NAC Transcription Factor GmNAC12 Improved Drought Stress Tolerance in Soybean
Abstract
1. Introduction
2. Results
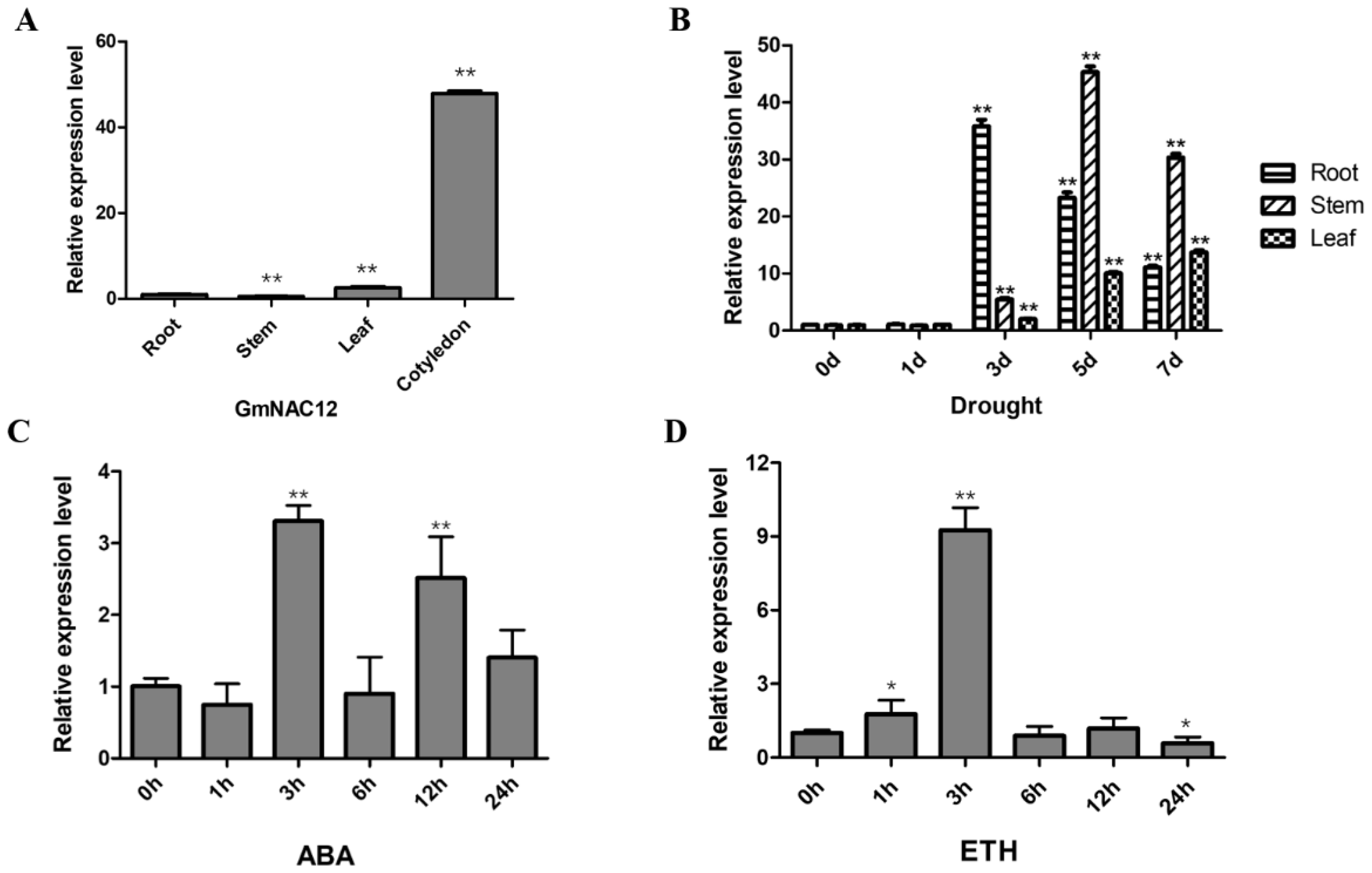
2.1. GmNAC12 Responded to Drought Stress as Well as ABA and ETH Hormone Treatments
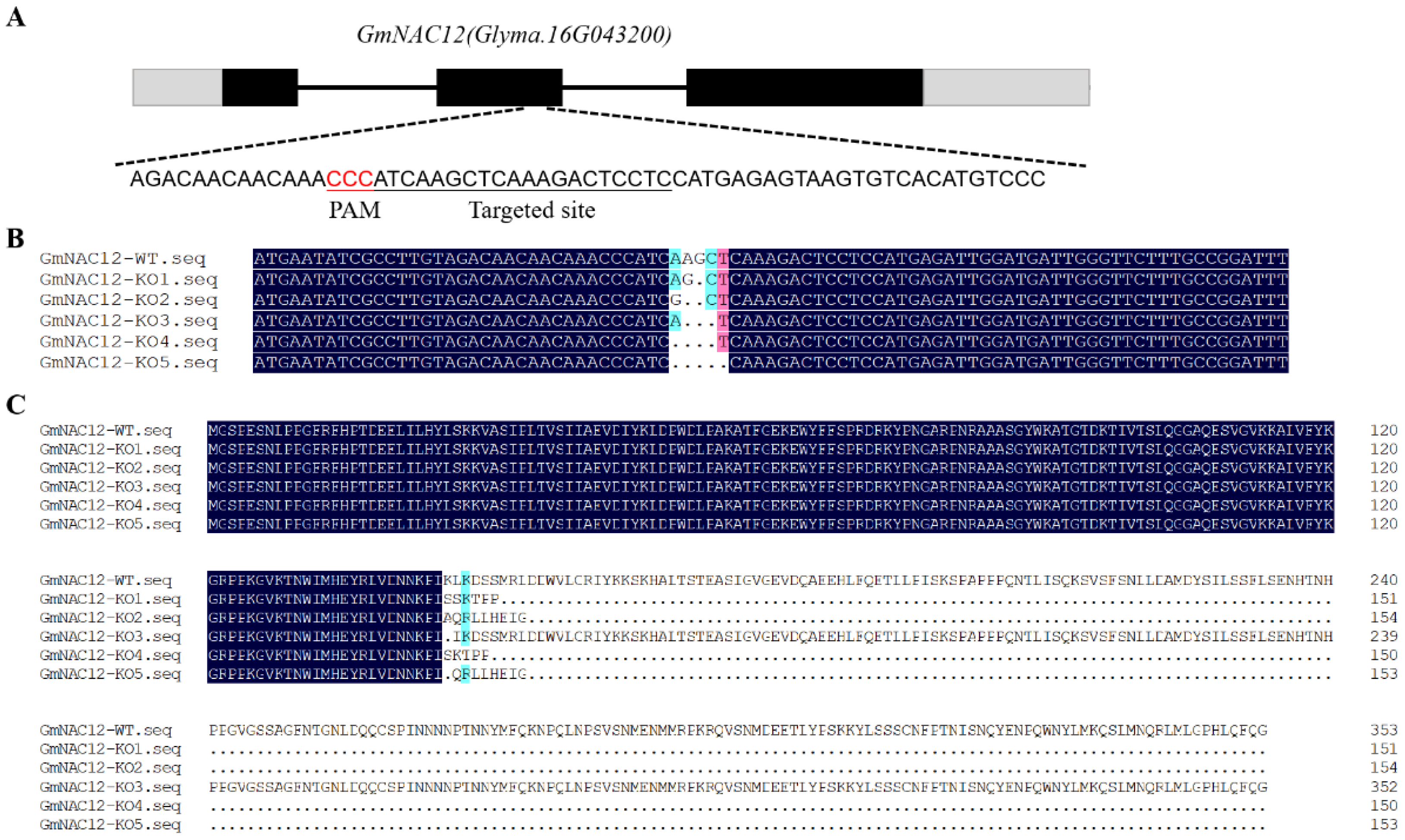
2.2. Targeted Mutagenesis of GmNAC12 Produced Using the CRISPR/Cas9 System
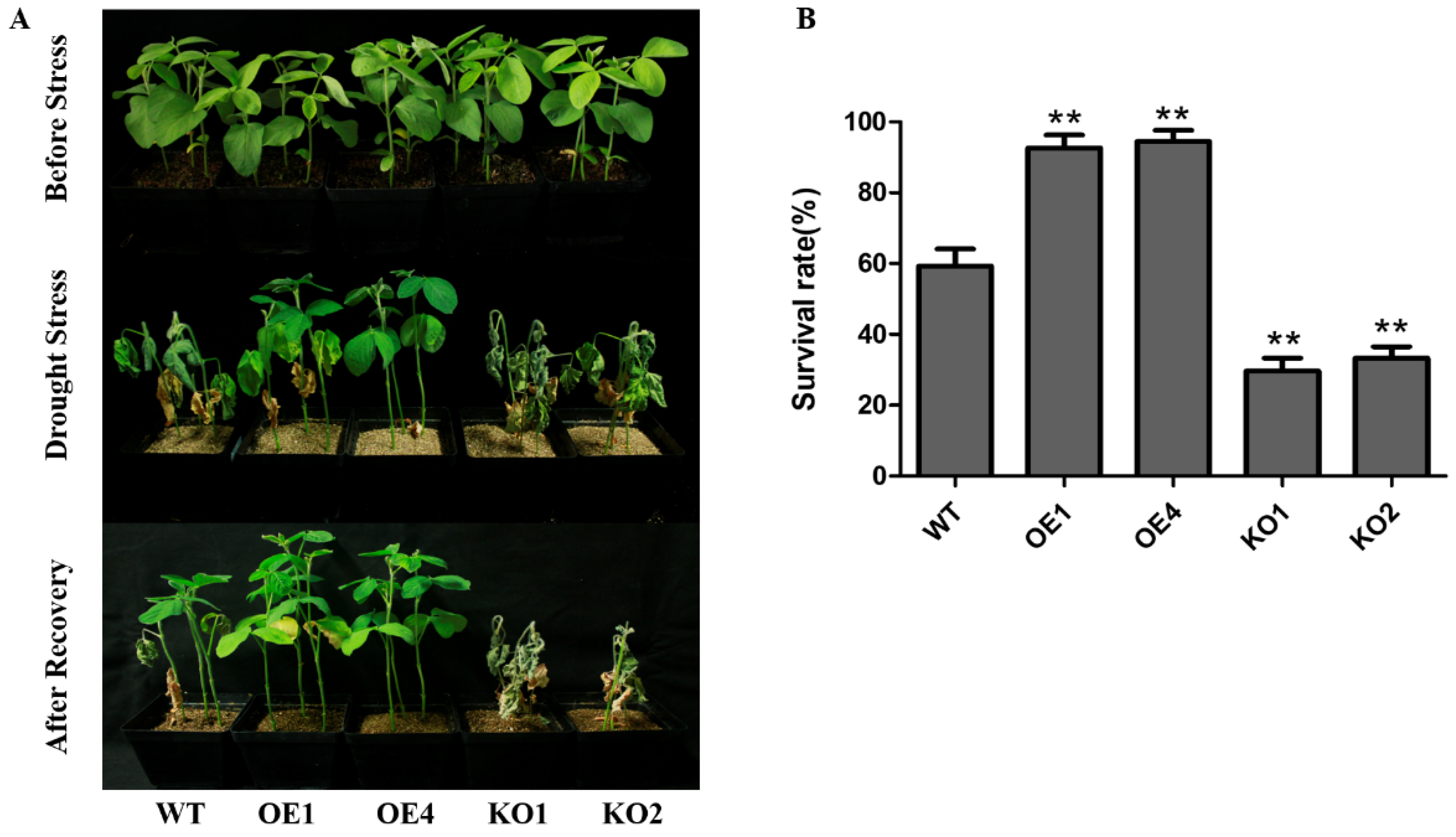
2.3. GmNAC12 Enhanced Drought Tolerance in Soybean Plants
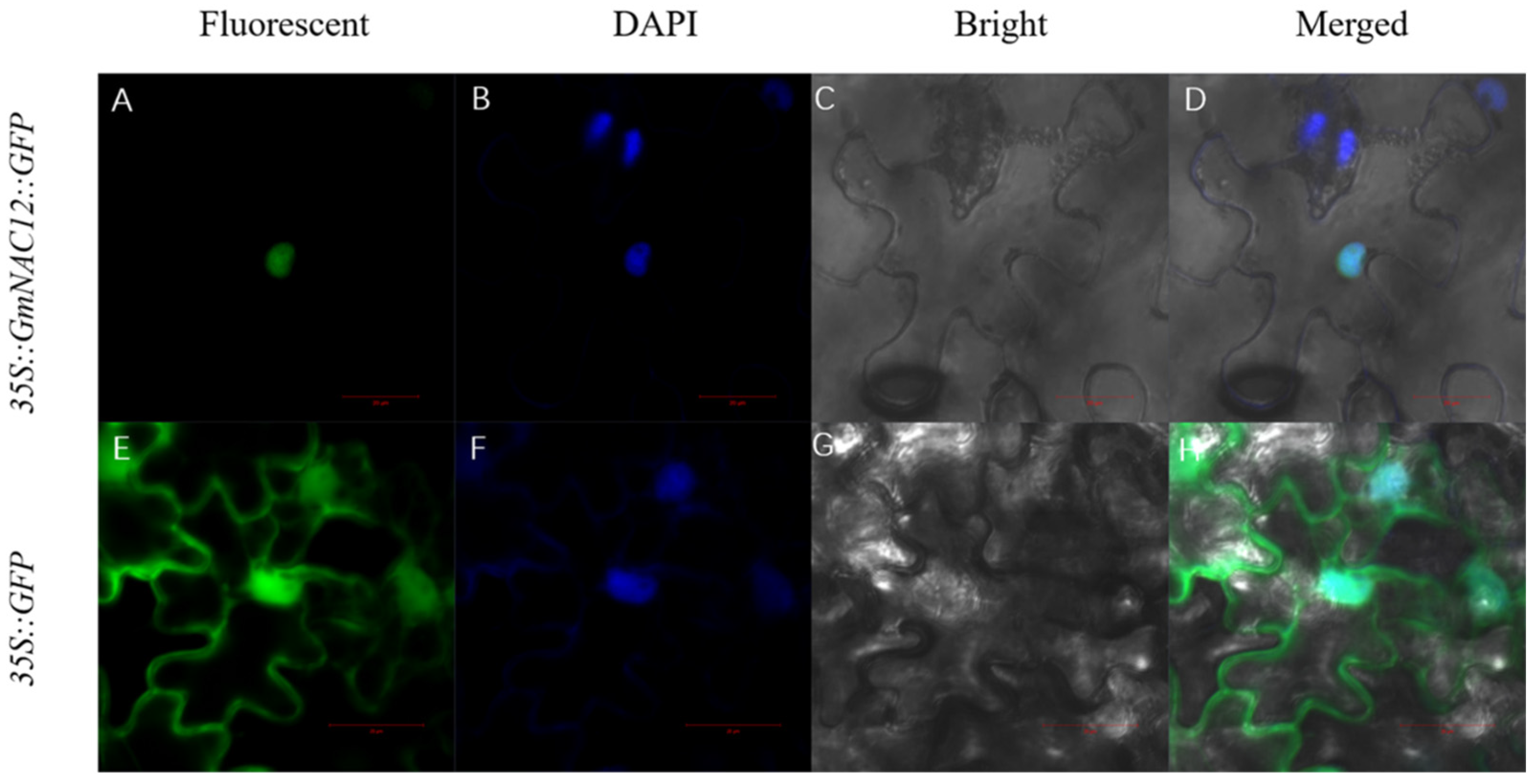
2.4. GmNAC12 Encoded a Nuclear Localisation Protein
2.5. Candidate Interaction Protein Analysis of GmNAC12
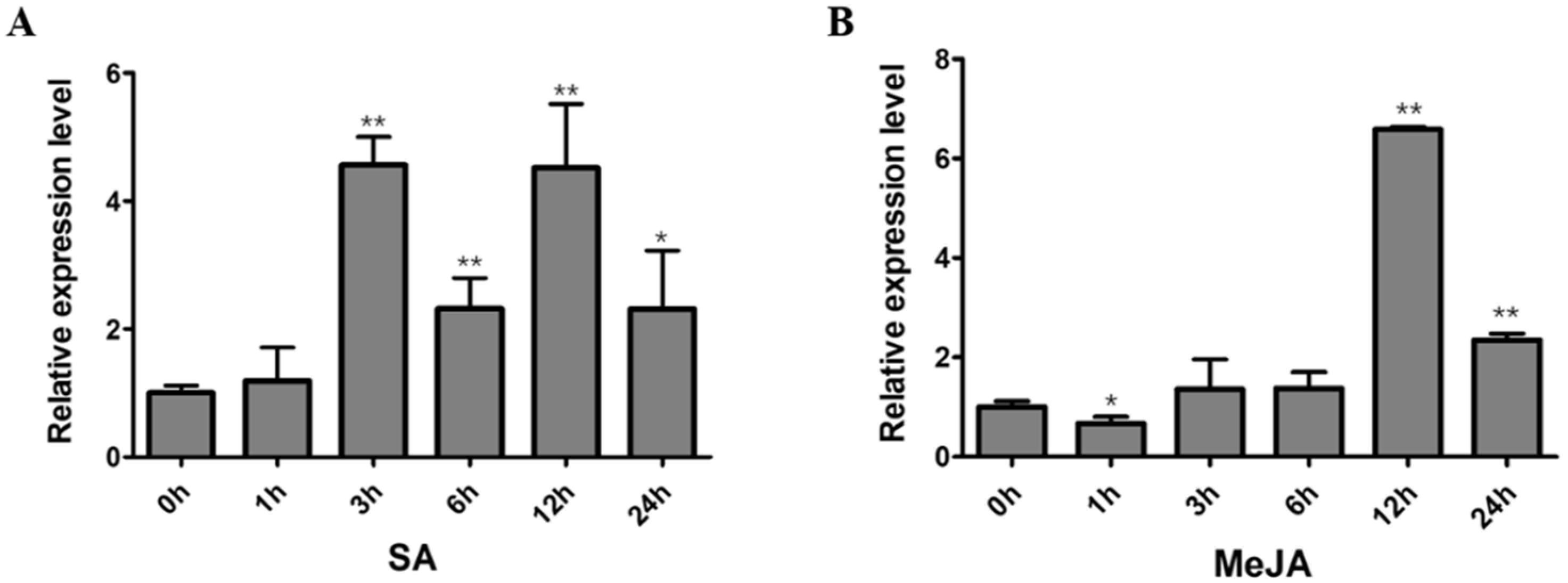
2.6. Expression Analysis of GmNAC12 under Biotic-Stress-Related Hormones
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and Stress Treatments
4.2. Quantitative Real-Time PCR Assay
4.3. Subcellular Localisation of GmNAC12 Protein
4.4. GmNAC12 Knockout in Soybean Plants Using the CRISPR/Cas9 System
4.5. GmNAC12 Overexpression in Soybean Plants
4.6. Yeast Two-Hybrid Assay
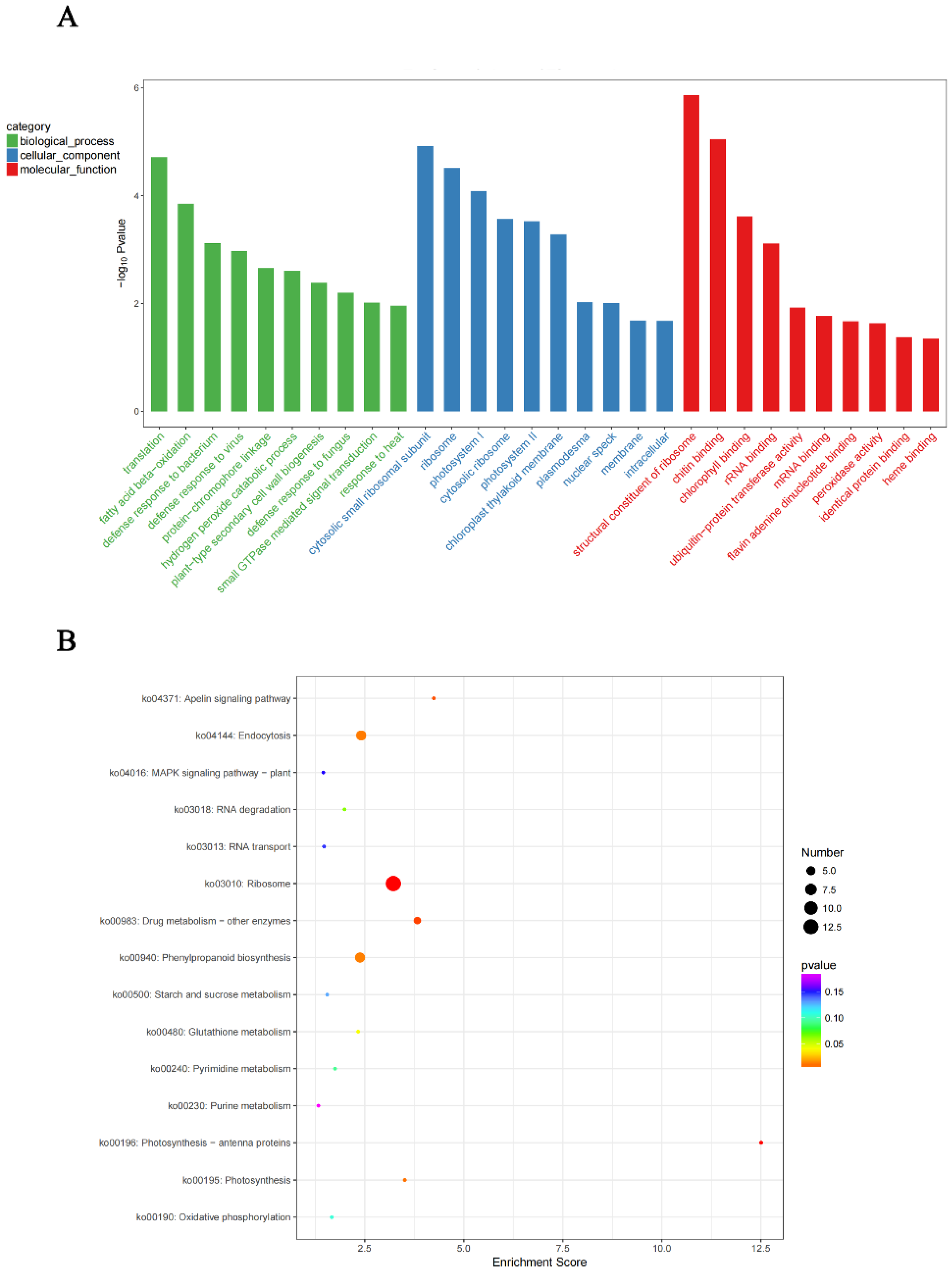
4.7. GO Annotation and KEGG Enrichment Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Shao, H.; Wang, H.; Tang, X. NAC transcription factors in plant multiple abiotic stress responses: Progress and prospects. Front. Plant Sci. 2015, 6, 902. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Zulfiqar, F.; Raza, A.; Mohsin, M.S.; Mahmud, A.J.; Fujita, M.; Fotopoulos, V. Reactive oxygen species and antioxidant defense in plants under abiotic stress: Revisiting the crucial role of a universal defense regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef] [PubMed]
