Integration of QTL Mapping and Whole Genome Sequencing Identifies Candidate Genes for Alkalinity Tolerance in Rice (Oryza sativa)
Abstract
1. Introduction
2. Results
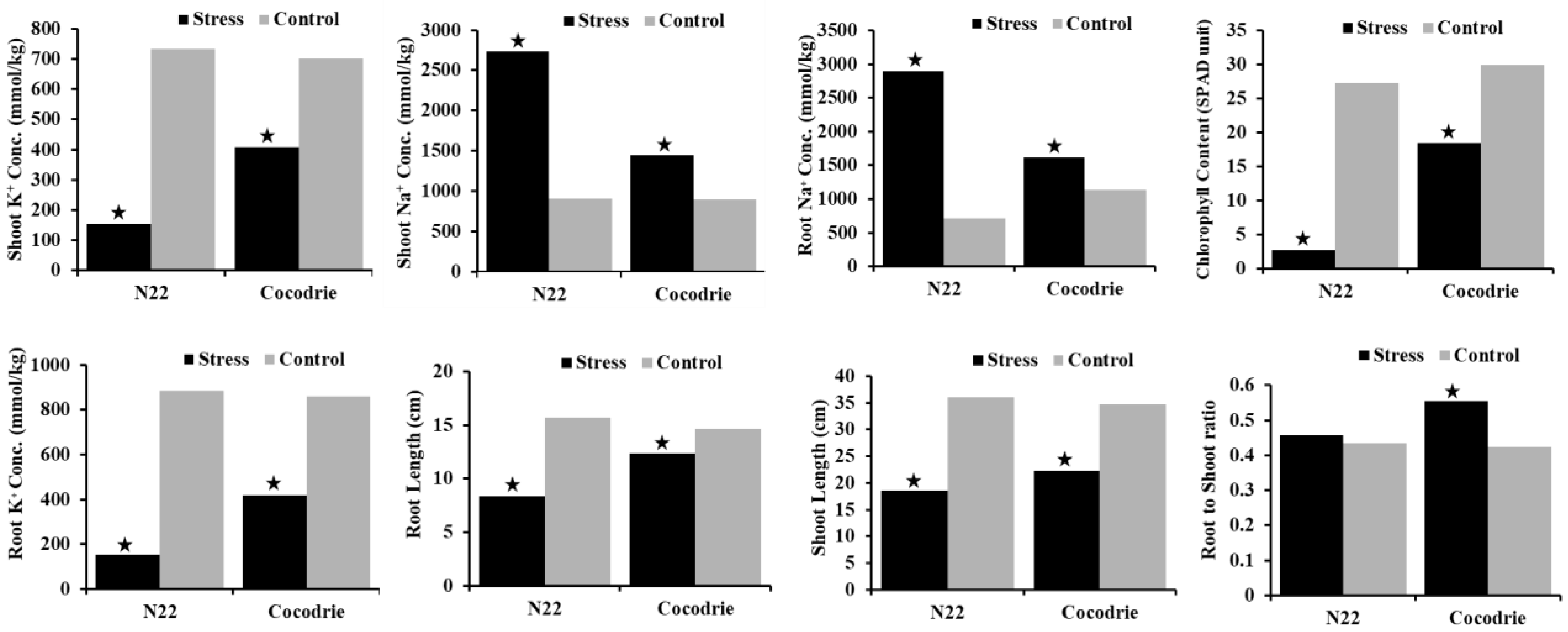
2.1. Phenotypic Evaluation
2.2. Correlations among Traits
2.3. QTL Analysis
2.3.1. Alkalinity Tolerance Score (AKT)
2.3.2. Chlorophyll Content (CHL)
2.3.3. Shoot Length (SHL), Root Length (RTL), and Root-to-Shoot Ratio (RSR)
2.3.4. Shoot Na+ Concentration (SNC) and Root Na+ Concentration (RNC)
2.3.5. Shoot K+ Concentration (SKC) and Root K+ Concentration (RKC)
2.3.6. Shoot and Root Na/K Ratio (SNK and RNK)
2.4. Co-Localization of QTLs
2.5. Gene Ontology Analysis
2.6. Discovery of Genotype-Specific SNPs, Indels, and Candidate Genes in the Selected QTL Regions
2.7. Expression Pattern of Candidate Genes Located in the Alkalinity Stress Tolerance QTL Intervals Using qRT-PCR
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Choice of Parents and Mapping Population
5.2. Evaluation of Rice Seedlings for Alkalinity Tolerance
5.3. Statistical Analysis
5.4. Linkage Mapping and QTL Analysis
5.5. Whole Genome Resequencing of Parents
5.6. Read-Mapping and Detection of SNPs and Indels
5.7. Gene Ontology and Annotation
5.8. Validation of Selected Genes in the QTL Interval by Quantitative Reverse Transcription PCR (qRT-PCR)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rao, G.J.N.; Reddy, J.N.; Variar, M.; Mahender, A. Molecular breeding to improve plant resistance to abiotic stresses. In Advances in Plant Breeding Strategies: Agronomic, Abiotic and Biotic Stress Traits; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 283–326. [Google Scholar]
- Bray, E.A.; Bailey-Serres, J.; Weretilnyk, E. Responses to abiotic stress. In Biochemistry & Molecular Biology of Plants; Gruissem, W., Jones, R., Eds.; American Society of Plant Biologists: Rockville, MD, USA, 2000; pp. 1158–1249. [Google Scholar]
- Martinez-Beltran, J.; Manzur, C.L. Overview of salinity problems in the world and FAO strategies to address the problem. In Proceedings of the International Salinity Forum, Riverside, CA, USA, 25–27 April 2005; pp. 311–313. [Google Scholar]
- Qadir, M.; Quillerou, E.; Nangia, V.; Murtaza, G.; Singh, M.; Thomas, R.; Drechsel, P.; Noble, A. Economics of salt-induced land degradation and restoration. Nat. Resour. Forum 2014, 38, 282–295. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Jagendorf, A.; Zhu, J. Understanding and improving salt tolerance in plants. Crop Sci. 2020, 45, 437–448. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, X.L.; Zhang, R.X.; Yuan, H.Y.; Wang, M.M.; Yang, H.Y.; Ma, H.Y.; Liu, D.; Jiang, C.J.; Liang, Z.W. Root damage under alkaline stress is associated with reactive oxygen species accumulation in rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 1580. [Google Scholar] [CrossRef]
- Lv, B.S.; Li, X.W.; Ma, H.Y.; Sun, Y.; Wei, L.X.; Jiang, C.J.; Liang, Z.W. Differences in growth and physiology of rice in response to different saline-alkaline stress factors. Agron. J. 2013, 105, 1119–1128. [Google Scholar] [CrossRef]
- Tian, Z.J.; Li, J.P.; Jia, X.Y.; Yang, F.; Wang, Z.C. Assimilation and translocation of dry matter and phosphorus in rice genotypes affected by salt-alkaline stress. Sustainability 2016, 8, 568. [Google Scholar] [CrossRef]
- Wang, H.; Takano, T.; Liu, S. Screening and evaluation of saline-alkaline tolerant germplasm of rice (Oryza sativa L.) in soda saline-alkali soil. Agronomy 2018, 8, 205. [Google Scholar] [CrossRef]
- De Leon, T.B.; Linscombe, S.; Gregorio, G.; Subudhi, P.K. Genetic variation in Southern USA rice genotypes for seedling salinity tolerance. Front. Plant Sci. 2015, 6, 374. [Google Scholar] [CrossRef]
