Abstract
Powdery mildew caused by Erysiphe pisi DC. is a major disease affecting pea worldwide. This study aimed to confirm the resistance genes contained in three powdery mildew-resistant Chinese pea landraces (Suoshadabaiwan, Dabaiwandou, and Guiwan 1) and to develop the functional markers of the novel resistance genes. The resistance genes were identified by genetic mapping and PsMLO1 gene sequence identification. To confirm the inheritance of powdery mildew resistance in the three Landraces, the susceptible cultivars Bawan 6, Longwan 1, and Chengwan 8 were crossed with Suoshadabaiwan, Dabaiwandou, and Guiwan 1 to produce F1, F2, and F2:3 populations, respectively. All F1 plants were susceptible to E. pisi, and phenotypic segregation patterns in all the F2 and F2:3 populations fit the 3:1 (susceptible: resistant) and 1:2:1 (susceptible homozygotes: heterozygotes: resistant homozygotes) ratios, respectively, indicating powdery mildew resistance in the three Landraces were controlled by a single recessive gene, respectively. The analysis of er1-linked markers and genetic mapping in the F2 populations suggested that the recessive resistance genes in three landraces could be er1 alleles. The cDNA sequences of 10 homologous PsMLO1 cDNA clones from the contrasting parents were obtained. A known er1 allele, er1-4, was identified in Suoshadabaiwan. Two novel er1 alleles were identified in Dabaiwandou and Guiwan 1, which were designated as er1-13 and er1-14, respectively. Both novel alleles were characterized with a 1-bp deletion (T) in positions 32 (exon 1) and 277 (exon 3), respectively, which caused a frame-shift mutation to result in premature termination of translation of PsMLO1 protein. The co-dominant functional markers specific for er1-13 and er1-14, KASPar-er1-13, and KASPar-er1-14 were developed and effectively validated in populations and pea germplasms. Here, two novel er1 alleles were characterized and their functional markers were validated. These results provide powerful tools for marker-assisted selection in pea breeding.
1. Introduction
Pea (Pisum sativum L.) is an important and widely distributed cool season legume crop, which frequently suffers from abiotic and biotic stresses during the whole growth season [1,2]. Among the biotic factors, the disease is the main cause affecting pea production [2]. Powdery mildew caused by Erysiphe pisi DC. is a major constraint for pea yield and quality worldwide [3]. E. pisi infections of peas can lead to yield losses of up to 80% in regions that are suitable for disease infection [3]. To date, the use of resistant cultivars carrying the E. pisi-resistant gene er1 has been considered to be the most effective and environmentally friendly way to control this disease [4,5].
E. pisi-resistance in pea has been proved to be controlled by three different genes in different germplasms, including two single recessive genes (er1 and er2) and one dominant gene (Er3) [6,7,8,9]. The er1, er2, and Er3 genes have been mapped on different linkage groups of peas using linked markers [10,11,12,13,14,15,16,17,18]. The two recessive genes er1 and er2 were mapped to pea linkage groups (LGs) VI and III, respectively [10,19]. The dominant gene Er3 isolated from wild pea (Pisum fulvum) was located to pea LG IV recently [20].
To date, the recessive gene er1 is the most widely used gene in pea production due to er1 conferring high resistance or immunity to E. pisi in most pea germplasms [21]. In contrast, resistance conferred by er2 is unstable and easily affected by leaf development stage and plant location [7,21,22,23]. er2 is only found in a few pea germplasms [21]. Er3 was known from wild pea (P. fulvum), and there have not been any extensive studies conducted to date [8,24].
Gene er1 inhibits the incursion of E. pisi into pea epidermal cells, which confers stable and durable resistance to E. pisi [23]. Recent studies have shown that the er1-resistant phenotype is caused by loss-of-function mutations in the pea MLO (Mildew Resistance Locus O) homolog (PsMLO1). The MLO gene family has been identified in both dicots (e.g., Arabidopsis thaliana, and tomato—Solanum lycopersicum) and monocots (e.g., barley—Hordeum vulgare) [9,25,26,27,28,29].
To date, total of 12 er1 alleles have been identified conferring resistance to E. pisi by natural mutation or obtained by mutagenesis in pea germplasms: er1-1 (also known as er1mut1) [9,13,16,30,31], er1-2 [9,15,16], er1-3 [9], er1-4 [9], er1-5 [28], er1-6 [18], er1-7 [17], er1-8, er1-9 [32], er1-10 (also known as er1mut2) [13,30,33], and er1-11 [33,34]; er1-12 was more recently identified in pea germplasm JI2019 from India [35]. Each er1 allele corresponds to a different PsMLO1 mutation site and pattern. Among the 12 er1 alleles identified, only er1-1 and er1-2 are commonly applied in pea breeding programs [9,28]. Previous studies revealed that the functional markers of the known er1 alleles have been developed and applied for the rapid selection of pea germplasms resistant to E. pisi in pea breeding [15,17,18,28,33,34,36].
E. pisi severely affects the yield and quality of pea crops in China [2]. The disease infects up to 100% of pea plants in some regions of planting susceptible cultivars. In our previous studies, we have focused on the identification of pea germplasms resistant to E. pisi [31,37]. A novel er1 allele er1-6 had been identified in a Chinese pea germplasm [17] and new alleles er1-7, er1-8, and er1-9 were identified in pea germplasms from India, Afghanistan, and Australia, respectively [17,32]. er1-6 was also identified in some pea landraces from Yunnan Province of China [18]. Thus, a natural mutation of the er1 gene conferring E. pisi-resistance has been observed in some Chinese pea landraces, which provides rich resistant sources that can be used to improve the E. pisi resistance of Chinese pea cultivars. The allelic diversity of this locus in the cultivated pea has been well characterized; however, relatively few studies have investigated and characterized the E. pisi-resistant gene in Chinese pea landraces. Thus, this study aimed to identify and characterize the E. pisi-resistant gene in three E. pisi-resistant Chinese pea landraces by genetic mapping and homologous PsMLO1 gene sequence cloning. Additionally, any novel er1 alleles were performed to develop their functional markers to improve marker-assisted selection in E. pisi-resistant pea breeding programs.
