Integrative Analysis of the Nasal Microbiota and Serum Metabolites in Bovines with Respiratory Disease by 16S rRNA Sequencing and Gas Chromatography/Mass Selective Detector-Based Metabolomics
Abstract
1. Introduction
2. Results
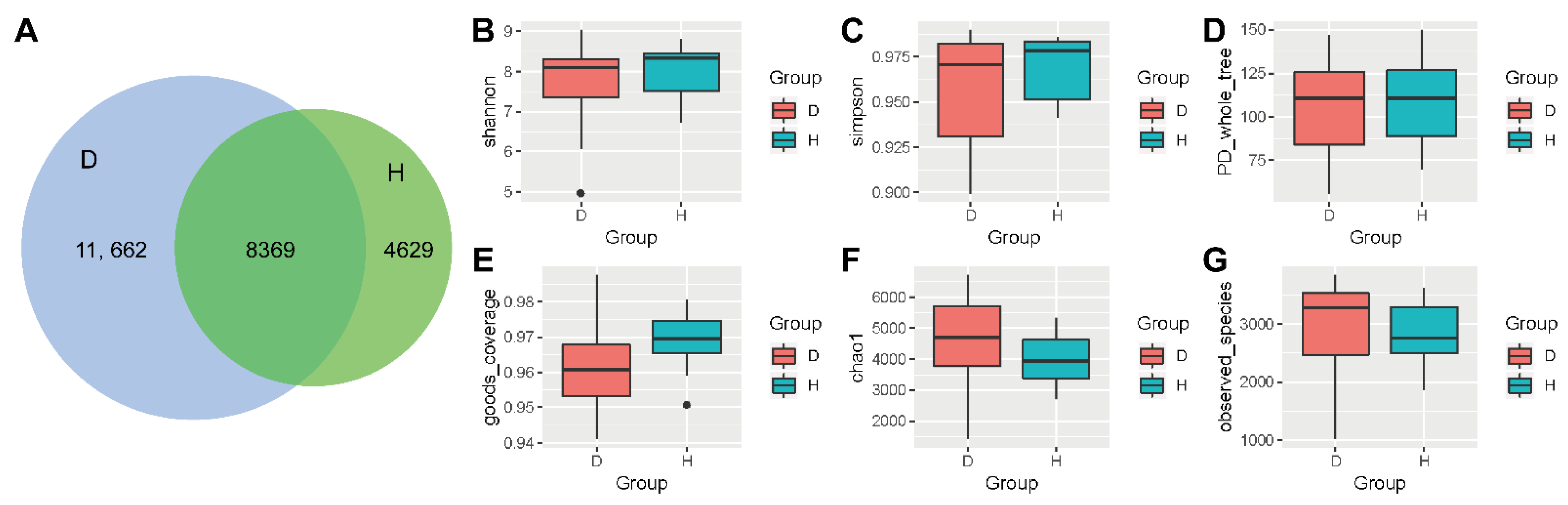
2.1. Alpha-Diversity Analysis Comparing Healthy Bovines and Those with Respiratory Disease
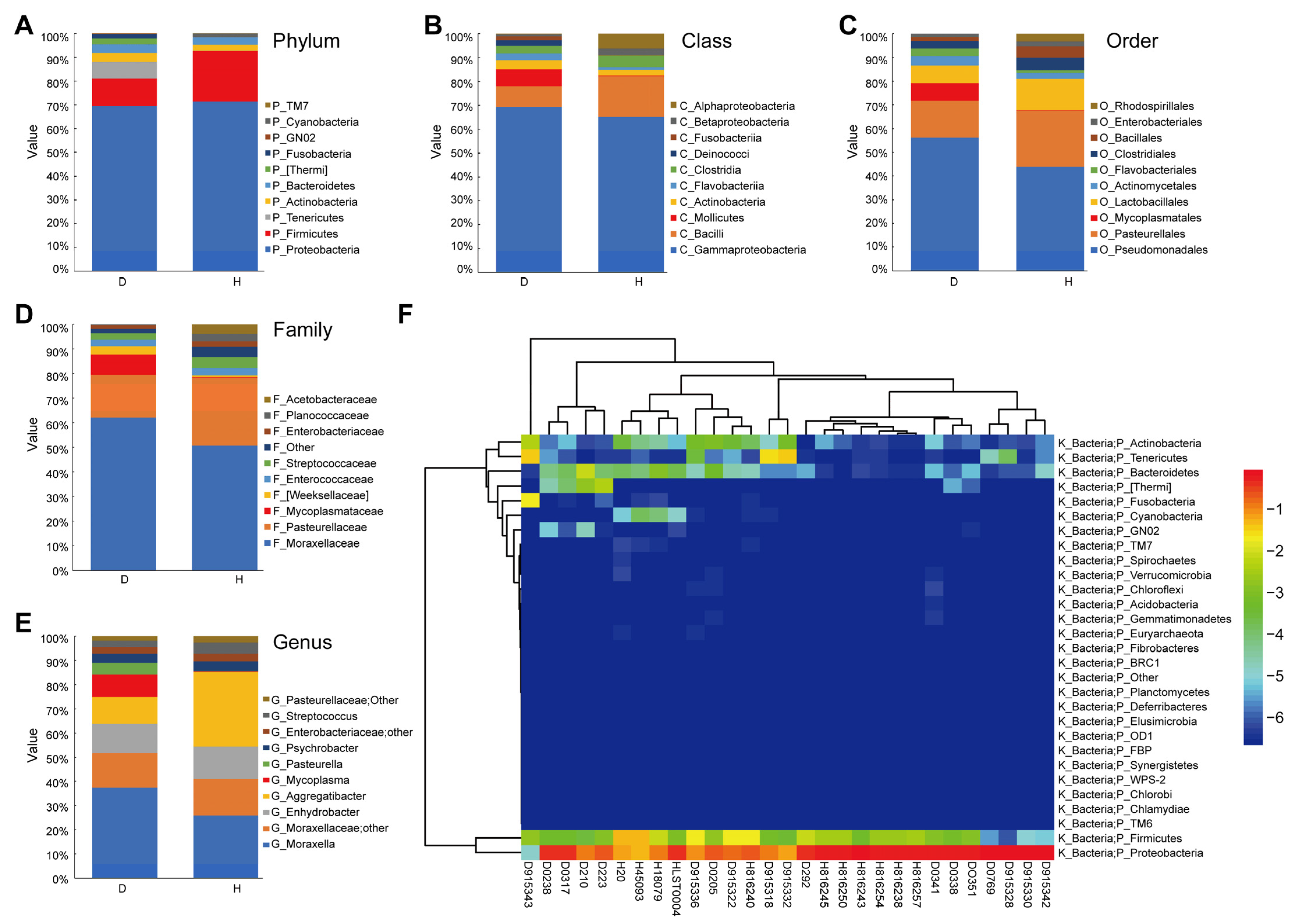
2.2. Characterization of the Nasal Microbial Communities in Healthy and Diseased Cattle
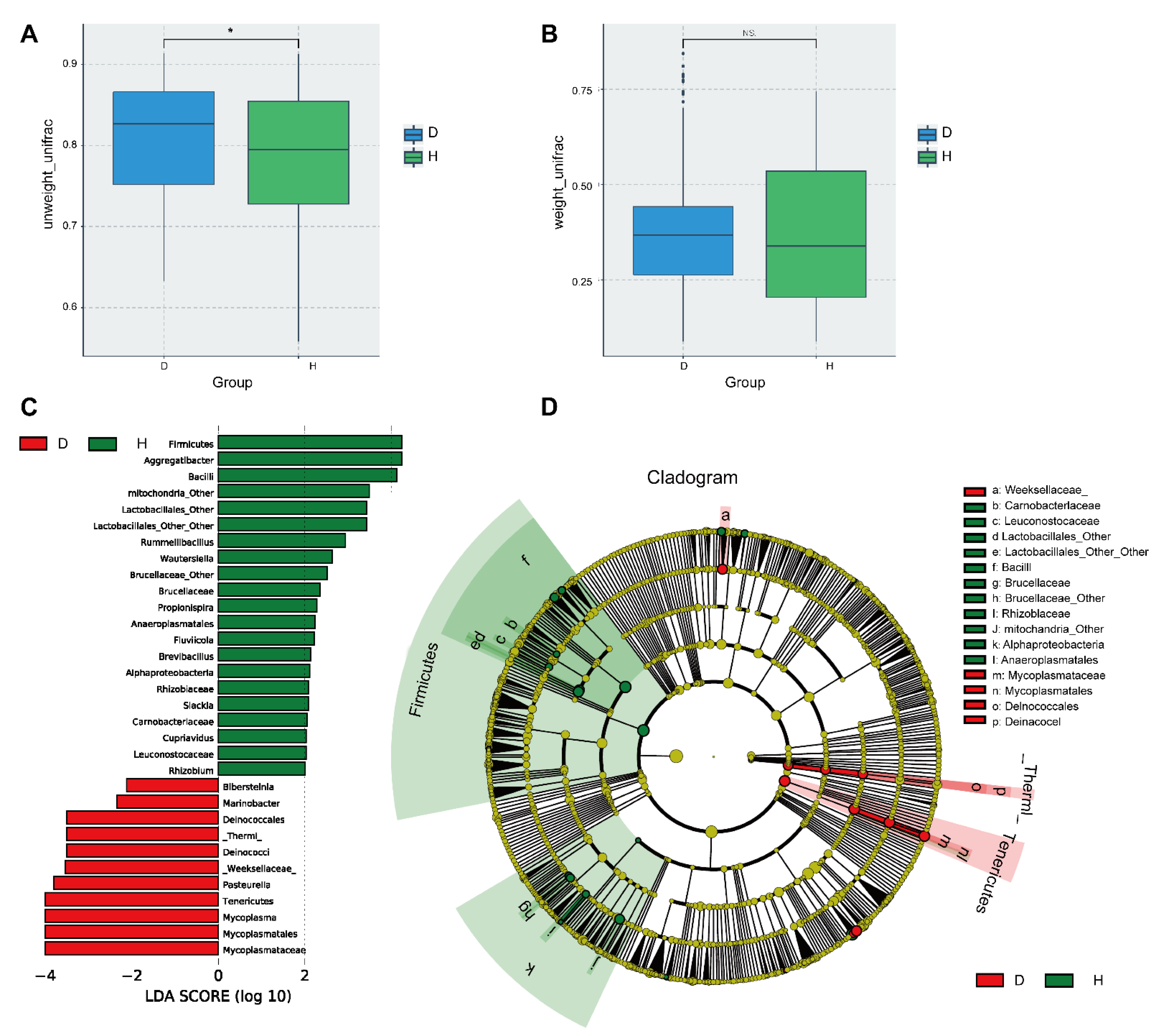
2.3. The Diversity and Composition of Nasal Microbiota Are Significantly Altered in Cattle with BRD
2.4. Evaluation of Serum Metabolites Reveals Alterations in the Metabolome of BRD Cattle
2.5. Correlations between Nasal Microbes and Serum Metabolites
3. Discussion
4. Materials and Methods
4.1. Specimens
4.2. 16S rRNA Sequencing
4.3. DNA Extraction and Metagenome Sequencing
4.4. Microbial Composition and Diversity Analysis
4.5. Metabolomics Processing
4.6. Statistical Analysis of Metabolomics
4.7. Correlation Profiling between the Metabolites and Nasal Microbiotas
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Krehbiel, C.R. Bovine Respiratory Disease Influences on Nutrition and Nutrient Metabolism. Vet. Clin. N. Am. Food Anim. Pract. 2020, 36, 361–373. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, K.P.; McGuirk, S.M. Respiratory disease of the bovine neonate. Vet. Clin. N. Am. Food Anim. Pract. 2009, 25, 121–137, vi–vii. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Schneider, L.G.; Hubbard, K.J.; Grotelueschen, D.M.; Daly, R.F.; Stokka, G.S.; Smith, D.R. Beef producer survey of the cost to prevent and treat bovine respiratory disease in preweaned calves. J. Am. Vet. Med. Assoc. 2018, 253, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Klima, C.L.; Holman, D.B.; Ralston, B.J.; Stanford, K.; Zaheer, R.; Alexander, T.W.; McAllister, T.A. Lower Respiratory Tract Microbiome and Resistome of Bovine Respiratory Disease Mortalities. Microb. Ecol. 2019, 78, 446–456. [Google Scholar] [CrossRef] [PubMed]