- De Leon, T.B.; Linscombe, S.; Subudhi, P.K. Identification and validation of QTLs for seedling salinity tolerance in introgression lines of a salt tolerant rice landrace ‘Pokkali’. PLoS ONE 2017, 12, e0175361. [Google Scholar] [CrossRef]
- Chandran, A.K.N.; Kim, J.; Yoo, Y.H.; Park, H.L.; Kim, Y.J.; Cho, M.H.; Jung, K.H. Transcriptome analysis of rice-seedling roots under soil-salt stress using RNA-Seq method. Plant Biotechnol. Rep. 2019, 13, 567–578. [Google Scholar] [CrossRef]
- Ren, Z.H.; Gao, J.P.; Li, L.G.; Cai, X.L.; Huang, W.; Chao, D.Y.; Zhu, M.Z.; Wang, Z.Y.; Luan, S.; Lin, H.X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005, 37, 1141–1146. [Google Scholar] [CrossRef]
- Huang, X.; Chao, D.; Gao, J.; Zhu, M.; Shi, M.; Lin, H. A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 2009, 23, 1805–1817. [Google Scholar] [CrossRef]
- Li, X.; Zheng, H.; Wu, W.; Liu, H.; Wang, J.; Jia, Y.; Li, J.; Yang, L.; Lei, L.; Zou, D.; et al. QTL mapping and candidate gene analysis for alkali tolerance in japonica rice at the bud stage based on linkage mapping and genome-wide association study. Rice N. Y. 2020, 13, 48. [Google Scholar] [CrossRef]
- Verslues, P.E.; Agarwal, M.; Katiyar-Agarwal, S.; Zhu, J.; Zhu, J.K. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 2006, 46, 1092. [Google Scholar] [CrossRef]
- Allah, A.A.; Badawy, S.A.; Zayed, B.; Gohary, A.A. The role of root system traits in the drought tolerance of rice (Oryza sativa L.). J. Plant Prod. 2011, 80, 700–704. [Google Scholar] [CrossRef]
- Li, Q.; Yang, A.; Zhang, W.H. Efficient acquisition of iron confers greater tolerance to saline-alkaline stress in rice (Oryza sativa L.). J. Exp. Bot. 2016, 67, 6431–6444. [Google Scholar] [CrossRef]
- Ogo, Y.; Itai, R.N.; Kobayashi, T.; Aung, M.S.; Nakanishi, H.; Nishizawa, N.K. OsIRO2 is responsible for iron utilization in rice and improves growth and yield in calcareous soil. Plant Mol. Biol. 2011, 75, 593–605. [Google Scholar] [CrossRef]
- Ji, H.; Pardo, J.; Batelli, G.; Oosten, M.; Bressan, R.; Li, X. The Salt Overly Sensitive (SOS) pathway: Established and emerging roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef]
- Cheng, H.T.; Jiang, H.; Xue, D.W.; Guo, L.B.; Zeng, D.L.; Zhang, G.H.; Qian, Q. Mapping of QTL underlying tolerance to alkali at germination and early seedling stages in rice. Acta Agron. Sin. 2008, 34, 1719–1727. [Google Scholar] [CrossRef]
- Qi, D.L.; Guo, G.Z.; Lee, M.C.; Zhang, J.G.; Cao, G.L.; Zhang, S.; Suh, S.; Zhou, Q.; Han, L. Identification of quantitative trait loci for the dead leaf rate and the seedling dead rate under alkaline stress in rice. J. Genet. Genom. 2008, 35, 299–305. [Google Scholar] [CrossRef]
- Liang, J.; Qu, Y.; Yang, C.; Ma, X.; Cao, G.; Zhao, Z.; Zhang, S.; Zhang, T.; Han, L. Identification of QTLs associated with salt or alkaline tolerance at the seedling stage in rice under salt or alkaline stress. Euphytica 2015, 201, 441–452. [Google Scholar] [CrossRef]
- Li, N.; Sun, J.; Wang, J.; Liu, H.; Zheng, H.; Yang, L.; Liang, Y.; Li, X.; Zou, D. QTL analysis for alkaline tolerance of rice and verification of a major QTL. Plant Breed. 2017, 136, 881–891. [Google Scholar] [CrossRef]
- Li, N.; Zheng, H.; Cui, J.; Wang, J.; Liu, H.; Sun, J.; Liu, T.; Zhao, H.; Lai, Y.; Zou, D. Genome-wide association study and candidate gene analysis of alkalinity tolerance in japonica rice germplasm at the seedling stage. Rice 2019, 12, 24. [Google Scholar] [CrossRef]
- Luo, X.; Deng, H.; Wang, P.; Zhang, X.; Li, C.; Li, C.; Tan, J.; Wu, G.; Wang, Y.; Cheng, Q.; et al. Genetic analysis of germinating ability under alkaline and neutral salt stress by a high-density bin genetic map in rice. Euphytica 2020, 216, 107. [Google Scholar] [CrossRef]
- Subudhi, P.K.; Shankar, R.; Jain, M. Whole genome sequence analysis of rice genotypes with contrasting response to salinity stress. Sci. Rep. 2020, 10, 21259. [Google Scholar] [CrossRef]