2. Results
2.1. Phenotypic Evaluation and Inheritence Analysis for Resistance
Six parental cultivars and contrasting controls were evaluated for their resistance to the E. pisi isolate EPYN. At 10 days post-inoculation, the E. pisi disease severity of the susceptible control was rated as score 4, indicating susceptibility to E. pisi. As expected, the three resistant pea parents, Suoshadabaiwan, Dabaiwandou, and Guiwan 1, and resistant control (Xucai 1) were immune to E. pisi infection (disease severity 0), while the three susceptible parents (Bawan 6, Longwan 1, and Chengwan 8) were susceptible to E. pisi (disease severity 4) (Figure 1). The segregation patterns of E. pisi resistance in the F1, F2, and F2:3 populations derived from the crosses Bawan 6 × Suoshadabaiwan, Longwan 1 × Dabaiwandou, and Chengwan 8 × Guiwan 1 are presented in Table 1.

Figure 1.
Phenotypic evaluation of the three Erysiphe pisi-resistant pea cultivars Suoshadabaiwan, Dabaiwandou, and Guiwan 1, as well as the three E. pisi-susceptible cultivars Bawan 6, Longwan 1, and Chengwan 8, after inoculation with E. pisi isolate EPYN. (A) Suoshadabaiwan and E. pisi-susceptible cultivar Bawan 6 (BW6). (B) Dabaiwandou and E. pisi-susceptible cultivar Longwan 1. (C) Guiwan 1 and E. pisi-susceptible cultivar Chengwan 8.

Table 1.
Segregation patterns of pea resistance to powdery mildew in the F1, F2, and F2:3 populations derived from three crosses, Bawan 6 × Suoshadabaiwan, Longwan 1 × Dabaiwandou, Chengwan 8 × Guiwan 1.
Five F1 plants produced from the cross Bawan 6 × Suoshadabaiwan were susceptible to E. pisi (Table 1). One of the five plants generated 102 F2 and F2:3 offspring through self-pollination. Of these 102 F2 plants, 26 were resistant (R) to E. pisi, and 76 were susceptible (S) to E. pisi. This indicates that the segregation ratio (resistance: susceptibility) in the F2 population was 1:3 (χ2 = 0.02; p = 0.88), indicating the inheritance of a single recessive gene. Moreover, a segregation ratio of 26 (homozygous resistant):51 (segregating):25 (homozygous susceptible) in the F2:3 population fitted well with the genetic model of 1:2:1 ratio (χ2 = 0.03, p = 0.99) (Table 1), confirming that the E. pisi resistance in Suoshadabaiwan was controlled by a single recessive gene.
The cross of Longwan 1 × Dabaiwandou generated six F1 plants, which showed E. pisi-susceptibility (Table 1). One of six F1 plants generated 121 F2 offspring. Of 121, 29 were resistant, and 92 of 121 were susceptible to E. pisi. The segregation ratio in the F2 population of resistance to susceptibility fitted a genetic model ratio of 1:3 (χ2 = 0.07; p = 0.79), also indicating the inheritance of a single recessive gene. Moreover, a segregation ratio of 29 (homozygous resistant):56 (segregating):36 (homozygous susceptible) in the F2:3 population (121 families) fitted well with the genetic model of 1:2:1 ratio (χ2 = 1.41; p = 0.49), indicating that E. pisi resistance in Dabaiwandou was also controlled by a single recessive gene (Table 1).
The cross of Chengwan 8 × Guiwan 1 generated eight F1 plants which showed E. pisi-susceptibility (Table 1). One of eight F1 plants generated 131 F2 offspring. Of 131, 36 were resistant, and 95 of 131 were susceptible to E. pisi. The segregation ratio in the F2 population of resistance to susceptibility fitted a genetic model ratio of 1:3 (χ2 = 0.43; p = 0.51), also indicating the recessive inheritance of a single gene. Moreover, a segregation ratio of 36 (homozygous resistant):61 (segregating):34 (homozygous susceptible) in the F2:3 population (131 families) fitted well with the genetic model of 1:2:1 ratio (χ2 = 0.67; p = 0.71), indicating that E. pisi resistance in Guiwan 1 was also controlled by a single recessive gene (Table 1).
2.2. Mapping of Resistance Genes
Of the molecular markers tested, six (c5DNAmet, AD160, AC74, AD51, AD59, and AD60) were polymorphic between contrasting parents Bawan 6 and Suoshadabaiwan, and three (c5DNAmet, AA220, and AD51) were polymorphic between Longwan 1 and Dabaiwandou, Unfortunately, no polymorphic marker appeared between Longwan 1 and Dabaiwandou among the above markers tested. Thus, the additional eight SSR markers (16410, 28516, 26140, 23309, 29872, 26514, 23949, and 22903) developed recently were used to test the polymorphism between Longwan 1 and Dabaiwandou [38]. Two (26514 and 22903) were polymorphic between the contrasting parents, Longwan 1 and Dabaiwandou. All polymorphic markers between the parents were likely linked to the E. pisi resistance gene, respectively. Thus, the six, three, and the two parental polymorphic markers were used to confirm the genotypes of each F2 plant derived from Bawan 6 × Suoshadabaiwan, Longwan 1 × Dabaiwandou, and Chengwan 8 × Guiwan 1, respectively. This genetic linkage analysis suggested that six markers (c5DNAmet, AD160, AC74, AD51, AD59, and AD60), three markers (c5DNAmet, AA220, and AD51), and two markers (26514 and 22903) were linked to the resistance gene er1 in Suoshadabaiwan, Dabaiwandou, and Guiwan 1, respectively (Figure 2). Our results also indicated that the resistance genes in the three resistant cultivars were located in the er1 region. In Suoshadabaiwan, the linkage map indicated that the markers (AD59 and AD60) were mapped on both sides of the target gene with 3.4 cM and 8.3 cM genetic distances, respectively (Figure 2A). In Dabaiwandou, two other markers (c5DNAmet and AA220) were located on both sides of the target gene with 2.6 cM and 11.6 cM genetic distances, respectively (Figure 2B). In Guiwan 1, two markers (26514 and 22903) were located on both sides of the target gene with 12.8 cM and 19.3 cM genetic distances, respectively (Figure 2C). Our linkage and genetic map analyses confirmed that an er1 allele controlled E. pisi resistance in Suoshadabaiwan, Dabaiwandou, and Guiwan 1, respectively (Figure 2).
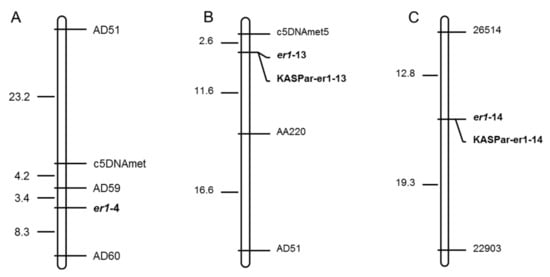
Figure 2.