- Bhat, M.A.; Mir, R.A.; Kumar, V.; Shah, A.A.; Zargar, M.A.; Rahman, S.; Jan, T.A. Mechanistic insights of CRISPR/Cas-mediated genome editing towards enhancing abiotic stress tolerance in plants. Physiol. Plant 2021, 172, 1255–1268. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, P.; Wang, D.; Shannon, G.; Zeng, A.; Orazaly, M.; Wu, C. Identification and mapping of stable QTL for protein content in soybean seeds. Mol. Breed. 2015, 35, 92. [Google Scholar] [CrossRef]
- Karikari, B.; Li, S.; Bhat, J.A.; Cao, Y.; Kong, J.; Yang, J.; Gai, J.; Zhao, T. Genome-wide detection of major and epistatic effect QTLs for seed protein and oil content in soybean under multiple environments using high-density bin map. Int. J. Mol. Sci. 2019, 20, 979. [Google Scholar] [CrossRef]
- Zhang, W.; Liao, X.; Cui, Y.; Ma, W.; Zhang, X.; Du, H.; Ma, Y.; Ning, L.; Wang, H.; Huang, F.; et al. A cation diffusion facilitator, GmCDF1, negatively regulates salt tolerance in soybean. PLoS Genet. 2019, 15, e1007798. [Google Scholar] [CrossRef] [PubMed]
- Chimungu, J.G.; Brown, K.M.; Lynch, J.P. Reduced root cortical cell file number improves drought tolerance in maize. Plant Physiol. 2014, 166, 1943–1955. [Google Scholar] [CrossRef] [PubMed]
- Per, T.S.; Khan, N.A.; Reddy, P.S.; Masood, A.; Hasanuzzaman, M.; Khan, M.I.R.; Anjum, N.A. Approaches in modulating proline metabolism in plants for salt and drought stress tolerance: Phytohormones, mineral nutrients and transgenics. Plant Physiol. Biochem. 2017, 115, 126–140. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Xuan, H.; Yang, C.; Guo, N.; Wang, H.; Zhao, J.; Xing, H. GmHsp90A2 is involved in soybean heat stress as a positive regulator. Plant Sci. 2019, 285, 26–33. [Google Scholar] [CrossRef]