- Puram, V.R.R.; Ontoy, J.; Linscombe, S.; Subudhi, P.K. Genetic dissection of seedling stage salinity tolerance in rice using introgression lines of a salt tolerant landrace Nona Bokra. J. Hered. 2017, 108, 658–670. [Google Scholar] [CrossRef]
- Sabouri, H.; Sabouri, A. New evidence of QTLs attributed to salinity tolerance in rice. Afr. J. Biotechnol. 2008, 7, 4376–4383. [Google Scholar]
- De Leon, T.B.; Linscombe, S.; Subudhi, P.K. Molecular dissection of seedling salinity tolerance in rice (Oryza sativa) using a high density GBS based SNP linkage map. Rice 2016, 9, 52. [Google Scholar] [CrossRef]
- Bimpong, I.K.; Manneh, B.; El-Namaky, R.; Diaw, F.; Amoah, N.K.A.; Sanneh, B.; Ghislain, K.; Sow, A.; Singh, R.; Gregorio, G.; et al. Mapping QTLs related to salt tolerance in rice at the young seedling stage using 384-plex single nucleotide polymorphism SNP marker sets. Mol. Plant Breed. 2013, 5, 47–62. [Google Scholar] [CrossRef]
- Lin, H.X.; Zhu, M.Z.; Yano, M.; Gao, J.P.; Liang, Z.W.; Su, W.A.; Hu, X.H.; Ren, Z.H.; Chao, D.Y. QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theor. Appl. Genet. 2004, 108, 253–260. [Google Scholar] [CrossRef]
- Lv, B.S.; Ma, H.Y.; Li, X.W.; Wei, L.X.; Lv, H.Y.; Yang, H.Y.; Jiang, C.J.; Liang, Z.W. Proline accumulation is not correlated with saline-alkaline stress tolerance in rice seedlings. Agron. J. 2015, 107, 51–60. [Google Scholar] [CrossRef]
- Li, N.; Liu, H.; Sun, J.; Zheng, H.; Wang, J.; Yang, L.; Zhao, H.; Zou, D. Transcriptome analysis of two contrasting rice cultivars during alkaline stress. Sci. Rep. 2018, 8, 9586. [Google Scholar] [CrossRef]
- Wang, H.; Lin, X.; Cao, S.; Wu, Z. Alkali tolerance in rice (Oryza sativa L.): Growth, photosynthesis, nitrogen metabolism, and ion homeostasis. Photosynthetica 2015, 53, 55–65. [Google Scholar] [CrossRef]
- Reddy, I.N.B.L.; Kim, B.K.; Yoon, I.S.; Kim, K.H.; Kwon, T.R. Salt tolerance in rice: Focus on mechanisms and approaches. Rice Sci. 2017, 24, 123–144. [Google Scholar] [CrossRef]
- Chuamnakthong, S.; Nampei, M.; Ueda, A. Characterization of Na+ exclusion mechanism in rice under saline-alkaline stress conditions. Plant Sci. 2019, 287, 110171. [Google Scholar] [CrossRef]
- Razzaque, S.; Elias, S.M.; Haque, T.; Biswas, S.; Jewel, N.A.; Rahman, S.; Weng, X.; Ismail, A.M.; Walia, H.; Juenger, T.E.; et al. Gene expression analysis associated with salt stress in a reciprocally crossed rice population. Sci. Rep. 2019, 9, 8249. [Google Scholar] [CrossRef]
- Yan, Y.S.; Chen, X.Y.; Yang, K.; Sun, Z.X.; Fu, Y.P.; Zhang, Y.M.; Fang, R.X. Overexpression of an F-box protein gene reduces abiotic stress tolerance and promotes root growth in rice. Mol. Plant 2011, 4, 190–197. [Google Scholar] [CrossRef]
- Dametto, A.; Buffon, G.; Dos Reis Blasi, É.A.; Sperotto, R.A. Ubiquitination pathway as a target to develop abiotic stress tolerance in rice. Plant Signal. Behav. 2015, 10, e1057369. [Google Scholar] [CrossRef]
- Goswami, K.; Tripathi, A.; Sanan-Mishra, N. Comparative miRomics of salt-tolerant and salt-sensitive rice. J. Integr. Bioinform. 2017, 14, 20170002. [Google Scholar] [CrossRef]
- Ye, Y.; Yuan, J.; Chang, X.; Yang, M.; Zhang, L.; Lu, K.; Lian, X. The phosphate transporter gene OsPht1;4 is involved in phosphate homeostasis in rice. PLoS ONE 2015, 10, e0126186. [Google Scholar] [CrossRef]
- Katsuhara, M.; Koshio, K.; Shibasaka, M.; Hayashi, Y.; Hayakawa, T.; Kasamo, K. Over-expression of a barley aquaporin increased the shoot/root ratio and raised salt sensitivity in transgenic rice plants. Plant Cell Physiol. 2003, 44, 1378–1383. [Google Scholar] [CrossRef]
