.; Gliwa, A.S.; et al. Transmission, Infectivity, and Antibody Neutralization of an Emerging SARS-CoV-2 Variant in California Carrying a L452R Spike Protein Mutation. medRxiv 2021. [Google Scholar] [CrossRef]
- Long, B.; Carius, B.M.; Chavez, S.; Liang, S.Y.; Brady, W.J.; Koyfman, A.; Gottlieb, M. Clinical Update on COVID-19 for the Emergency Clinician: Presentation and Evaluation. Am. J. Emerg. Med. 2022, 54, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Li, X.; Zhang, L.; Wan, S.; Zhang, L.; Zhou, F. SARS-CoV-2 Omicron Variant: Recent Progress and Future Perspectives. Signal Transduct. Target. Ther. 2022, 7, 141. [Google Scholar] [CrossRef] [PubMed]
- Houston, M.C. Pleural Fluid PH: Diagnostic, Therapeutic, and Prognostic Value. Am. J. Surg. 1987, 154, 333–337. [Google Scholar] [CrossRef]
- Fischer, H.; Widdicombe, J.H. Mechanisms of Acid and Base Secretion by the Airway Epithelium. J. Membr. Biol. 2006, 211, 139. [Google Scholar] [CrossRef] [PubMed]
- Kosolapova, A.O.; Antonets, K.S.; Belousov, M.V.; Nizhnikov, A.A. Biological Functions of Prokaryotic Amyloids in Interspecies Interactions: Facts and Assumptions. Int. J. Mol. Sci. 2020, 21, 7240. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Li, W.; Peng, G.; Li, F. Crystal Structure of NL63 Respiratory Coronavirus Receptor-Binding Domain Complexed with Its Human Receptor. Proc. Natl. Acad. Sci. USA 2009, 106, 19970–19974. [Google Scholar] [CrossRef]
- Xu, C.; Wang, Y.; Liu, C.; Zhang, C.; Han, W.; Hong, X.; Wang, Y.; Hong, Q.; Wang, S.; Zhao, Q.; et al. Conformational Dynamics of SARS-CoV-2 Trimeric Spike Glycoprotein in Complex with Receptor ACE2 Revealed by Cryo-EM. Sci. Adv. 2021, 7, eabe5575. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904.e9. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 Spike Receptor-Binding Domain Bound to the ACE2 Receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020, 94, e00127-20. [Google Scholar] [CrossRef]
- Cong, Y.; Feng, Y.; Ni, H.; Zhi, F.; Miao, Y.; Fang, B.; Zhang, L.; Zhang, J.Z.H. Anchor-Locker Binding Mechanism of the Coronavirus Spike Protein to Human ACE2: Insights from Computational Analysis. J. Chem. Inf. Model. 2021, 61, 3529–3542. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.M.; Lau, E.H.Y.; Wong, J.Y.; et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Lin, Q.; Ran, J.; Musa, S.S.; Yang, G.; Wang, W.; Lou, Y.; Gao, D.; Yang, L.; He, D.; et al. The Basic Reproduction Number of Novel Coronavirus (2019-NCoV) Estimation Based on Exponential Growth in the Early Outbreak in China from 2019 to 2020: A Reply to Dhungana. Int. J. Infect. Dis. 2020, 94, 148–150. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Rocklöv, J. The Reproductive Number of the Delta Variant of SARS-CoV-2 Is Far Higher Compared to the Ancestral SARS-CoV-2 Virus. J. Travel Med. 2021, 28, taab124. [Google Scholar] [CrossRef]
- Kumar, S.; Thambiraja, T.S.; Karuppanan, K.; Subramaniam, G. Omicron and Delta Variant of SARS-CoV-2: A Comparative Computational Study of Spike Protein. J. Med. Virol. 2022, 94, 1641–1649. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wei, G.-W. Omicron BA.2 (B.1.1.529.2): High Potential for Becomingthe Next Dominant Variant. J. Phys. Chem. Lett. 2022, 13, 3840. [Google Scholar] [CrossRef]
- Zaraket, H.; Bridges, O.A.; Duan, S.; Baranovich, T.; Yoon, S.-W.; Reed, M.L.; Salomon, R.; Webby, R.J.; Webster, R.G.; Russell, C.J. Increased Acid Stability of the Hemagglutinin Protein Enhances H5N1 Influenza Virus Growth in the Upper Respiratory Tract but Is Insufficient for Transmission in Ferrets. J. Virol. 2013, 87, 9911–9922. [Google Scholar] [CrossRef]
- Reed, M.L.; Bridges, O.A.; Seiler, P.; Kim, J.-K.; Yen, H.-L.; Salomon, R.; Govorkova, E.A.; Webster, R.G.; Russell, C.J. The PH of Activation of the Hemagglutinin Protein Regulates H5N1 Influenza Virus Pathogenicity and Transmissibility in Ducks. J. Virol. 2010, 84, 1527–1535. [Google Scholar] [CrossRef]
- Focosi, D.; Novazzi, F.; Genoni, A.; Dentali, F.; Gasperina, D.D.; Baj, A.; Maggi, F. Emergence of SARS-COV-2 Spike Protein Escape Mutation Q493R after Treatment for COVID-19. Emerg. Infect. Dis. 2021, 27, 2728–2731. [Google Scholar] [CrossRef]
- Assessment Report Eli Lilly. Company Limited use of bamlanivimab and etesevimab for the treatment of COVID-19, 2021 Bamlanivimab and Etesevimab for the Treatment of COVID-19 (Eli Lilly) Art. 5(3)—AR EMA/177113/2021 Page. Eur. Med. Agency 2021. Available online: https://www.ema.europa.eu/en/documents/referral/eli-lilly-company-limited-antibody-combination-bamlanivimab/etesevimab-covid19-article-53-procedure-assessment-report_en.pdf (accessed on 30 October 2022).