Genetic linkage maps constructed using the er1-linked markers and the functional markers for er1-13 and er1-14, based on the F2 populations derived from (A) Bawan 6 × Suoshadabaiwan, (B) Longwan 1 × Dabaiwandou, and (C) Chengwan 8 × Guiwan 1. Map distances and loci order were determined with MAPMAKER v3.0. Estimated genetic distances between loci are shown to the left of the maps in centiMorgans (cM).
2.3. PsMLO1 Sequence Analysis
The PsMLO1 cDNA sequences of Bawan 6, Longwan 1, Chengwan 8, and the susceptible parents, were consistent with that of the wild-type PsMLO1 cDNA.
In landrace Suoshadabaiwan, a 1-bp deletion (A) was identified in a previously reported position 91 in exon 1 of the PsMLO1 cDNA sequence. This result is consistent with the mutation in the er1 gene carried by germplasm YI (JI1591), named er1-4. In landrace Dabaiwandou, a novel mutation pattern was found in the Dabaiwandou cDNA fragment homologous to PsMLO1: a 1-bp deletion (T) corresponding to positions 32 in exon 1 (the first exon) of the PsMLO1 cDNA sequence. This deletion caused a substitution of the amino acid leucine with tryptophan at position 11 of the PsMLO1 protein sequence (Figure 3A). This change caused the early termination of protein translation, probably also resulting in a functional change of PsMLO1 (Figure 3A). In Guiwan 1, a 1-bp deletion (T) was also identified in a previously unreported position 277 in exon 3 of the PsMLO1 cDNA sequence. This deletion caused a substitution of the amino acid tryptophan with glycine at position 93 of the PsMLO1 protein sequence (Figure 3B). This change caused the early termination of protein translation, probably also resulting in a functional change of PsMLO1 (Figure 3B). The two natural mutations differed from all known er1 alleles, indicating that the E. pisi resistance of Dabaiwandou and Guiwan 1 was controlled by the novel alleles of er1. These novel alleles were designated er1-13 and er1-14, respectively, following the accepted nomenclature [9,17,18,32,33,35,36]. Thus, a known and two novel er1 alleles were discovered in the three resistant cultivars, Suoshadabaiwan (from Chongqing), Dabaiwandou (from Yunnan), and Guiwan 1 (from Guangxi), respectively.
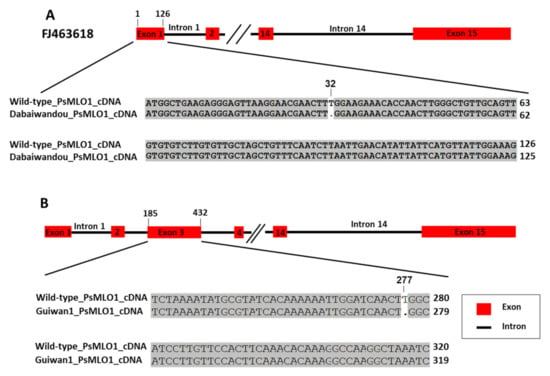
Figure 3.
PsMLO1 cDNA sequence comparisons of those from the powdery mildew-resistant pea landrace Dabaiwandou and Guiwan 1 with the wild-type pea cultivar Sprinter (GenBank accession number: FJ463618.1). (A) There is a single base deletion (T) in the PsMLO1 cDNA of Dabaiwandou at positions 32 of exon 1. (B) There is a single base deletion (T) in the PsMLO1 cDNA sequence of Guiwan 1 at position277 in exon 3. The figure shows the difference of nucleotide sequence from Dabaiwandou, Guiwan 1, and the wild-type pea cultivar Sprinter. The two mutation sites are indicated in the respective cDNA sequences.
2.4. Development of Functional Markers for er1-13 and er1-14
The KASPar markers flanking the 1-bp (T) deletion sequences from Dabaiwandou and Guiwan 1 were designed as functional markers specific for KASPar-er1-13 and KASPar-er1-14, respectively.
As expected, KASPar-er1-13 and KASPar-er1-14 successfully distinguished the contrasting parents (Longwan 1 and Dabaiwandou, Chengwan 8 and Guiwan 1) into two different clusters corresponding to the FAM-labeled and HEX-labeled groups in the Kompetitive allele-specific PCR (KASPar) assay, respectively. When KASPar-er1-13 and KASPar-er1-14 were used to analyze the 121 and 131 F2 progeny derived from Longwan 1 × Dabaiwandou and Chengwan 8 × Guiwan 1, the KASPar markers clearly separated the F2 progeny into three clusters corresponding to three genotypes: homozygous resistant, homozygous susceptible, and heterozygous (Figure 4). In the F2 population derived from Longwan 1 × Dabaiwandou, 29 plants were identified as homozygous resistant, 56 were heterozygous, and 36 were homozygous susceptible. In the F2 population derived from Chengwan 8 × Guiwan 1, 36 plants were homozygous resistant, 61 were heterozygous, and 34 were homozygous susceptible. These results were completely consistent with the phenotypes of both F2:3 populations, suggesting that KASPar-er1-13 and KASPar-er1-14 co-segregated with er1-13 and er1-14, respectively. A chi-squared (χ2) test showed that both segregation ratios of KASPar-er1-13 and KASPar-er1-14 in respective F2 populations fit 1:2:1 (KASPar-er1-13: χ2 = 1.41, p = 0.49; KASPar-er1-14: χ2 = 0.67; p = 0.71), indicating co-dominant markers.
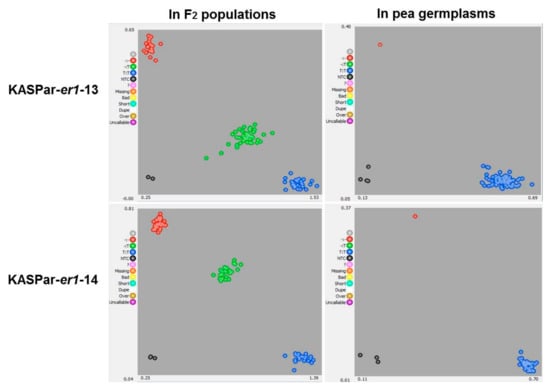
Figure 4.
KASPar genotyping detection with markers KASPar-er1-13 and KASPar-er1-14 in the F2 populations derived from Longwan 1 × Dabaiwandou and Chengwan 8 × Guiwan 1, as well as in other pea germplasms. Red dots indicate er1-13/er1-14 homozygous Erysiphe pisi-resistant lines/germplasms, blue dots indicate Er1-13/Er1-14 homozygous E. pisi-susceptible lines/germplasms, and green dots indicate er1-13/Er1-13 or er1-14/Er1-14 heterozygotes. Grey dots are blank samples used as controls. In our KASPar assay, the co-dominant markers KASPar-er1-13 and KASPar-er1-14 correctly categorized all F2 individuals into three clusters corresponding to homozygous resistant (red dots), homozygous susceptible (blue dots), and heterozygous (green dots) lines, and categorized tested pea germplasms into two clusters corresponding to homozygous resistant (red dots) and homozygous susceptible (blue dots).