- Lin, R.; Zheng, J.; Pu, L.; Wang, Z.; Mei, Q.; Zhang, M.; Jian, S. Genome-wide identification and expression analysis of aquaporin family in Canavalia rosea and their roles in the adaptation to saline-alkaline soils and drought stress. BMC Plant Biol. 2021, 21, 333. [Google Scholar] [CrossRef]
- Li, D.; Zhang, H.; Hong, Y.; Huang, L.; Li, X.; Zhang, Y.; Ouyang, Z.; Song, F. Genome-wide identification, biochemical characterization, and expression analyses of the YTH domain-containing RNA-binding protein family in Arabidopsis and rice. Plant Mol. Biol. Rep. 2014, 32, 1169–1186. [Google Scholar] [CrossRef]
- Chen, G.; Zou, Y.; Hu, J.; Ding, Y. Genome-wide analysis of the rice PPR gene family and their expression profiles under different stress treatments. BMC Genom. 2018, 19, 720. [Google Scholar] [CrossRef]
- Shim, J.S.; Park, S.H.; Lee, D.K.; Kim, Y.S.; Park, S.C.; Redillas, M.R.; Seo, J.S.; Kim, J.K. The rice GLYCINE-RICH PROTEIN 3 confers drought tolerance by regulating mRNA stability of ROS scavenging-related genes. Rice 2021, 14, 31. [Google Scholar] [CrossRef]
- Huang, W.; Ma, X.; Wang, Q.; Gao, Y.; Xue, Y.; Niu, X.; Yu, G.; Liu, Y.S. Significant improvement of stress tolerance in tobacco plants by overexpressing a stress-responsive aldehyde dehydrogenase gene from maize (Zea mays). Plant Mol. Biol. 2008, 68, 451–463. [Google Scholar] [CrossRef]
- Gautam, R.; Meena, R.K.; Woch, N.; Kirti, P. Ectopic expression of BrALDH7B2 gene encoding an antiquitin from Brassica rapa confers tolerance to abiotic stresses and improves photosynthetic performance under salt stress in tobacco. Environ. Exp. Bot. 2020, 180, 104223. [Google Scholar] [CrossRef]
- Guo, J.; Sun, W.; Liu, H.; Chi, J.; Odiba, A.S.; Li, G.; Li-Ping, J.; Xin, C. Aldehyde dehydrogenase plays crucial roles in response to lower temperature stress in Solanum tuberosum and Nicotiana benthamiana. Plant Sci. 2020, 297, 110525. [Google Scholar] [CrossRef]
- Yuan, X.; Sun, H.; Tang, Z.; Tang, H.; Zhang, H.; Huang, J. A novel little membrane protein confers salt tolerance in rice (Oryza sativa L.). Plant Mol. Biol. Rep. 2016, 34, 524–532. [Google Scholar] [CrossRef]
- Guan, Q.; Ma, H.; Wang, Z.J.; Wang, Z.Y.; Bu, Q.Y.; Liu, S.K. A rice LSD1-like-type ZFP gene OsLOL5 enhances saline-alkaline tolerance in transgenic Arabidopsis thaliana, yeast and rice. BMC Genom. 2016, 17, 142. [Google Scholar] [CrossRef]
- Qiao, B.; Zhang, Q.; Liu, D.; Wang, H.; Yin, J.; Wang, R.; He, M.; Cui, M.; Shang, Z.; Wang, D.; et al. A calcium-binding protein, rice annexin OsANN1, enhances heat stress tolerance by modulating the production of H2O2. J. Exp. Bot. 2015, 66, 5853–5866. [Google Scholar] [CrossRef]
- Jangam, A.P.; Pathak, R.R.; Raghuram, N. Microarray analysis of rice d1 (RGA1) mutant reveals the potential role of G-protein alpha subunit in regulating multiple abiotic stresses such as drought, salinity, heat, and cold. Front. Plant Sci. 2016, 7, 11. [Google Scholar] [CrossRef]
- Cui, Y.; Wang, M.; Zhou, H.; Li, M.; Huang, L.; Yin, X.; Zhao, G.; Lin, F.; Xia, X.; Xu, G. OsSGL, a novel DUF1645 domain-containing protein, confers enhanced drought tolerance in transgenic rice and Arabidopsis. Front. Plant Sci. 2016, 7, 2001. [Google Scholar] [CrossRef]
- Singh, B.; Khurana, P.; Khurana, J.P.; Singh, P. Gene encoding vesicle-associated membrane protein-associated protein from Triticum aestivum (TaVAP) confers tolerance to drought stress. Cell Stress Chaperones 2018, 23, 411–428. [Google Scholar] [CrossRef]
- Jia, B.; Sun, M.; DuanMu, H.; Ding, X.; Liu, B.; Zhu, Y.; Sun, X. GsCHX19.3, a member of cation/H+ exchanger superfamily from wild soybean contributes to high salinity and carbonate alkaline tolerance. Sci. Rep. 2017, 7, 9423. [Google Scholar] [CrossRef]
- Wei, S.; Hu, W.; Deng, X.; Zhang, Y.; Liu, X.; Zhao, X.; Luo, Q.; Jin, Z.; Zhou, S.; Sun, T.; et al. A rice calcium-dependent protein kinase OsCPK9 positively regulates drought stress tolerance and spikelet fertility. BMC Plant Biol. 2014, 14, 133. [Google Scholar] [CrossRef]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef]