- Scott, M.A.; Woolums, A.R.; Swiderski, C.E.; Perkins, A.D.; Nanduri, B. Genes and regulatory mechanisms associated with experimentally-induced bovine respiratory disease identified using supervised machine learning methodology. Sci. Rep. 2021, 11, 22916. [Google Scholar] [CrossRef]
- Chai, J.; Capik, S.F.; Kegley, B.; Richeson, J.T.; Powell, J.G.; Zhao, J. Bovine respiratory microbiota of feedlot cattle and its association with disease. Vet. Res. 2022, 53, 4. [Google Scholar] [CrossRef]
- Ellis, J.A. Update on viral pathogenesis in BRD. Anim. Health Res. Rev. 2009, 10, 149–153. [Google Scholar] [CrossRef]
- Mekata, H.; Hamabe, S.; Sudaryatma, P.E.; Kobayashi, I.; Kanno, T.; Okabayashi, T. Molecular epidemiological survey and phylogenetic analysis of bovine respiratory coronavirus in Japan from 2016 to 2018. J. Vet. Med. Sci. 2020, 82, 726–730. [Google Scholar] [CrossRef]
- Hashem, Y.M.; Mousa, W.S.; Abdeen, E.E.; Abdelkhalek, H.M.; Nooruzzaman, M.; El-Askary, A.; Ismail, K.A.; Megahed, A.M.; Abdeen, A.; Soliman, E.A.; et al. Prevalence and Molecular Characterization of Mycoplasma Species, Pasteurella multocida, and Staphylococcus aureus Isolated from Calves with Respiratory Manifestations. Animals 2022, 12, 312. [Google Scholar] [CrossRef]
- Confer, A.W.; Ayalew, S. Mannheimia haemolytica in bovine respiratory disease: Immunogens, potential immunogens, and vaccines. Anim. Health Res. Rev. 2018, 19, 79–99. [Google Scholar] [CrossRef]
- Neu, U.; Mainou, B.A. Virus interactions with bacteria: Partners in the infectious dance. PLoS Pathog. 2020, 16, e1008234. [Google Scholar] [CrossRef]
- Oliveira, V.H.S.; Dall Agnol, A.M.; Fritzen, J.T.T.; Lorenzetti, E.; Alfieri, A.A.; Alfieri, A.F. Microbial diversity involved in the etiology of a bovine respiratory disease outbreak in a dairy calf rearing unit. Comp. Immunol. Microbiol. Infect. Dis. 2020, 71, 101494. [Google Scholar] [CrossRef] [PubMed]
- Timsit, E.; Workentine, M.; van der Meer, F.; Alexander, T. Distinct bacterial metacommunities inhabit the upper and lower respiratory tracts of healthy feedlot cattle and those diagnosed with bronchopneumonia. Vet. Microbiol. 2018, 221, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Manna, S.; McAuley, J.; Jacobson, J.; Nguyen, C.D.; Ullah, M.A.; Sebina, I.; Williamson, V.; Mulholland, E.K.; Wijburg, O.; Phipps, S.; et al. Synergism and Antagonism of Bacterial-Viral Coinfection in the Upper Respiratory Tract. mSphere 2022, 7, e0098421. [Google Scholar] [CrossRef] [PubMed]