2.5. Validation and Application of Functional Markers
The 56 germplasms with the known resistance phenotype to E. pisi isolate EPYN that carrying the known er1 alleles (er1-1, er1-2, er1-4, er1-6, er1-7, er1-8, and er1-9) were selected to genotyping by KASPar-er1-13 and KASPar-er1-13 (Table S1). It included 49 that were phenotypically immune to E. pisi and contained known er1 alleles; seven were resistant. The three resistant and three susceptible parents were also tested at the same time (Table S1).
When the 56 germplasms were genotyped with KASPar-er1-13, two distinct clusters were recovered, with one gene (er1-13) corresponding to Dabaiwandou and the other (non-er1-13) to the other germplasms, respectively. Similarly, when the germplasms were genotyped with KASPar-er1-14, two distinct clusters were recovered, corresponding to Guiwan 1 and all of the other germplasms, respectively (Figure 4; Table S1). Thus, markers KASPar-er1-13 and KASPar-er1-14 effectively identified pea germplasms carrying the er1-13 and er1-14 alleles, respectively. Our results also showed that KASPar-er1-13 and KASPar-er1-14 could distinguish the know er1 alleles and Er1 from er1-13 or er1-14, respectively.
3. Discussion
Pea powdery mildew caused by E. pisi DC. is an important disease and reduces considerable yield in pea production worldwide. The deployment of resistant cultivars containing the er1 gene is the most effective way to control this disease The E. pisi resistance gene er1 is recessive in pea cultivars, which is the most widely deployed gene for controlling powdery mildew worldwide.
To date, there were 12 er1 alleles identified in resistant pea germplasms. Among the 12 known er1 alleles, er1-1 and er1-2 are most commonly used in pea breeding programs because they confer stable resistance to E. pisi [9,16,28,39,40]. Previously, er1-1 has been identified in four E. pisi-resistant pea cultivars (JI1559, Tara, and Cooper from Canada; and Yunwan 8 from China), while er1-2 has been identified in seven E. pisi-resistant pea cultivars (Stratagem, Franklin, Dorian, Nadir, X9002, Xucai 1, and G0005576) [9,15,16,18,28]. Recently, the er1 gene for E. pisi resistance was confirmed in 53 pea germplasms from a worldwide collection [32]. Here, more E. pisi-resistant germplasms carrying the er1-1, er1-2, er1-6, and er1-7 alleles were identified. To date, a dominant gene Er3 had been found in wild pea (Pisum fulvum) and mapped to pea LG IV [20]. It is possible that a rich diversity of E. pisi-resistant genes were contained in wild pea. Thus, searching for novel E. pisi-resistant genes from wild pea germplasms should be a good strategy [8].
To date, more than 40 MLO mutant alleles have been described in the monocotyledonous plant barley [41], and PsMLO1 allelic diversity has been widely studied in pea [9,13,16,17,18,28,30,31,32,33,34,35,36,40]. Wild-type PsMLO1 of pea consists of 15 exons and 14 introns (NCBI accession number: KC466597). To date, a total of 12 er1 alleles associated with the er1-resistance phenotype have been identified and 11 of 12 er1 alleles PsMLO1 mutations were caused by alterations of the coding sequence. There was 1, 1, 1, 1, 3, 1, and 2 allele mutations occurred in exons 1, 5, 6, 8, 10, 11, and 15 of wild-type PsMLO1, respectively. Eight alleles (er1-1/er1mut1, er1-3, er1-4, er1-5, er1-6, er1-9, er1-10/er1mut2, and er1-12) are the result of point mutations in the exons of wild-type PsMLO1. Four alleles result from single base substitutions in wild-type PsMLO1 cDNA: in er1-1, a C→G at position 680 (exon 6); in er1-5, a G→A at position 570 (exon 5); in er1-6, a T→C at position 1121 (exon 11); and in er1-10, a G→A at position 939 (exon 10) [9,18,28,30]. Three alleles result from single base deletions in wild-type PsMLO1 cDNA, including ΔG at position 862 (exon 8) in er1-3; ΔA at position 91 (exon 1) in er1-4; and ΔT at position 928 (exon 10) in er1-9 identified in this study [9]. Recently, er1-12 allele was found resulting from single base insertion (A) in front of the last exon (exon 15) in wild-type PsMLO1 cDNA [35]. Two alleles result from small fragment deletions in wild-type PsMLO1 cDNA, including a 10-bp deletion of positions 111–120 (exon 1) in er1-7 [17]; and a 3-bp deletion of positions 1339–1341 (exon 15) in er1-8 [32]. Only the er1-11 mutation is known to have resulted from an intron mutation in PsMLO1 (a 2-bp insertion in intron 14) [33,34], and only er1-2 results from a large insert of unknown size in wild-type PsMLO1 cDNA [9,15,18].
Several functional markers specific to the previously recognized er1 alleles have already been developed to facilitate the marker-assisted breeding of pea cultivars resistant to E. pisi [9,15,17,18,30,32,33,34,36]. Pavan et al. [28] developed a functional cleaved amplified polymorphic sequence (CAPS) marker for er1-5, while Pavan et al. [36] developed functional markers for the five er1 alleles, er1-1 through er1-5. Santo et al. [30] developed functional markers for er1mut1 and er1mut2, and Wang et al. [15] developed a dominant marker for er1-2. Sudheesh et al. [34] developed a functional marker for er1-11, while Sun et al. [17,18] developed co-dominant functional markers for er1-6 and er1-7. Ma et al. [33] developed eight KASPar markers for eight known er1 alleles, excluding er1-2, and renamed the er1-10 and er1-11. More recently, Sun et al. [32] identified the two novel er1 alleles, er1-8 and er1-9, and developed KASPar-er1-8 and KASPar-er1-9. In this study, the developed markers, KASPar-er1-13 and KASPar-er1-14, could accurately and effectively identify pea germplasms carrying the er1-13 and er1-14 alleles and distinguish them from the know er1 alleles or Er1, respectively.