- Han, G.; Lu, C.; Guo, J.; Qiao, Z.; Sui, N.; Qiu, N.; Wang, B. C2H2 zinc finger proteins: Master regulators of abiotic stress responses in plants. Front. Plant Sci. 2020, 11, 115. [Google Scholar] [CrossRef]
- Li, Y.; Liu, F.; Li, P.; Wang, T.; Zheng, C.; Hou, B. An Arabidopsis cytokinin-modifying glycosyltransferase UGT76C2 improves drought and salt tolerance in rice. Front. Plant Sci. 2020, 11, 560696. [Google Scholar] [CrossRef]
- Jeyasri, R.; Muthuramalingam, P.; Satish, L.; Adarshan, S.; Lakshmi, M.A.; Pandian, S.K.; Chen, J.T.; Ahmar, S.; Wang, X.; Mora-Poblete, F.; et al. The role of OsWRKY genes in rice when faced with single and multiple abiotic stresses. Agronomy 2021, 11, 1301. [Google Scholar] [CrossRef]
- Zheng, L.; Ying, Y.; Wang, L.; Wang, F.; Whelan, J.; Shou, H. Identification of a novel iron regulated basic helix-loop-helix protein involved in Fe homeostasis in Oryza sativa. BMC Plant Biol. 2010, 10, 166. [Google Scholar] [CrossRef]
- Linscombe, S.; Jodari, F.; Bollich, P.; Groth, D.; White, L.; Chu, Q.; Dunand, R.; Sanders, D. Registration of “Cocodrie” rice. Crop Sci. 2000, 40, 294. [Google Scholar] [CrossRef]
- Vikram, P.; Swamy, B.M.; Dixit, S.; Ahmed, H.U.; Cruz, M.T.S.; Singh, A.K.; Kumar. qDTY1.1, a major QTL for rice grain yield under reproductive-stage drought stress with a consistent effect in multiple elite genetic backgrounds. BMC Genet. 2011, 12, 89. [Google Scholar] [CrossRef]
- Jones, J.B.; Case, V.W. Sampling, handling, and analyzing plant tissue samples. In Soil Testing and Plant Analysis, 3rd ed.; Westerman, R.L., Ed.; Book Series No. 3; Soil Science Society of America: Madison, WI, USA, 1990; pp. 389–427. [Google Scholar]
- R Foundation. R: A Language and Environment for Statistical Computing, Reference Index Version 2.2.1; R Foundation: Vienna, Austria, 2005. [Google Scholar]
- SAS Institute Inc. SAS® 9.4 System Options: Reference, 2nd ed.; SAS Institute Inc.: Cary, NC, USA, 2012. [Google Scholar]
- Holland, J.B.; Nyquist, W.E.; Cervantes-Martínez, C.T. Estimating and interpreting heritability for plant breeding: An update. Plant Breed. Rev. 2003, 22, 9–111. [Google Scholar]
- Bhattarai, U.; Subudhi, P.K. Identification of drought responsive QTLs during vegetative growth stage of rice using a saturated GBS-based SNP linkage map. Euphytica 2018, 214, 38. [Google Scholar] [CrossRef]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef]
- Patel, R.K.; Jain, M. NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Giga Sci. 2021, 10, giab008. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; O’Connor, B.D. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra, 1st ed.; O’Reilly Media: Newton, MA, USA, 2020. [Google Scholar]
- Poplin, R.; Ruano-Rubio, V.; DePristo, M.A.; Fennel, T.J.; Carneiro, M.O.; Van der Auwera, G.A.; Kling, D.E.; Gauthier, L.D.; Levy-Moonshine, A.; Roazen, D.; et al. Scaling accurate genetic variant discovery of tens of thousands of samples. BioRxiv 2017. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w(1118); iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. AgriGO v2.0: A GO analysis toolkit for the agricultural community update. Nucl. Acids Res. 2017, 45, 122–129. [Google Scholar] [CrossRef]
- Subudhi, P.K.; Garcia, R.S.; Coronejo, S.; Tapia, R. Comparative transcriptional profiling of root tissues in two rice genotypes reveals differential expressed genes associated with root architecture under nitrogen stress. Int. J. Mol. Sci. 2020, 21, 5759. [Google Scholar] [CrossRef]
- Livak, K.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

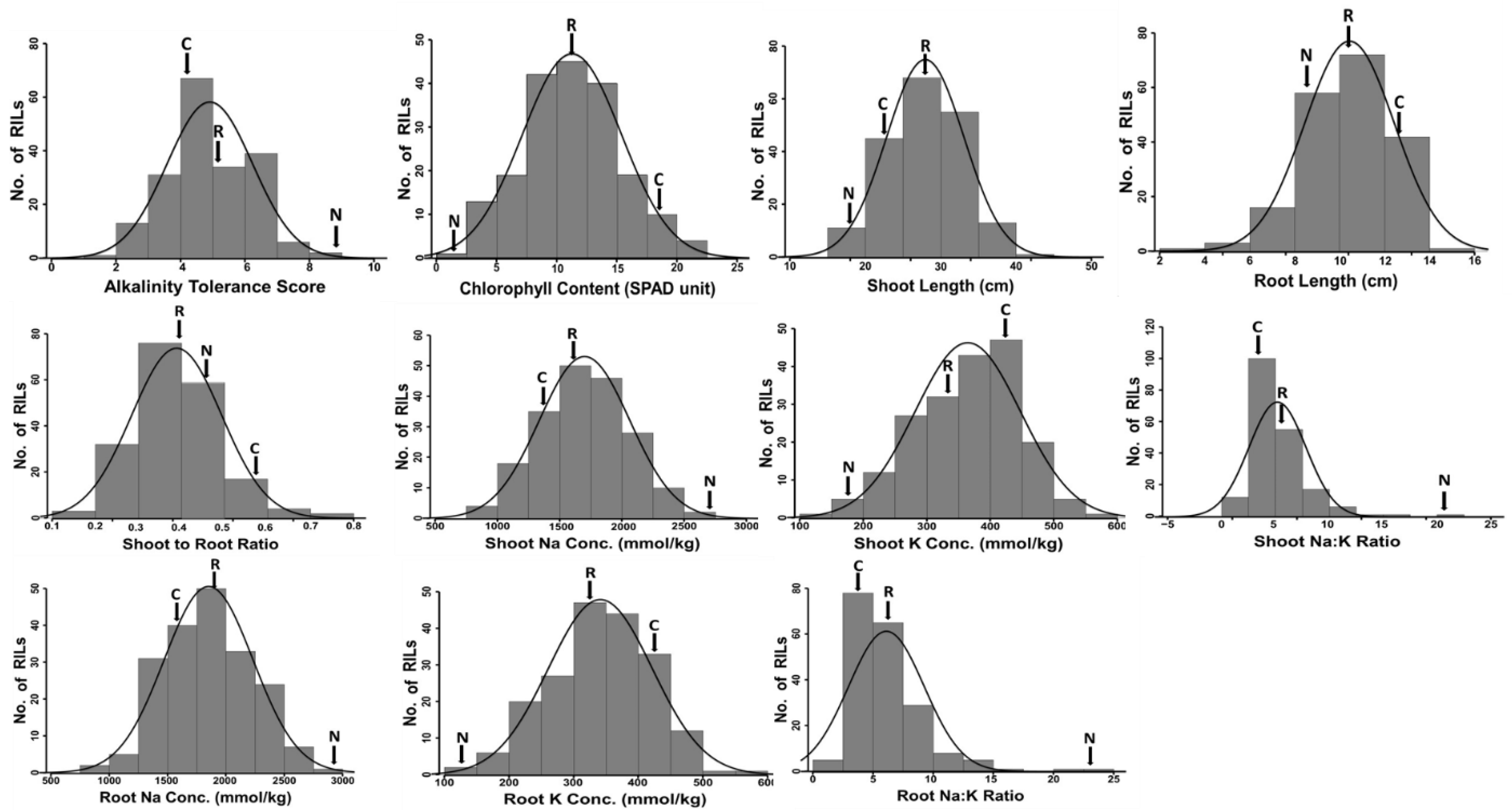