- McMullen, C.; Alexander, T.W.; Orsel, K.; Timsit, E. Progression of nasopharyngeal and tracheal bacterial microbiotas of feedlot cattle during development of bovine respiratory disease. Vet. Microbiol. 2020, 248, 108826. [Google Scholar] [CrossRef]
- Kanda, T.; Tanaka, S.; Suwanruengsri, M.; Sukmawinata, E.; Uemura, R.; Yamaguchi, R.; Sueyoshi, M. Bovine Endocarditis Associated with Mycoplasma bovis. J. Comp. Pathol. 2019, 171, 53–58. [Google Scholar] [CrossRef] [PubMed]
- McMullen, C.; Alexander, T.W.; Leguillette, R.; Workentine, M.; Timsit, E. Topography of the respiratory tract bacterial microbiota in cattle. Microbiome 2020, 8, 91. [Google Scholar] [CrossRef]
- Nicola, I.; Cerutti, F.; Grego, E.; Bertone, I.; Gianella, P.; D’Angelo, A.; Peletto, S.; Bellino, C. Characterization of the upper and lower respiratory tract microbiota in Piedmontese calves. Microbiome 2017, 5, 152. [Google Scholar] [CrossRef]
- McMullen, C.; Orsel, K.; Alexander, T.W.; van der Meer, F.; Plastow, G.; Timsit, E. Comparison of the nasopharyngeal bacterial microbiota of beef calves raised without the use of antimicrobials between healthy calves and those diagnosed with bovine respiratory disease. Vet. Microbiol. 2019, 231, 56–62. [Google Scholar] [CrossRef]
- Lee, J.; Han, G.; Kim, J.W.; Jeon, C.O.; Hyun, S. Taxon-Specific Effects of Lactobacillus on Drosophila Host Development. Microb. Ecol. 2020, 79, 241–251. [Google Scholar] [CrossRef]
- Snyder, E.; Credille, B. Mannheimia haemolytica and Pasteurella multocida in Bovine Respiratory Disease: How Are They Changing in Response to Efforts to Control Them? Vet. Clin. N. Am. Food Anim. Pract. 2020, 36, 253–268. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jiang, X.; Liu, X.; Zhao, X.; Liu, S.; Li, Y.; Zhang, Y. Growth, health, rumen fermentation, and bacterial community of Holstein calves fed Lactobacillus rhamnosus GG during the preweaning stage1. J. Anim. Sci. 2019, 97, 2598–2608. [Google Scholar] [CrossRef] [PubMed]