- He, M.; He, C.Q.; Ding, N.Z. Abiotic stresses: General defenses of land plants and chances for engineering multistress tolerance. Front. Plant Sci. 2018, 9, 1771. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Morran, S.; Eini, O.; Pyvovarenko, T.; Parent, B.; Singh, R.; Ismagul, A.; Eliby, S.; Langride, P.; Lopato, S. Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol. J. 2011, 9, 230–249. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.; Guo, Q.; Chen, L.; Ren, F.; Wang, Q.; Zheng, Y.; Li, X. Two Brassica napus genes encoding NAC transcription factors are involved in response to high-salinity stress. Plant Cell Rep. 2012, 31, 1991–2003. [Google Scholar] [CrossRef]
- Zhang, H.; Ma, F.; Wang, X.; Liu, S.; Saeed, U.H.; Hou, X.; Zhang, Y.; Luo, D.; Meng, Y.; Zhang, W.; et al. Molecular and functional characterization of CaNAC035, an NAC transcription factor from pepper (Capsicum annuum L.). Front. Plant Sci. 2020, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 2012, 217037. [Google Scholar] [CrossRef]
- Rauf, M.; Arif, M.; Fisahn, J.; Xue, G.P.; Balazadeh, S.; Mueller-Roeber, B. NAC transcription factor speedy hyponastic growth regulates flooding-induced leaf movement in Arabidopsis. Plant Cell 2013, 25, 4941–4955. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.K.; Rung, J.H.; Gregersen, P.L.; Gjetting, T.; Fuglsang, A.T.; Hansen, M.; Joehnk, N.; Lynkjaer, M.F.; Collinge, D.B. The Hv NAC6 transcription factor: A positive regulator of penetration resistance in barley and Arabidopsis. Plant Mol. Biol. 2007, 65, 37–150. [Google Scholar] [CrossRef] [PubMed]
- Ernst, H.A.; Olsen, N.A.; Skriver, K.; Larsen, S.; Leggio, L.L. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 2004, 5, 297–303. [Google Scholar] [CrossRef]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Osato, N.; Kawai, J.; Carninci, P.; Hayashizaki, Y.; et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef]