This study discovered a known and two novel er1 alleles, resulting from new mutations of wild-type PsMLO1 cDNA: er1-13 and er1-14 was generated by a 1-bp deletion in exon 1 and 3, respectively. The co-dominant functional markers specific to er1-13 (KASPar-er1-13) and to er1-14 (KASPar-er1-14) were developed. These markers were validated in genetic populations and pea germplasms. These results will support future studies to reveal the powdery mildew resistance mechanisms. The two novel er1 alleles and the developed co-dominant functional markers could be powerful tools for the breeding of pea cultivars resistance to E. pisi.
4. Materials and Methods
4.1. Plant Material and E. pisi Inoculum
Previously, many Chinese pea germplasms had been screened for E. pisi and some were found to be E. pisi-resistant [31,37,39]. In this study, the three E. pisi-resistant Chinese pea landraces, Suoshadabaiwan, Dabaiwandou, and Guiwan 1, respectively, from the Chongqing, Yunnan, and Guangxi provinces of China were conducted to reveal their E. pisi-resistant genes. The three E. pisi-susceptible Chinese pea cultivars, Bawan 6, Longwan 1, and Chengwan8, were used as susceptible controls or cross susceptible parents for genetic analysis [15,40]. The Chinese pea cultivar Xucai 1 containing er1-2 was used as E. pisi-resistant control [16].
The E. pisi isolate EPYN from Yunnan Province of China was used as the inoculum, which is highly virulent on pea [17,18]. The EPYN isolate was maintained through the continuous re-inoculation of healthy seedlings of Longwan 1 under controlled conditions. The inoculated plants were incubated in a growth chamber with controlled conditions [16].
4.2. Powdery Mildew Resistance Evaluation
Thirty-five seeds were planted from each of the three E. pisi-resistant germplasms (Suoshadabaiwan, Dabaiwandou, and Guiwan 1), three E. pisi-susceptible pea cultivars (Bawan 6, Longwan 1, and Chengwan 8), and from the resistant and susceptible controls (Bawan 6, Longwan 1 and Chengwan 8, and Xucai 1) [18]. The healthy seedlings were thinned to 30 per pot before the phenotypic evaluation. Three replications were planted. Seeded pots were placed in a greenhouse maintained at 18 to 26 °C. At the same time, the E. pisi inoculum was prepared by inoculating the 10-day-old seedlings of Longwan 1, which were incubated in a growth chamber at 20 ± 1 °C with a 12-h photoperiod. Two weeks later, all seedlings of the germplasms and controls were inoculated by gently shaking off the conidia of the Longwan 1 plants. Inoculated plants were incubated in a growth chamber at 20 ± 1 °C with a 12-h photoperiod. Ten days later, disease severity was rated based on a scale (0–4 scale) [17,18]. Plants with a score of 0 were considered E. pisi-immune, while those with scores of 1, 2 and 3, 4 were considered E. pisi-resistant and E. pisi-susceptible, respectively. For those identified as immune or resistant to E. pisi, repeated identification was performed.
4.3. Inheritance Analysis of Resistant Pea Cultivars
To reveal the inheritance controlled by E. pisi resistance genes in the three E. pisi-resistant Chinese pea landrace, Suoshadabaiwan, Dabaiwandou, and Guiwan 1, they were crossed with the E. pisi-susceptible cultivars Bawan 6, Longwan 1, and Chengwan 8, respectively, to generate genetic populations. The derived F1, F2, and F2:3 populations from three crosses (Bawan 6 × Suoshadabaiwan, Longwan 1 × Dabaiwandou, and Chengwan 8 × Guiwan 1), which were used for the powdery mildew resistance genetic analysis of Suoshadabaiwan, Dabaiwandou, and Guiwan 1. The six parents and the derived F1 and F2 populations were planted in a propagation greenhouse to generate F2 and F2:3 family seeds, respectively.
The plants of the F1 and F2 populations at the fourth or fifth leaf stage were inoculated with the E. pisi isolate EPYN using the detached leaf method [16,17,18]. After inoculation, the treated leaves were placed in a growth chamber at 20 °C with a 14-h photoperiod. The six parents were also inoculated as controls. Ten days after inoculation, disease severity was rated based on a scale of 0–4 as described above. Plants with scores of 0–2 and 3–4 were classified as resistant and susceptible, respectively [16,17,18]. Those plants identified as E. pisi-resistant were tested again to confirm their resistance.
Twenty-five seeds were selected randomly from each of the 102, 121, and 131 F2:3 families derived from Bawan 6 × Suoshadabaiwan, Longwan 1 × Dabaiwandou, and Chengwan 8 × Guiwan 1, respectively. These seeds were planted and cultivated together with their parents, following previously published protocols [25,26,27]. Disease severity was scored 10 days after inoculation using the 0–4 scale, as described above for the phenotypic identification of the pea germplasms. The F2:3 families with scores of 0–2 and 3–4 were classified as homozygous resistant and homozygous susceptible, respectively. Families with scores of 0–2 and 3–4 were considered segregated to E. pisi resistance. The families identified as homozygous-resistant or resistance segregated were subjected to repeated testing.
A chi-squared (χ2) analysis was used to evaluate the goodness-of-fit to Mendelian segregation ratio of the F2 and F2:3 phenotypes derived from Bawan 6 × Suoshadabaiwan, Longwan 1 × Dabaiwandou, and Chengwan 8 × Guiwan 1, respectively.
4.4. Genetic Mapping of the Resistance Gene
The Genomic DNA was isolated from the leaves of the F2 populations and of their parents using a slightly modified cetyltrimethylammonium bromide (CTAB) extraction method [42]. The DNA solution was diluted and stored at −20 °C until use.
Previous studies suggested that E. pisi resistance was controlled by er1 in most of all pea germplasms except for lines SVP952 and JI 2480 [7,21]. Thus, this study was performed to map the E. pisi-resistance gene using the known er1-linked markers on the pea LG VI, including five simple sequence repeat (SSR) markers (PSMPSAD51, PSMPSA5, PSMPSAD60, i.e., AD60, PSMPSAA374e, and PSMPSAA369); a gene marker (Cytosine-5, DNA-methyltransferase (c5DNAmet)) [12,15,16,17,18,37,43]; and 10 molecular markers on the pea LG VI (AD160, AC74, AC10_1, AA224, AA200, AD159, AD59, AB71, AA335, and AB86). Firstly, these markers were used to screen for polymorphisms between the crossed parents, Bawan 6 × Suoshadabaiwan, Longwan 1 × Dabaiwandou, and Chengwan 8 × Guiwan 1 [44]. The parental polymorphic markers were then used to confirm the genotypes of each F2 plant for genetic linkage analysis. PCR amplification was conducted in a total volume of 20 µL [16,17,18]. PCR reactions were performed in a thermal cycler (Biometra, Göttingen, Germany). The PCR products were separated on 6%–8% polyacrylamide gels.