- Amat, S.; Holman, D.B.; Timsit, E.; Schwinghamer, T.; Alexander, T.W. Evaluation of the Nasopharyngeal Microbiota in Beef Cattle Transported to a Feedlot, With a Focus on Lactic Acid-Producing Bacteria. Front. Microbiol. 2019, 10, 1988. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, S.P.; Sindt, J.J.; Greenquist, M.A.; Miller, W.F.; Pike, J.N.; Loe, E.R.; Sulpizio, M.J.; Drouillard, J.S. Plasma metabolites of receiving heifers and the relationship between apparent bovine respiratory disease, body weight gain, and carcass characteristics. J. Anim. Sci. 2009, 87, 328–333. [Google Scholar] [CrossRef] [PubMed]
- De Boeck, I.; Spacova, I.; Vanderveken, O.M.; Lebeer, S. Lactic acid bacteria as probiotics for the nose? Microb. Biotechnol. 2021, 14, 859–869. [Google Scholar] [CrossRef]
- Amat, S.; Timsit, E.; Baines, D.; Yanke, J.; Alexander, T.W. Development of Bacterial Therapeutics against the Bovine Respiratory Pathogen Mannheimia haemolytica. Appl. Environ. Microbiol. 2019, 85, e01359-19. [Google Scholar] [CrossRef]
- Chong, H.X.; Yusoff, N.A.A.; Hor, Y.Y.; Lew, L.C.; Jaafar, M.H.; Choi, S.B.; Yusoff, M.S.B.; Wahid, N.; Abdullah, M.; Zakaria, N.; et al. Lactobacillus plantarum DR7 improved upper respiratory tract infections via enhancing immune and inflammatory parameters: A randomized, double-blind, placebo-controlled study. J. Dairy Sci. 2019, 102, 4783–4797. [Google Scholar] [CrossRef]
- Wescombe, P.A.; Heng, N.C.; Burton, J.P.; Chilcott, C.N.; Tagg, J.R. Streptococcal bacteriocins and the case for Streptococcus salivarius as model oral probiotics. Future Microbiol. 2009, 4, 819–835. [Google Scholar] [CrossRef]
- Rabinowitz, J.D.; Enerback, S. Lactate: The ugly duckling of energy metabolism. Nat. Metab. 2020, 2, 566–571. [Google Scholar] [CrossRef]
- Offermanns, S. Hydroxy-Carboxylic Acid Receptor Actions in Metabolism. Trends Endocrinol. Metab. 2017, 28, 227–236. [Google Scholar] [CrossRef]