- Na, C.; Shuanghua, W.; Jinglong, F.; Bihao, C.; Jianjun, L.; Changming, C.; Jin, J. Overexpression of the eggplant (Solanum melongena) NAC family transcription factor S mNAC suppresses resistance to bacterial wilt. Sci. Rep. 2016, 6, 31568. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Yin, Y.; Chen, F.; Xu, Y.; Dixon, R.A. A bioinformatic analysis of NAC genes for plant cell wall development in relation to lignocellulosic bioenergy production. Bioenergy Res. 2009, 2, 217–232. [Google Scholar] [CrossRef]
- Aida, M.; Ishida, T.; Fukaki, H.; Fujisawa, H.; Tasaka, M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 1997, 9, 841–857. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.K.; Mostofa, G.M.; Li, W.; Ha, V.C.; Watanabe, Y.; Le, D.T.; Thao, N.P.; Tran, L.S.P. The soybean transcription factor GmNAC085 enhances drought tolerance in Arabidopsis. Environ. Exp. Bot. 2018, 151, 12–20. [Google Scholar] [CrossRef]
- Le, D.T.; Nishiyama, R.; Watanabe, Y.; Mochida, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S.P. Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 2011, 18, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Ju, Y.; Yue, X.; Min, Z.; Wang, X.; Fang, Y.; Zhang, J. VvNAC17, a novel stress-responsive grapevine (Vitis vinifera L.) NAC transcription factor, increases sensitivity to abscisic acid and enhances salinity, freezing, and drought tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2020, 146, 98–111. [Google Scholar] [CrossRef] [PubMed]
- Negi, S.; Tak, H.; Ganapathi, T.R. A banana NAC transcription factor (MusaSNAC1) impart drought tolerance by modulating stomatal closure and H2O2 content. Plant Mol. Biol. 2018, 96, 57–471. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Wei, W.; Song, Q.; Chen, H.; Zhang, Y.; Wang, F.; Zou, H.; Lei, G.; Tian, A.; Zhang, W.; et al. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 2011, 68, 302–313. [Google Scholar] [CrossRef] [PubMed]