- Starr, T.N.; Greaney, A.J.; Dingens, A.S.; Bloom, J.D. Complete Map of SARS-CoV-2 RBD Mutations That Escape the Monoclonal Antibody LY-CoV555 and Its Cocktail with LY-CoV016. Cell Rep. Med. 2021, 2, 100255. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Iketani, S.; Guo, Y.; Chan, J.F.W.; Wang, M.; Liu, L.; Luo, Y.; Chu, H.; Huang, Y.; Nair, M.S.; et al. Striking Antibody Evasion Manifested by the Omicron Variant of SARS-CoV-2. Nature 2022, 602, 676–681. [Google Scholar] [CrossRef] [PubMed]
- Iketani, S.; Liu, L.; Guo, Y.; Liu, L.; Chan, J.F.W.; Huang, Y.; Wang, M.; Luo, Y.; Yu, J.; Chu, H.; et al. Antibody Evasion Properties of SARS-CoV-2 Omicron Sublineages. Nature 2022, 604, 553–556. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Jalali, S.; Nilsson, B.L.; Dias, C.L. Binding Mechanisms of Amyloid-like Peptides to Lipid Bilayers and Effects of Divalent Cations. ACS Chem. Neurosci. 2021, 12, 2027–2035. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, J.M. The Association of Lipids with Amyloid Fibrils. J. Biol. Chem. 2022, 298, 102108. [Google Scholar] [CrossRef]
- Cai, Y.; Zhang, J.; Xiao, T.; Peng, H.; Sterling, S.M.; Walsh, R.M.; Rawson, S.; Rits-Volloch, S.; Chen, B. Distinct Conformational States of SARS-CoV-2 Spike Protein. Science 2020, 369, 1586–1592. [Google Scholar] [CrossRef]
- Kirchdoerfer, R.N.; Wang, N.; Pallesen, J.; Wrapp, D.; Turner, H.L.; Cottrell, C.A.; Corbett, K.S.; Graham, B.S.; McLellan, J.S.; Ward, A.B. Stabilized Coronavirus Spikes Are Resistant to Conformational Changes Induced by Receptor Recognition or Proteolysis. Sci. Rep. 2018, 8, 15701. [Google Scholar] [CrossRef]
- Grobbelaar, L.M.; Venter, C.; Vlok, M.; Ngoepe, M.; Laubscher, G.J.; Lourens, P.J.; Steenkamp, J.; Kell, D.B.; Pretorius, E. SARS-CoV-2 Spike Protein S1 Induces Fibrin(Ogen) Resistant to Fibrinolysis: Implications for Microclot Formation in COVID-19. Biosci. Rep. 2021, 41, BSR20210611. [Google Scholar] [CrossRef] [PubMed]
- Christ, W.; Kapell, S.; Mermelekas, G.; Evertsson, B.; Sork, H.; Bazaz, S.; Gustafsson, O.; Sobkowiak, M.J.; Cardenas, E.I.; Villa, V.; et al. SARS-CoV-2 and HSV-1 Induce Amyloid Aggregation in Human CSF. bioRxiv 2022. [Google Scholar] [CrossRef]
- Spyropoulos, A.C.; Goldin, M.; Giannis, D.; Diab, W.; Wang, J.; Khanijo, S.; Mignatti, A.; Gianos, E.; Cohen, M.; Sharifova, G.; et al. Efficacy and Safety of Therapeutic-Dose Heparin vs Standard Prophylactic or Intermediate-Dose Heparins for Thromboprophylaxis in High-Risk Hospitalized Patients with COVID-19: The HEP-COVID Randomized Clinical Trial. JAMA Intern. Med. 2021, 181, 1612–1620. [Google Scholar] [CrossRef]
- Spyropoulos, A.C.; Levy, J.H.; Ageno, W.; Connors, J.M.; Hunt, B.J.; Iba, T.; Levi, M.; Samama, C.M.; Thachil, J.; Giannis, D.; et al. Scientific and Standardization Committee Communication: Clinical Guidance on the Diagnosis, Prevention, and Treatment of Venous Thromboembolism in Hospitalized Patients with COVID-19. J. Thromb. Haemost. 