- Adeva-Andany, M.; Lopez-Ojen, M.; Funcasta-Calderon, R.; Ameneiros-Rodriguez, E.; Donapetry-Garcia, C.; Vila-Altesor, M.; Rodriguez-Seijas, J. Comprehensive review on lactate metabolism in human health. Mitochondrion 2014, 17, 76–100. [Google Scholar] [CrossRef] [PubMed]
- Duan, K.; Liu, Z.J.; Hu, S.Q.; Huo, H.Y.; Xu, Z.R.; Ruan, J.F.; Sun, Y.; Dai, L.P.; Yan, C.B.; Xiong, W.; et al. Lactic acid induces lactate transport and glycolysis/OXPHOS interconversion in glioblastoma. Biochem. Biophys. Res. Commun. 2018, 503, 888–894. [Google Scholar] [CrossRef] [PubMed]
- Ying, M.; You, D.; Zhu, X.; Cai, L.; Zeng, S.; Hu, X. Lactate and glutamine support NADPH generation in cancer cells under glucose deprived conditions. Redox Biol. 2021, 46, 102065. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Li, H.; Chen, J.; Qian, Q. Lactic Acid: No Longer an Inert and End-Product of Glycolysis. Physiology 2017, 32, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Wagner, W.; Ciszewski, W.M.; Kania, K.D. L- and D-lactate enhance DNA repair and modulate the resistance of cervical carcinoma cells to anticancer drugs via histone deacetylase inhibition and hydroxycarboxylic acid receptor 1 activation. Cell Commun. Signal. 2015, 13, 36. [Google Scholar] [CrossRef]
- Brooks, G.A. Lactate as a fulcrum of metabolism. Redox Biol. 2020, 35, 101454. [Google Scholar] [CrossRef]
- Blakebrough-Hall, C.; Dona, A.; D’Occhio, M.J.; McMeniman, J.; Gonzalez, L.A. Diagnosis of Bovine Respiratory Disease in feedlot cattle using blood (1)H NMR metabolomics. Sci. Rep. 2020, 10, 115. [Google Scholar] [CrossRef]
- MahmoudianDehkordi, S.; Ahmed, A.T.; Bhattacharyya, S.; Han, X.; Baillie, R.A.; Arnold, M.; Skime, M.K.; John-Williams, L.S.; Moseley, M.A.; Thompson, J.W.; et al. Alterations in acylcarnitines, amines, and lipids inform about the mechanism of action of citalopram/escitalopram in major depression. Transl. Psychiatry 2021, 11, 153. [Google Scholar] [CrossRef]

| Align ID | Name | Synon |
|---|---|---|
| 8 | Lactic acid (2TMS) | (S)-Lactic acid |
| 83 | Phosphoric acid (3TMS) | [PO(OH)3] |
| 11 | Sarcosine (2TMS) | (methylamino)acetic acid |
| 10 | Valine (1TMS) | (2S)-2-amino-3-methylbutanoic acid |