The segregation data of the polymorphic markers in the F2 populations were evaluated for goodness-of-fit to Mendelian segregation patterns with a chi-squared (χ2) test. Genetic linkage analyses were completed using MAPMAKER/EXP version 3.0b. A logarithm of odds (LOD) score > 3.0 and a distance < 50 cM were used as the thresholds to determine the linkage groups [45]. Genetic distances were determined using the Kosambi mapping function [46]. The genetic linkage map was constructed using the Microsoft Excel macro MapDraw [47].
4.5. RNA Extraction and PsMLO1 Sequence Analysis
The extraction of total RNA and synthesis of cDNA from Suoshadabaiwan, Dabaiwandou, and Guiwan 1 and controls were completed according to our previous studies [16,17,18].
To identify the resistance gene er1 alleles, the full-length cDNAs of the PsMLO1 homologs were amplified using the primers specific to PsMLO1 [9]. The PCR cycling conditions were as follows: 95 °C for 5 min; then 35 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 45 s, and extension at 72 °C for 1 min; and a final extension at 72 °C for 10 min. The purified amplicons were cloned with a pEasy-T5 vector (TransGen Biotech, Beijing, China). The sequencing reactions of 10 clones per parental cultivars and controls were performed by the Shanghai Shenggong Biological Engineering Co., Ltd. (Shanghai, China). The resulting sequences were aligned with the wild-type PsMLO1 of pea (NCBI accession number: FJ463618.1) using DNAMAN v6.0 (Lynnon Biosoft, Vaudreuil, QC, Canada).
4.6. Development of Functional Markers for the Novel er1 Alleles
Primers flanking the mutation site were designed based on the PsMLO1 gene sequence (GenBank accession number KC466597), using Primer Premier v5.0, to develop a functional marker specific to allele er1-13 and er1-14 (Table 2).

Table 2.
Sequence information for the indel and Kompetitive allele-specific PCR (KASPar) markers specific to er1-13 and for the KASPar marker specific to er1-14.
The Kompetitive allele-specific PCR (KASPar) markers (KASPar-er1-13 and KASPar-er1-14) specific to the two novel er1 alleles were developed based on allele er1-13 SNPs (1-bp deletion) and er1-14 SNPs (1-bp deletion) in PsMLO1. The forward primers and the common reverse primers specific to er1-13 and er1-14 were designed for KASPar markers by LGC KBioscience (KBioscience, Hoddesdon, UK), respectively. In brief, two KASPar markers (KASPar-er1-13 and KASPar-er1-14) were used to detect parental polymorphisms (Longwan 1 × Dabaiwandou, and Chengwan 8 × Guiwan 1), and then used to analyze the genotypes of each F2 offspring (Longwan 1 × Dabaiwandou: 121 F2 individuals; Chengwan 8 × Guiwan: 131 F2 individuals).
KASPar markers were amplified with a Douglas Scientific Array Tape Platform (China Golden Marker Biotech Co., Ltd., (Beijing, China)) in a 0.8 µL Array Tape reaction volume with 10 ng dry DNA, 0.8 µL 2 × KASP master mix, and 0.011 µL primer mix (KBioscience, Hoddesdon, UK). A Nexar Liquid handling instrument was used to add the PCR solution to the Array Tape (Douglas Scientific). PCRs were performed on a Soellex PCR Thermal Cycler with the following conditions: initial denaturation at 94 °C for 15 min; followed by 10 cycles of denaturation at 94 °C for 20 s and 65 °C for 60 s at an annealing temperature that decreased by 0.8 °C per cycle; and then 26 cycles of denaturation at 94 °C for 20 s and 57 °C for 60 s; and a final cooling to 4 °C. A fluorescent end-point reading was completed with the Araya fluorescence detection system (part of the Douglas Scientific Array Tape Platform). Genotypes and clusters were visualized with Kraken (http://ccb.jhu.edu/software/kraken/MANUAL.html (accessed on 5 August 2022)).
4.7. Validation and Application of Functional Markers
To test the efficacy of the novel functional markers specific to er1-13 (KASPar-er1-13) and er1-14 (KASPar-er1-14), 56 pea germplasms with known phenotypic resistance to E. pisi isolate EPYN and carrying known er1 alleles, and six parents were tested for whether they carried the er1 alleles er1-13 or er1-14 (Table S1) [9,15,17,18,32,40]. The six parents (Suoshadabaiwan, Dabaiwandou and Guiwan 1, Bawan 6, Longwan 1, and Chengwan 8) were used as contrasting controls.
DNA was extracted from the 56 selected pea germplasms (resistant cultivars with known er1 alleles) and the six parents using the CTAB method [42]. The PCR amplification of the KASPar markers were performed as described above (in the section “Development of Functional Markers for the novel er1 alleles”).
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms231912016/s1.