2020, 18, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Bikdeli, B.; Madhavan, M.V.; Jimenez, D.; Chuich, T.; Dreyfus, I.; Driggin, E.; Der Nigoghossian, C.; Ageno, W.; Madjid, M.; Guo, Y.; et al. COVID-19 and Thrombotic or Thromboembolic Disease: Implications for Prevention, Antithrombotic Therapy, and Follow-Up: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2020, 75, 2950–2973. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, A.K.; Nutter-Upham, A.; Lindquist, S.; King, O.D. PLAAC: A Web and Command-Line Application to Identify Proteins with Prion-like Amino Acid Composition. Bioinformatics 2014, 30, 2501–2502. [Google Scholar] [CrossRef] [PubMed]
- Du, Z. The Complexity and Implications of Yeast Prion Domains. Prion 2011, 5, 311. [Google Scholar] [CrossRef] [PubMed]
- Garbuzynskiy, S.O.; Lobanov, M.Y.; Galzitskaya, O.V. FoldAmyloid: A Method of Prediction of Amyloidogenic Regions from Protein Sequence. Bioinformatics 2010, 26, 326–332. [Google Scholar] [CrossRef]
- Galzitskaya, O.V.; Garbuzynskiy, S.O.; Lobanov, M.Y. Is It Possible to Predict Amyloidogenic Regions from Sequence Alone? J. Bioinform. Comput. Biol. 2006, 4, 373–388. [Google Scholar] [CrossRef]
- Galzitskaya, O.V.; Garbuzynskiy, S.O.; Lobanov, M.Y. Prediction of Amyloidogenic and Disordered Regions in Protein Chains. PLoS Comput. Biol. 2006, 2, 1639–1648. [Google Scholar] [CrossRef]
- Lemak, A.S.; Balabaev, N.K. A Comparison between Collisional Dynamics and Brownian Dynamics. Mol. Simul. 1995, 15, 223–231. [Google Scholar] [CrossRef]
- Lemak, A.S.; Balabaev, N.K. Molecular Dynamics Simulation of a Polymer Chain in Solution by Collisional Dynamics Method. J. Comput. Chem. 1996, 17, 1685–1695. [Google Scholar] [CrossRef]
- Wang, J.; Cieplak, P.; Kollman, P.A. How Well Does a Restrained Electrostatic Potential (RESP) Model Perform in Calculating Conformational Energies of Organic and Biological Molecules? J. Comput. 2000, 21, 1049–1074. [Google Scholar] [CrossRef]
- Mahoney, M.W.; Jorgensen, W.L. A Five-Site Model for Liquid Water and the Reproduction of the Density Anomaly by Rigid, Nonpolarizable Potential Functions. J. Chem. Phys. 2000, 112, 8910. [Google Scholar] [CrossRef]
- Likhachev, I.V.; Balabaev, N.K.; Galzitskaya, O.V. Available Instruments for Analyzing Molecular Dynamics Trajectories. Open Biochem. J. 2016, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Likhachev, I.Y.V.; Balabaev, N.K. Trajectory Analyzer of Molecular Dynamics. Mat. Biol. Bioinform. 2007, 2, 120–129. [Google Scholar] [CrossRef]
- Likhachev, I.V.; Balabaev, N.K. Construction of Extended Dynamical Contact Maps by Molecular-Dynamics Simulation Data. Mat. Biol. Bioinform. 2009, 4, 36–45. [Google Scholar] [CrossRef]
- Sehnal, D.; Bittrich, S.; Deshpande, M.; Svobodová, R.; Berka, K.; Bazgier, V.; Velankar, S.; Burley, S.K.; Koča, J.; Rose, A.S. Mol* Viewer: Modern Web App for 3D Visualization and Analysis of Large Biomolecular Structures. Nucleic Acids Res. 2021, 49, W431–W437. [Google Scholar] [CrossRef]
- Oliveberg, M. Waltz, an Exciting New Move in Amyloid Prediction. Nat. Methods 2010, 7, 187–188. [Google Scholar] [CrossRef]