| Alkaline Stress | Control | Reduction in Trait Mean (%) Under Stress d | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait a | Cocodrie Mean | N22 Mean b | RIL Mean c | RIL Range | h2 | Cocodrie Mean | N22 Mean b | RIL Mean | RIL Range | Cocodrie | N22 |
| AKT | 4.3 | 9.0 ** | 5.0 ** | 2.0–9.0 | 0.7 | 1.0 | 1.0 ns | 1.1 | 1.0–3.0 | −76.7 | −88.9 |
| CHL (SPAD) | 18.4 | 2.7 * | 11.3 * | 2.2–22.1 | 0.5 | 29.9 | 27.2 ns | 25.5 | 18.2–34.2 | 38.5 | 90.1 |
| SHL (cm) | 22.2 | 18.5 * | 28.1 ** | 17.2–40.1 | 0.8 | 34.7 | 36.0 ns | 37.8 | 20.3–55.3 | 35.9 | 48.6 |
| RTL (cm) | 12.3 | 8.3 * | 10.4 ** | 3.9–14.1 | 0.7 | 14.7 | 15.7 ns | 12.7 | 5.5–16.5 | 16.1 | 47.0 |
| RSR | 0.6 | 0.4 ** | 0.4 ** | 0.2–0.8 | 0.7 | 0.4 | 0.5 ns | 0.4 | 0.2–0.6 | −35.7 | 2.2 |
| SNC (mmol/Kg) | 1448.2 | 2734.2 ** | 1690.4 ** | 832.4–2734.2 | 0.9 | 866.7 | 900.0 ns | 944.0 | 508.3–1383.3 | −67.1 | −203.8 |
| SKC (mmol/Kg) | 408.6 | 153.6 ** | 365.1 ** | 149.2–592.3 | 0.8 | 711.3 | 720.0 ns | 755.2 | 406.7–1106.7 | 42.6 | 78.7 |
| RNC (mmol/Kg) | 1619.2 | 2900.1 ** | 1851.7 ** | 903.3–2900.1 | 0.8 | 1106.0 | 657.1 ** | 1048.4 | 564.5–1536.3 | −46.4 | −341.3 |
| RKC (mmol/Kg) | 431.0 | 131.3 ** | 342.6 ** | 131.3–556.2 | 0.9 | 779.3 | 809.3 ns | 848.8 | 124.3–457.1 | 44.7 | 83.8 |
| SNK | 3.5 | 20.8 ** | 5.1 ** | 1.4–20.8 | 0.9 | 1.2 | 1.3 ns | 1.5 | 1.1–2.9 | −192.6 | −1565.6 |
| RNK | 3.8 | 23.0 ** | 6.0 ** | 1.6–23.0 | 0.9 | 1.4 | 0.8 ns | 1.4 | 0.6–3.1 | −166.0 | −2739.5 |
| Trait a | AKT | CHL | SHL | RTL | RSR | SNC | SKC | RNC | RKC | SNK | RNK |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AKT | 1.00 | ||||||||||
| CHL | −0.77 ** | 1.00 | |||||||||
| SHL | −0.02 | −0.11 | 1.00 | ||||||||
| RTL | −0.16 * | 0.11 | 0.05 | 1.00 | |||||||
| RSR | −0.09 | 0.17 * | −0.64 ** | 0.69 ** | 1.00 | ||||||
| SNC | 0.98 ** | −0.76 ** | −0.01 | −0.15 * | −0.09 | 1.00 | |||||
| SKC | −0.98 ** | 0.76 ** | 0.02 ** | 0.18 ** | 0.11 | −0.96 ** | 1.00 | ||||
| RNC | 0.97 ** | −0.75 ** | −0.01 * | −0.14 * | −0.09 | 0.99 ** | −0.96 ** | 1.00 | |||
| RKC | −0.96 ** | 0.74 ** | −0.00 ** | 0.19 ** | 0.14 * | −0.94 ** | 0.98 ** | −0.94 ** | 1.00 | ||
| SNK | 0.92 ** | −0.68 ** | −0.06 | −0.21 ** | −0.10 | 0.91 ** | −0.92 ** | 0.90 ** | −0.91 ** | 1.00 | |
| RNK | 0.92 ** | −0.68 ** | −0.05 | −0.20 ** | −0.11 | 0.91 ** | −0.92 ** | 0.90 ** | −0.92 ** | 0.99 ** | 1.00 |
| Trait a | QTL | Chr | Position(cM) | Left Marker | Right Marker | Interval (bp) | LOD b | PVE (%) c | Additive Effect | No. of Genes in QTL Interval | Parental Allele with Increasing Effect |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AKT | qAKT3.03 | 3 | 167 | S3_3798053 | S3_3978854 | 180,801 | 2.40 | 4.5 | 0.28 | 26 | N22 |
| qAKT5.008 | 5 | 5 | S5_865267 | S5_891285 | 26,018 | 2.30 | 4.0 | −0.26 | 4 | Cocodrie | |
| qAKT8.002 | 8 | 125 | S8_261276 | S8_498009 | 236,733 | 3.38 | 6.4 | 0.33 | 37 | N22 | |
| qAKT9.19 qAKT10.18 | 9 10 | 22 70 | S9_19333995 S10_18053155 | S9_19696641 S10_19335416 | 362,646 1,282,261 | 5.89 2.02 | 11.6 4.2 | 0.45 0.26 | 59 187 | N22 N22 | |
| CHL | qCHL1.37 | 1 | 186 | S1_37740707 | S1_37777636 | 36,929 | 3.04 | 6.3 | 0.97 | 4 | N22 |
| qCHL8.002 | 8 | 125 | S8_261276 | S8_498009 | 236,733 | 2.64 | 5.3 | −0.88 | 37 | Cocodrie | |
| qCHL9.20 | 9 | 17 | S9_20470185 | S9_20519258 | 49,073 | 8.27 | 17.8 | 1.63 | 3 | N22 | |
| SHL | qSHL1.03 | 1 | 20 | S1_3695146 | S1_3708821 | 13,675 | 2.31 | 1.8 | −0.67 | 2 | Cocodrie |
| qSHL1.38 | 1 | 190 | S1_38286772 | S1_38611845 | 325,073 | 33.95 | 43.8 | 3.37 | 44 | N22 | |
| qSHL3.13 | 3 | 110 | S3_13712517 | S3_13934642 | 222,125 | 3.25 | 2.7 | 0.84 | 20 | N22 | |
| qSHL6.26 | 6 | 29 | S6_26499660 | S6_26675063 | 175,403 | 2.58 | 2.1 | −0.74 | 19 | Cocodrie | |
| qSHL7.05 | 7 | 33 | S7_5312649 | S7_5549169 | 236,520 | 2.52 | 2.0 | 0.73 | 28 | N22 | |