Author Contributions
Z.Z. conceived and designed the experiments. S.S., D.D., W.W., Y.H., G.L., C.D. (Chengzhang Du) and C.D. (Canxing Duan), performed the experiments. S.S. analyzed the data and wrote the manuscript. Z.Z. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Key R&D Program of China (2019YFD1001300, 2019YFD1001301), the China Agriculture Research System of MOF and MARA (CARS-08), National Crop Germplasm Resources Center (NCGRC-2022-09), Species and Variety Resource Protection from the Ministry of Agriculture and Rural of China, and the Scientific Innovation Program of the Chinese Academy of Agricultural Sciences from the Institute of Crop Sciences, Chinese Academy of Agricultural Sciences.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
We sincerely thank Xuxiao Zong, Xiaoming Yang, Dongxu Xu, and Dongmei Yu for providing the pea cultivars and germplasms.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| SSR | Simple sequence repeat |
| SNP | Single nucleotide polymorphism |
| KASPar | Kompetitive allele-specific PCR |
References
- Ali, S.M.; Sharma, B.; Ambrose, M.J. Current status and future strategy in breeding pea to improve resistance to biotic and abiotic stresses. Euphytica 1993, 1, 115–126. [Google Scholar] [CrossRef]
- Wang, X.; Zhu, Z.; Duan, C.; Zong, X. Identification and Control Technology of Disease and Pest on Faba Bean and Pea; Chinese Agricultural Science and Technology Press: Beijing, China, 2007. [Google Scholar]
- Smith, P.H.; Foster, E.M.; Boyd, L.A.; Brown, J.K.M. The early development of Erysiphe pisi on Pisum sativum L. Plant Pathol. 1996, 45, 302–309. [Google Scholar] [CrossRef]
- Ghafoor, A.; McPhee, K. Marker assisted selection (MAS) for developing powdery mildew resistant pea cultivars. Euphytica 2012, 186, 593–607. [Google Scholar] [CrossRef]
- Fondevilla, S.; Rubiales, D. Powdery mildew control in pea, a review. Agron. Sustain. Dev. 2012, 32, 401–409. [Google Scholar] [CrossRef]
- Harland, S.C. Inheritance of immunity to mildew in Peruvian forms of Pisum sativum. Heredity 1948, 2, 263–269. [Google Scholar] [CrossRef]
- Heringa, R.J.; Van Norel, A.; Tazelaar, M.F. Resistance to powdery mildew (Erysiphe polygoni D.C.) in peas (Pisum sativum L.). Euphytica 1969, 18, 163–169. [Google Scholar] [CrossRef]
- Fondevilla, S.; Torres, A.M.; Moreno, M.T.; Rubiales, D. Identification of a new gene for resistance to powdery mildew in Pisum fulvum, a wild relative of pea. Breed. Sci. 2007, 57, 181–184. [Google Scholar] [CrossRef]
- Humphry, M.; Reinstädler, A.; Ivanov, S.; Bisseling, T.; Panstruga, R. Durable broad-spectrum powdery mildew resistance in pea er1 plants is conferred by natural loss-of-function mutations in PsMLO1. Mol. Plant Pathol. 2011, 12, 866–878. [Google Scholar] [CrossRef]
- Timmerman, G.M.; Frew, T.J.; Weeden, N.F. Linkage analysis of er1, a recessive Pisum sativum gene for resistance to powdery mildew fungus (Erysiphe pisi DC). Theor. Appl. Genet. 1994, 88, 1050–1055. [Google Scholar] [CrossRef]
- Tiwari, K.R.; Penner, G.A.; Warkentin, T.D. Identification of coupling and repulsion phase RAPD markers for powdery mildew resistance gene er1 in pea. Genome 1998, 41, 440–444. [Google Scholar] [CrossRef]
- Ek, M.; Eklund, M.; von Post, R.; Dayteg, C.; Henriksson, T.; Weibull, P.; Ceplitis, A.; Isaac, P.; Tuvesson, S. Microsatellite markers for powdery mildew resistance in pea (Pisum sativum L.). Hereditas 2005, 142, 86–91. [Google Scholar] [CrossRef]
- Pereira, G.; Marques, C.; Ribeiro, R.; Formiga, S.; Dâmaso, M.; Sousa, T.; Farinhó, M.; Leitão, J.M. Identification of DNA markers linked to an induced mutated gene conferring resistance to powdery mildew in pea (Pisum sativum L.). Euphytica 2010, 171, 327–335. [Google Scholar] [CrossRef]
- Srivastava, R.K.; Mishra, S.K.; Singh, K.; Mohapatra, T. Development of a coupling-phase SCAR marker linked to the powdery mildew resistance gene er1 in pea (Pisum sativum L.). Euphytica 2012, 86, 855–866. [Google Scholar] [CrossRef]
- Wang, Z.; Fu, H.; Sun, S.; Duan, C.; Wu, X.; Yang, X.; Zhu, Z. Identification of powdery mildew resistance gene in pea line X9002. Acta Agron. Sin. 2015, 41, 515–523, (In Chinese with English abstract). [Google Scholar] [CrossRef]
- Sun, S.; Wang, Z.; Fu, H.; Duan, C.; Wang, X.; Zhu, Z. Resistance to powdery mildew in the pea cultivar Xucai 1 is conferred by the gene er1. Crop. J. 2015, 3, 489–499. [Google Scholar] [CrossRef]
- Sun, S.; Deng, D.; Wang, Z.; Duan, C.; Wu, X.; Wang, X.; Zong, X.; Zhu, Z. A novel er1 allele and the development and validation of its functional marker for breeding pea (Pisum sativum L.) resistance to powdery mildew. Appl. Genet. 2016, 129, 909–919. [Google Scholar]
- Sun, S.; Fu, H.; Wang, Z.; Duan, C.; Zong, X.; Zhu, Z. Discovery of a novel er1 allele conferring powdery mildew resistance in Chinese pea (Pisum sativum L.) landraces. PLoS ONE 2016, 11, e0147624. [Google Scholar] [CrossRef]
- Katoch, V.; Sharma, S.; Pathania, S.; Banayal, D.K.; Sharma, S.K.; Rathour, R. Molecular mapping of pea powdery mildew resistance gene er2 to pea linkage group III. Mol. Breed. 2010, 25, 229–237. [Google Scholar] [CrossRef]
- Cobos, M.J.; Satovic, Z.; Rubiales, D.; Fondevilla, S. Er3 gene, conferring resistance to Erysiphe pisi, is located in pea LGIV. Euphytica 2018, 214, 203. [Google Scholar] [CrossRef]
- Tiwari, K.R.; Penner, G.A.; Warkentin, T.D. Inheritance of powdery mildew resistance in pea. Can. J. Plant Sci. 1997, 77, 307–310. [Google Scholar] [CrossRef]
- Vaid, A.; Tyagi, P.D. Genetics of powdery mildew resistance in pea. Euphytica 1997, 96, 203–206. [Google Scholar] [CrossRef]
- Fondevilla, S.; Carver, T.L.W.; Moreno, M.T.; Rubiales, D. Macroscopic and histological characterisation of genes er1 and er2 for powdery mildew resistance in pea. Eur. J. Plant Pathol. 2006, 115, 309–321. [Google Scholar] [CrossRef]
- Fondevilla, S.; Cubero, J.I.; Rubiales, D. Confirmation that the Er3 gene, conferring resistance to Erysiphe pisi in pea, is a different gene from er1 and er2 genes. Plant Breed. 2011, 130, 281–282. [Google Scholar] [CrossRef]