- Kou, X.; Liu, C.; Han, L.; Wang, S.; Xue, Z. NAC transcription factors play an important role in ethylene biosynthesis, reception and signaling of tomato fruit ripening. Mol. Genet. Genom. 2016, 291, 1205–1217. [Google Scholar] [CrossRef] [PubMed]
- Pimenta, M.R.; Silva, P.A.; Mendes, G.C.; Alves, J.R.; Caetano, H.D.N.; Machado, J.P.B.; Brustolini, O.J.B.; Carpinetti, P.A.; Melo, B.P.; Silva, J.C.F.; et al. The stress-induced soybean NAC transcription factor GmNAC81 plays a positive role in developmentally programmed leaf senescence. Plant Cell Physiol. 2016, 57, 1098–1114. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Deng, Z.; Lai, J.; Zhang, Y.; Yang, C.; Yin, B.; Zhao, Q.; Zhang, L.; Li, Y.; Yang, C.; et al. Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 2009, 19, 1279–1290. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.S.P.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Tran, L.S.P.; Nguyen, V.D.; Fujita, M.; Maruyama, K.; Todaka, D.; Ito, Y.; Hayashi, N.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007, 51, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Dai, M.; Yao, J.; Xiao, B.; Li, X.; Zhang, Q.; Xiong, L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 2987–12992. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.S.; Kim, Y.S.; Baek, K.H.; Jung, H.; Ha, S.H.; Choi, Y.D.; Kim, M.; Reuzeau, C.; Kim, J.K. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 2010, 153, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Takasaki, H.; Maruyama, K.; Kidokoro, S.; Ito, Y.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K.; Nakashima, K. The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol. Genet. Genomics 2010, 284, 173–183. [Google Scholar] [CrossRef]