- Conchillo-Solé, O.; de Groot, N.S.; Avilés, F.X.; Vendrell, J.; Daura, X.; Ventura, S. AGGRESCAN: A Server for the Prediction and Evaluation of “Hot Spots” of Aggregation in Polypeptides. BMC Bioinform. 2007, 8, 65. [Google Scholar] [CrossRef]
- Walsh, I.; Seno, F.; Tosatto, S.C.E.; Trovato, A. PASTA 2.0: An Improved Server for Protein Aggregation Prediction. Nucleic Acids Res. 2014, 42, W301–W307. [Google Scholar] [CrossRef]

| SARS-CoV-2 Variant | RBD | RBM | Transmissibility (Average R0 Increase) | ||||
|---|---|---|---|---|---|---|---|
| RBD | pI | Charge at pH 7 | RBM | pI | Charge at pH 7 | ||
| Wuhan-Hu-1 (WT) | 323–531 | 8.27 | 3.9 | 438–506 | 7.94 | 0.8 | 1 |
| Alpha | 320–528 | 8.27 | 3.9 | 435–503 | 7.94 | 0.8 | ~29% increased over WT [69] |
| Beta | 320–528 | 8.42 | 4.9 | 435–503 | 8.83 | 2.8 | ~25% increased over WT [69] |
| Epsilon | 323–531 | 8.42 | 4.9 | 438–506 | 8.45 | 1.9 | 18.6–24% increased over WT [70] |
| Gamma | 323–531 | 8.42 | 4.9 | 438–506 | 8.83 | 2.8 | ~38% increased over WT [69] |
| Delta | 321–529 | 8.57 | 5.9 | 436–504 | 8.85 | 2.8 | ~97% increased over WT [69] |
| Omicron BA.1 | 320–528 | 8.70 | 7.2 | 435–503 | 9.55 | 6.1 | ~3.2 times more than Delta [71,72] |
| Omicron BA.2 | 320–528 | 8.70 | 7.2 | 435–503 | 9.55 | 6.1 | ~1.4 times more than BA.1 [72] |
| RBD | Amyloidogenicity * (%) |
|---|---|
| SARS_CoV (310–517) | 26.9% |
| Delta (321–529) | 29.7% |
| Epsilon (323–531) | 29.7% |
| Wuhan-Hu-1 (323–531) | 34.4% |
| Alpha (320–528) | 34.4% |
| Beta (320–528) | 34.4% |
| Gamma (323–521) | 34.4% |
| Omicron BA.1 (320–528) | 38.3% |
| Omicron BA.2 (320–528) | 40.7% |
| HCoV-NL63 (481–616) ** | 47.1% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aksenova, A.Y.; Likhachev, I.V.; Grishin, S.Y.; Galzitskaya, O.V. The Increased Amyloidogenicity of Spike RBD and pH-Dependent Binding to ACE2 May Contribute to the Transmissibility and Pathogenic Properties of SARS-CoV-2 Omicron as Suggested by In Silico Study. Int. J. Mol. Sci. 2022, 23, 13502. https://doi.org/10.3390/ijms232113502
Aksenova AY, Likhachev IV, Grishin SY, Galzitskaya OV. The Increased Amyloidogenicity of Spike RBD and pH-Dependent Binding to ACE2 May Contribute to the Transmissibility and Pathogenic Properties of SARS-CoV-2 Omicron as Suggested by In Silico Study. International Journal of Molecular Sciences. 2022; 23(21):13502. https://doi.org/10.3390/ijms232113502
Chicago/Turabian StyleAksenova, Anna Y., Ilya V. Likhachev, Sergei Y. Grishin, and Oxana V. Galzitskaya. 2022. "The Increased Amyloidogenicity of Spike RBD and pH-Dependent Binding to ACE2 May Contribute to the Transmissibility and Pathogenic Properties of SARS-CoV-2 Omicron as Suggested by In Silico Study" International Journal of Molecular Sciences 23, no. 21: 13502. https://doi.org/10.3390/ijms232113502
APA StyleAksenova, A. Y., Likhachev, I. V., Grishin, S. Y., & Galzitskaya, O. V. (2022). The Increased Amyloidogenicity of Spike RBD and pH-Dependent Binding to ACE2 May Contribute to the Transmissibility and Pathogenic Properties of SARS-CoV-2 Omicron as Suggested by In Silico Study. International Journal of Molecular Sciences, 23(21), 13502. https://doi.org/10.3390/ijms232113502