| qSHL7.28 | 7 | 139 | S7_28852810 | S7_28875695 | 22,885 | 5.79 | 5.0 | 1.13 | 3 | N22 | |
| qSHL8.27 | 8 | 2 | S8_27384352 | S8_27875737 | 491,385 | 4.00 | 3.5 | −0.95 | 79 | Cocodrie | |
| RTL | qRTL3.28 | 3 | 52 | S3_28513305 | S3_28809504 | 296,199 | 2.66 | 6.8 | −0.46 | 41 | Cocodrie |
| qRTL7.26 | 7 | 11 | S7_26075952 | S7_26090857 | 14,905 | 3.03 | 7.7 | 0.50 | 1 | N22 | |
| RSR | qRSR1.30 | 1 | 147 | S1_30155765 | S1_30162203 | 6,438 | 2.79 | 4.0 | −0.02 | 1 | Cocodrie |
| qRSR1.35 | 1 | 181 | S1_35776217 | S1_37068548 | 1,292,331 | 13.43 | 21.2 | −0.04 | 94 | Cocodrie | |
| qRSR3.15 | 3 | 101 | S3_15513823 | S3_15747509 | 233,686 | 4.39 | 6.1 | −0.02 | 25 | Cocodrie | |
| qRSR8.17 | 8 | 67 | S8_17338253 | S8_17443562 | 105,309 | 2.47 | 3.4 | −0.01 | 9 | Cocodrie | |
| SNC | qSNC4.16 | 4 | 40 | S4_16612171 | S4_16880788 | 268,617 | 2.12 | 4.4 | 73.20 | 26 | N22 |
| qSNC8.002 | 8 | 125 | S8_261276 | S8_498009 | 236,733 | 3.41 | 8.2 | 94.03 | 37 | N22 | |
| qSNC9.19 qSNC12.19 | 9 12 | 22 68 | S9_19333995 S12_19968349 | S9_19696641 S12_20375777 | 362,646 407,428 | 4.80 2.04 | 11.8 4.2 | 113.8 71.79 | 59 33 | N22 N22 | |
| RNC | qRNC8.002 | 8 | 125 | S8_261276 | S8_498009 | 236,733 | 3.60 | 8.5 | 103.34 | 37 | N22 |
| qRNC9.19 | 9 | 22 | S9_19333995 | S9_19696641 | 362,646 | 4.87 | 11.6 | 122.41 | 59 | N22 | |
| qRNC12.19 | 12 | 68 | S12_19968349 | S12_20375777 | 407,428 | 2.07 | 4.5 | −77.19 | 33 | Cocodrie | |
| SKC | qSKC5.008 | 5 | 5 | S5_865267 | S5_891285 | 26,018 | 2.07 | 3.9 | −15.81 | 4 | Cocodrie |
| qSKC8.002 | 8 | 125 | S8_261276 | S8_498009 | 236,733 | 4.10 | 8.4 | −23.04 | 37 | Cocodrie | |
| qSKC10.18 | 10 | 70 | S10_18053155 | S10_19335416 | 1,282,261 | 6.01 | 12.6 | −28.49 | 187 | Cocodrie | |
| RKC | qRKC3.02 qRKC3.32 | 3 3 | 13 106 | S3_2264990 S3_32785101 | S3_2295597 S3_36366411 | 30,607 3,581,307 | 5.33 6.57 | 6.5 8.3 | −19.03 −27.12 | 5 559 | Cocodrie Cocodrie |
| qRKC8.002 qRKC9.19 | 8 9 | 125 22 | S8_261276 S9_19333995 | S8_498009 S9_19696641 | 236,733 362,646 | 5.66 7.66 | 7.0 9.8 | −24.51 −29.33 | 37 59 | Cocodrie Cocodrie | |
| SNK | qSNK3.03 | 3 | 21 | S3_3978853 | S3_4050070 | 71,217 | 2.85 | 6.3 | 0.57 | 8 | N22 |
| qSNK8.002 | 8 | 125 | S8_261276 | S8_498009 | 236,733 | 2.55 | 5.6 | 0.54 | 37 | N22 | |
| qSNK9.19 | 9 | 22 | S9_19333995 | S9_19696641 | 362,646 | 3.77 | 8.5 | 0.68 | 59 | N22 | |
| RNK | qRNK3.03 | 3 | 167 | S3_3798053 | S3_3978854 | 180,801 | 2.87 | 6.3 | 0.69 | 26 | N22 |
| qRNK4.16 | 4 | 40 | S4_16612171 | S4_16880788 | 268,617 | 3.16 | 6.7 | 0.79 | 26 | N22 | |
| qRNK8.002 | 8 | 125 | S8_261276 | S8_498009 | 236,733 | 2.69 | 5.9 | 0.67 | 37 | N22 | |
| qRNK9.19 | 9 | 22 | S9_19333995 | S9_19696641 | 362,646 | 3.87 | 8.6 | 0.82 | 59 | N22 |
| Current Study | ||||
|---|---|---|---|---|
| QTL a | Physical Position | QTL | Position (Flanking Markers) | References |
| qSHL1.38 | 38,286,772–38,611,845 | qSHL1.38 qPH1.2 | 38286772–38611845 164.4–170.3 cM (id1024972–id1025983) | [30,31] |
| qSHL3.13 | 13,712,517–13,934,642 | qDWSH-3 | 7,232,837–16,968,975 (RM1022–RM6283) | [29] |
| qSHL6.26 | 26,499,660–26,675,063 | qNAUP-6 qRSH6 | 22,297,146–28,599.319 (RM3287–RM340) 26,554,756–28,532,453 (RM454-RM528) | [21,29] |
| qSHL7.05 | 5,312,649–5,549,169 | qSDS7 | 4,573,316–7,739,951 (R2401–L538T7) | [32] |
| qSHL8.27 | 27,384,352–27,875,737 | qRTL8.27 | 27,238,050–27,304,101 | [30] |
| qRSR1.35 | 35,776,217–37,068,548 | qRNTQ-1, qSDS1 | 33,956,950–37,713,775 (C813–C86) | [32] |
| qSHL3.13 qRSR3.15 | 13,712,517–13,934,642 15,513,823–15,747,509 | qDLR3 | 13,221,482–20,244,184 (RM338–RM2453) | [22] |
| qRKC3.32 | 32,785,101–36,366,411 | qSNC3 | 33,386,334–35,669,797 (RM1221–RM130 | [24] |
| qAKT3.03, qRNK3.03, qSNK3.03 | 3,798,053–3,798,854 3,978,853–4,050,070 | qRTL3.1 | 3,803,115–5,337,745 (RM5474–RM5480) | [28] |