- Bai, Y.; Pavan, S.; Zheng, Z.; Zappel, N.F.; Reinstadler, A.; Lotti, C.; DeGiovanni, C.; Ricciardi, L.; Lindhout, P.; Visser, R.; et al. Naturally occurring broad-spectrum powdery mildew resistance in a central American tomato accession is caused by loss of MLO1 function. Mol. Plant Microbe Interact. 2008, 21, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Büschges, R.; Hollricher, K.; Panstruga, R.; Simons, G.; Wolter, M.; Frijters, A.; van Daelen, R.; van der Lee, T.; Diergaarde, P.; Groenendijk, J. The barley MLO gene, a novel control element of plant pathogen resistance. Cell 1997, 88, 695–705. [Google Scholar] [CrossRef]
- Devoto, A.; Hartmann, H.A.; Piffanelli, P.; Elliott, C.; Simmons, C.; Taramino, G.; Goh, C.S.; Cohen, F.E.; Emerson, B.C.; Schulze-Lefert, P.; et al. Molecular phylogeny and evolution of the plant-specific seven-transmembrane MLO family. J. Mol. Evol. 2003, 56, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Pavan, S.; Schiavulli, A.; Appiano, M.; Marcotrigiano, A.R.; Cillo, F.; Visser, R.G.F.; Bai, Y.; Lotti, C.; Luigi Ricciardi, L. Pea powdery mildew er1 resistance is associated to loss-of-function mutations at a MLO homologous locus. Appl. Genet. 2011, 123, 1425–1431. [Google Scholar] [CrossRef]
- Rispail, N.; Rubiales, D. Genome-wide identification and comparison of legume MLO gene family. Sci. Rep. 2016, 6, 32673. [Google Scholar] [CrossRef]
- Santo, T.; Rashkova, M.; Alabaca, C.; Leitao, J. The ENU–induced powdery mildew resistant mutant pea (Pisum sativum L.) lines S (er1mut1) and F (er1mut2) harbour early stop codons in the PsMLO1 gene. Mol. Breed. 2013, 32, 723–727. [Google Scholar] [CrossRef]
- Fu, H.; Sun, S.; Zhu, Z.; Duan, C.; Yang, X. Phenotypic and genotypic identification of powdery mildew resistance in pea cultivars or lines from Canada. J. Plant. Genet. Resour. 2014, 15, 1028–1033, (In Chinese with English abstract). [Google Scholar]
- Sun, S.; Deng, D.; Duan, C.; Zong, X.; Xu, D.; He, Y.; Zhu, Z. Two novel er1 alleles conferring powdery mildew (Erysiphe pisi) resistance identified in a worldwide collection of pea (Pisum sativum L.) germplasms. Int. J. Mol. Sci. 2019, 20, 5071. [Google Scholar] [CrossRef]
- Ma, Y.; Coyne, C.J.; Main, D.; Pavan, S.; Sun, S.; Zhu, Z.; Zong, X.; Leitão, J.; McGee, R.J. Development and validation of breeder-friendly KASPar markers for er1, a powdery mildew resistance gene in pea (Pisum sativum L.). Mol. Breed. 2017, 37, 151. [Google Scholar] [CrossRef]
- Sudheesh, S.; Lombardi, M.; Leonforte, A.; Cogan, N.O.I.; Materne, M.; Forster, J.W.; Kaur, S. Consensus Genetic Map Construction for Field Pea (Pisum sativum L.), Trait dissection of biotic and abiotic stress tolerance and development of a diagnostic marker for the er1 powdery mildew resistance gene. Plant. Mol. Biol. Rep. 2015, 33, 1391–1403. [Google Scholar] [CrossRef]
- Sulima, A.S.; Zhukov, V.A. War and Peas: Molecular bases of resistance to powdery mildew in pea (Pisum sativum L.) and other legumes. Plants 2022, 11, 339. [Google Scholar] [CrossRef]
- Pavan, S.; Schiavulli, A.; Appiano, M.; Miacola, C.; Visser, R.G.F.; Bai, Y.L.; Lotti, C.; Ricciardi, L. Identification of a complete set of functional markers for the selection of er1 powdery mildew resistance in Pisum sativum L. Mol. Breed. 2013, 31, 247–253. [Google Scholar] [CrossRef]
- Wang, Z.; Bao, S.; Duan, C.; Zong, X.; Zhu, Z. Screening and molecular identification of resistance to powdery mildew in pea germplasm. Acta Agron. Sin. 2013, 39, 1030–1038, (In Chinese with English abstract). [Google Scholar] [CrossRef]
- Liu, R.; Wang, F.; Fang, L.; Yang, T.; Zhang, H.; Huang, Y.; Wang, D.; Ji, Y.; Xu, D.; Li, G.; et al. An integrated high-density SSR genetic linkage map from two F2 population in Chinese pea. Acta Agron. Sin. 2020, 46, 1496–1506, (In Chinese with English abstract). [Google Scholar]
- Fu, H. Phenotyping and Genotyping Powdery Mildew Resistance in Pea. Ph.D. Thesis, Gansu Agricultural University, Gansu, China, 2014. (In Chinese with English abstract). [Google Scholar]
- Sun, S.; He, Y.; Dai, C.; Duan, C.; Zhu, Z. Two major er1 alleles confer powdery mildew resistance in three pea cultivars bred in Yunnan Province, China. Crop. J. 2016, 4, 353–359. [Google Scholar] [CrossRef][Green Version]
- Kusch, S.; Panstruga, R. Mlo–based resistance, an apparently universal “Weapon” to defeat powdery mildew disease. MPMI 2017, 30, 179–189. [Google Scholar] [CrossRef]
- Shure, M.; Wessler, S.; Fedoroff, N. Molecular-identification and isolation of the waxy locus in maize. Cell 1983, 35, 225–233. [Google Scholar] [CrossRef]
- Bordat, A.; Savois, V.; Nicolas, M.; Salse, J.; Chauveau, A.; Bourgeois, M.; Potier, J.; Houtin, H.; Rond, C.; Murat, F.; et al. Translational genomics in legumes allowed placing in silico 5460 unigenes on the pea functional map and identified candidate genes in Pisum sativum L. Genes Genome Genet. 2011, 1, 93–103. [Google Scholar] [CrossRef]
- Loridon, K.; McPhee, K.; Morin, J.; Dubreuil, P.; Pilet-Nayel, M.L.; Aubert, G.; Rameau, C.; Baranger, A.; Coyne, C.; Lejeune-Hénaut, I.; et al. Microsatellite marker polymorphism and mapping in pea (Pisum sativum L.). Theor. Appl. Genet. 2005, 111, 1022–1031. [Google Scholar] [CrossRef]
- Lander, E.S.; Daly, M.J.; Lincoln, S.E. Constructing genetic linkage maps with MAPMAKER/EXP Version 3.0, a tutorial and reference manual. In Institute for Biomedical Research Technical Report, 3rd ed.; Whitehead, A., Ed.; Whitehead Institute for Biomedical Research: Cambridge, MA, USA, 1993. [Google Scholar]
- Kosambi, D.D. The estimation of map distances from recombination values. Ann. Eugen. 1944, 12, 172–175. [Google Scholar] [CrossRef]
- Liu, R.H.; Meng, J.L. MapDraw, A Microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas 2003, 25, 317–321, (In Chinese with English abstract). [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).