- So, H.A.; Lee, J.H. NAC transcription factors from soybean (Glycine max L.) differentially regulated by abiotic stress. J. Plant Biol. 2019, 62, 147–160. [Google Scholar] [CrossRef]
- Tak, H.; Negi, S.; Ganapathi, T.R. Banana NAC transcription factor MusaNAC042 is positively associated with drought and salinity tolerance. Protoplasma 2017, 254, 803–816. [Google Scholar] [CrossRef]
- Yang, C.; Huang, Y.; Lv, W.; Zhang, Y.; Bhat, J.A.; Kong, J.; Xing, H.; Zhao, J.; Zhao, T. GmNAC8 acts as a positive regulator in soybean drought stress. Plant Sci. 2020, 293, 110442. [Google Scholar] [CrossRef]
- Yang, X.; Kim, M.Y.; Ha, J.; Lee, S.H. Overexpression of the soybean NAC gene GmNAC109 increases lateral root formation and abiotic stress tolerance in transgenic Arabidopsis plants. Front. Plant Sci. 2019, 10, 1036. [Google Scholar] [CrossRef]
- Melo, B.P.; Fraga, O.T.; Silva, J.C.F.; Ferreira, D.O.; Brustolini, O.J.; Carpinetti, P.A.; Machado, J.P.B.; Reis, P.A.B.; Fontes, E.P.B. Revisiting the soybean GmNAC superfamily. Front. Plant Sci. 2018, 9, 1864. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Chen, L.; Liu, X.; Sun, S.; Wu, C.; Jiang, B.; Han, T.; Hou, W. CRISPR/Cas9-mediated genome editing in soybean hairy roots. PLoS ONE 2015, 10, e0136064. [Google Scholar] [CrossRef] [PubMed]
- Bao, A.; Chen, H.; Chen, L.; Chen, S.; Hao, Q.; Guo, W.; Qiu, D.; Shan, Z.; Yang, Z.; Yuan, S.; et al. CRISPR/Cas9-mediated targeted mutagenesis of GmSPL9 genes alters plant architecture in soybean. BMC Plant Biol. 2019, 19, 131. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Chen, L.; Liu, X.; Guo, C.; Sun, S.; Wu, C.; Jiang, B.; Han, T.; Hou, W. CRISPR/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean. Plant Biotechnol. J. 2018, 16, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Hussain, R.M.; Ali, M.; Feng, X.; Li, X. The essence of NAC gene family to the cultivation of drought-resistant soybean (Glycine max L. Merr.) cultivars. BMC Plant Biol. 2017, 17, 55. [Google Scholar] [CrossRef]
- Rao, V.S.; Srinivas, K.; Sujini, G.N.; Kumar, G.N.S. Protein-protein interaction detection: Methods and analysis. J. Proteom. 2014, 2014, 147648. [Google Scholar] [CrossRef] [PubMed]
- Wani, S.H.; Kumar, V.; Shriram, V.; Sah, S.K. Phytohormones and their metabolic engineering for abiotic stress tolerance in crop plants. Crop J 2016, 4, 162–176. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Qian, D.; Zhang, Z.; He, J.; Zhang, P.; Ou, X.; Li, T.; Niu, L.; Nan, Q.; Niu, Y.; He, W.; et al. Arabidopsis ADF5 promotes stomatal closure by regulating actin cytoskeleton remodeling in response to ABA and drought stress. J. Exp. Bot. 2019, 70, 435–446. [Google Scholar] [CrossRef]
- Jiang, W.; Zhou, H.; Bi, H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013, 41, e188. [Google Scholar] [CrossRef]
- Mushtaq, M.; Bhat, J.A.; Mir, Z.A.; Sakina, A.; Ali, S.; Singh, A.K.; Tyagi, A.; Salgotra, R.K.; Dar, A.A.; Bhat, R. CRISPR/Cas approach: A new way of looking at plant-abiotic interactions. J. Plant Physiol. 2018, 224, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Feng, M.; Chen, W.; Zhou, X.; Lu, J.; Wang, Y.; Li, Y.; Jiang, C.; Gan, S.; Ma, N.; et al. In rose, transcription factor PTM balances growth and drought survival via PIP2;1 aquaporin. Nat. Plants 2019, 5, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, Q.; Nan, H.; Li, X.; Lu, S.; Zhao, X.; Liu, B.; Guo, C.; Kong, F.; Cao, D. Overexpression of GmFDL19 enhances tolerance to drought and salt stresses in soybean. PLoS ONE 2017, 12, e0179554. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.D.; Volko, S.M.; Ledford, H.; Ausubel, F.M.; Dong, X. Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell 2000, 12, 2175–2190. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Huang, G.; Zou, D.; Yan, J.; Li, Y.; Hu, S.; Li, X. The cotton (Gossypium hirsutum) NAC transcription factor (FSN1) as a positive regulator participates in controlling secondary cell wall biosynthesis and modification of fibers. New Phytol. 2018, 217, 625–640. [Google Scholar] [CrossRef] [PubMed]
- Livak, J.K.; Schmittgen, D.T. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2 −ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.; Huang, Y.; Lv, P.; Antwi-Boasiako, A.; Begum, N.; Zhao, T.; Zhao, J. NAC Transcription Factor GmNAC12 Improved Drought Stress Tolerance in Soybean. Int. J. Mol. Sci. 2022, 23, 12029. https://doi.org/10.3390/ijms231912029
Yang C, Huang Y, Lv P, Antwi-Boasiako A, Begum N, Zhao T, Zhao J. NAC Transcription Factor GmNAC12 Improved Drought Stress Tolerance in Soybean. International Journal of Molecular Sciences. 2022; 23(19):12029. https://doi.org/10.3390/ijms231912029
Chicago/Turabian StyleYang, Chengfeng, Yanzhong Huang, Peiyun Lv, Augustine Antwi-Boasiako, Naheeda Begum, Tuanjie Zhao, and Jinming Zhao. 2022. "NAC Transcription Factor GmNAC12 Improved Drought Stress Tolerance in Soybean" International Journal of Molecular Sciences 23, no. 19: 12029. https://doi.org/10.3390/ijms231912029
APA StyleYang, C., Huang, Y., Lv, P., Antwi-Boasiako, A., Begum, N., Zhao, T., & Zhao, J. (2022). NAC Transcription Factor GmNAC12 Improved Drought Stress Tolerance in Soybean. International Journal of Molecular Sciences, 23(19), 12029. https://doi.org/10.3390/ijms231912029