| 63 | Norvaline, DL-(2TMS) | (2S)-2-aminopentanoic acid |
| 130 | Pyroglutamic acid (2TMS) | (S)-(-)-2-Pyrrolidone-5-carboxylic acid |
| 68 | Urea (2TMS) | Carbamide |
| 27 | Leucine (1TMS) | 2-amino-4-methylpentanoic acid |
| 95 | Norleucine (2TMS) | (2S)-2-aminohexanoic acid |
| 131 | Glutamic acid (2TMS) | (2S)-2-aminopentanedioic acid |
| 51 | Leucine, cyclo-(1TMS) | 1-Amino-1-cyclopentanecarboxylic acid |
| 52 | Isoleucine (1TMS) | Hile |
| 170 | Lactic acid, 3-(4-hydroxyphenyl)-(3TMS) | 2-hydroxy-3-(4-hydroxyphenyl) propanoic acid |
| 182 | Hexadecanoic acid (1TMS) | — |
| 122 | Glutamic acid, N-methyl-(2TMS) | (2S)-2-(methylamino) pentanedioic acid |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Ma, C.; Han, Y.; Jin, H.; Luo, H.; Hao, X.; Li, M. Integrative Analysis of the Nasal Microbiota and Serum Metabolites in Bovines with Respiratory Disease by 16S rRNA Sequencing and Gas Chromatography/Mass Selective Detector-Based Metabolomics. Int. J. Mol. Sci. 2022, 23, 12028. https://doi.org/10.3390/ijms231912028
Zhang Y, Ma C, Han Y, Jin H, Luo H, Hao X, Li M. Integrative Analysis of the Nasal Microbiota and Serum Metabolites in Bovines with Respiratory Disease by 16S rRNA Sequencing and Gas Chromatography/Mass Selective Detector-Based Metabolomics. International Journal of Molecular Sciences. 2022; 23(19):12028. https://doi.org/10.3390/ijms231912028
Chicago/Turabian StyleZhang, Ying, Chunji Ma, Yang Han, Hua Jin, Haixia Luo, Xiujing Hao, and Min Li. 2022. "Integrative Analysis of the Nasal Microbiota and Serum Metabolites in Bovines with Respiratory Disease by 16S rRNA Sequencing and Gas Chromatography/Mass Selective Detector-Based Metabolomics" International Journal of Molecular Sciences 23, no. 19: 12028. https://doi.org/10.3390/ijms231912028
APA StyleZhang, Y., Ma, C., Han, Y., Jin, H., Luo, H., Hao, X., & Li, M. (2022). Integrative Analysis of the Nasal Microbiota and Serum Metabolites in Bovines with Respiratory Disease by 16S rRNA Sequencing and Gas Chromatography/Mass Selective Detector-Based Metabolomics. International Journal of Molecular Sciences, 23(19), 12028. https://doi.org/10.3390/ijms231912028