| qAKT8.002, qSNK8.002, qRNK8.002, qSNC8.002, qRNC8.002, qSKC8.002, qRKC8.002 qCHL8.002, | 261,176–498,009 | qNA8.1, qCHL8.1 | 125,275–4,772,897 (RM408–RM1111) | [28] |
| qSNC9.19, qRNC9.19, qRKC9.19, qSNK9.19, qRNK9.19, qAKT9.19 | 19,333,995–19,696,641 | qNAUP-9a qDWRO-9a | 16,580,865–21,003,387 (RM1553–RM7424) | [29] |
| QTL Clusters a | Total No. of Genes | No. of Genes Annotated | Annotated Genes (%) | Numbers of Significant Ontology Terms | ||
|---|---|---|---|---|---|---|
| Biological Process | Cellular Components | Molecular Function | ||||
| qAKT3.03, qRNK3.03, qSNK3.03 | 26 | 8 | 31 | 18 | 6 | 3 |
| qAKT5.008, qSKC5.008 | 4 | 2 | 50 | 2 | 0 | 0 |
| qAKT8.002, qCHL8.002, qSNC8.002, qRNC8.002, qSKC8.002, qRKC8.002, qSNK8.002, qRNK8.002 | 37 | 26 | 70 | 31 | 13 | 6 |
| qAKT9.19, qSNC9.19, qRNC9.19, qRKC9.19, qSNK9.19, qRNK9.19 | 59 | 37 | 63 | 35 | 16 | 8 |
| qAKT10.18, qSKC10.18 | 187 | 109 | 58 | 48 | 9 | 2 |
| qSNC12.19, qRNC12.19 | 33 | 16 | 48 | 19 | 14 | 6 |
| qSNC4.16, qRNK4.16 | 26 | 14 | 54 | 13 | 5 | 4 |
| QTL Cluster $ | MSU Locus ID | Physical Position # | N22 Allele | Cocodrie Allele | SNP/Indel Annotation ¥ | Molecular Function |
|---|---|---|---|---|---|---|
| qAKT8.002 qCHL8.002 qSNC8.002 qRNC8.002 qRKC8.002 qSKC8.002 qSNK8.002 qRNK8.002 | LOC_Os08g01560 | 332092 | G | a 17-bp | FS | Expressed protein |
| 332260 | A | b 11-bp | FS | |||
| 332300 | T | c 14-bp | FS | |||
| 332354 | A | d 39-bp | FS | |||
| LOC_Os08g01720 | 439569 | A | G | SG | Expressed protein | |
| 439759 | G | GC | FS | |||
| qAKT9.19 qSNC9.19 qRNC9.19 qRKC9.19 qSNK9.19 qRNK9.19 | LOC_Os09g32550 | 19437532 | C | T | SA | Glucan endo-1,3-beta-glucosidase precursor, putative, expressed |
| LOC_Os09g32860 | 19591049 | T | A | SA | OsFBX335—F-box domain-containing protein, expressed | |
| 19592499 | CA | C | FS | |||
| LOC_Os09g32890 | 19609436 | A | C | SG | Expressed protein | |
| qAKT10.18 qSKC10.18 | LOC_Os10g33970 | 18106744 | C | G | SG | Double-stranded RNA-binding motif-containing protein, expressed |
| LOC_Os10g34000 | 18141437 | C | G | SA | Aquaporin protein, putative, expressed | |
| LOC_Os10g34300 | 18295111 | A | G | SG | OsFBX387—F-box domain-containing protein, expressed | |
| 18295616 | C | CGGCG | FS | |||
| 18296954 | T | e 17-bp | FS | |||
| LOC_Os10g34490 | 18402218 | G | A | SL | Phosphate translocator-related, putative, expressed | |
| 18402757 | C | CG | FS | |||
| 18403126 | GCGCTCAC | G | FS | |||
| LOC_Os10g34614 | 18452230 | C | CAA | FS | csAtPR5, putative, expressed | |
| 18453490 | A | C | SG | |||
| LOC_Os10g34960 | 18658668 | C | f 23-bp | FS | Ubiquitin family protein, putative, expressed | |
| 18659948 | TC | T | FS | |||
| LOC_Os10g34990 | 18666630 | T | TCTTC | FS | Ubiquitin-carboxyl extension, putative, expressed | |
| 18666647 | ATGCT | A | FS | |||
| 18666931 | A | G | SG | |||
| LOC_Os10g35040 | 18697585 | C | g 10-bp | FS | Receptor kinase-like protein, putative, expressed | |
| LOC_Os10g35160 | 18769659 | CTT | C | FS | Expressed protein | |
| LOC_Os10g35170 | 18774108 | A | G | SD | Semialdehyde dehydrogenase, NAD-binding domain-containing protein, putative, expressed | |
| LOC_Os10g35230 | 18823689 | C | A | SL | Rf1, mitochondrial precursor, putative, expressed | |
| LOC_Os10g35330 | 18885478 | A | G | SG | Expressed protein | |
| LOC_Os10g35640 | 19057187 | TCC | T | FS | Rf1, mitochondrial precursor, putative, expressed | |
| LOC_Os10g35940 | 19206693 | A | AGC | FS | Folylpolyglutamate synthetase, putative, expressed | |
| 19206697 | T | TGGTG | FS | |||
| LOC_Os10g36050 | 19263325 | GAC | G | FS | Hypothetical protein |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, L.; Coronejo, S.; Pruthi, R.; Chapagain, S.; Subudhi, P.K. Integration of QTL Mapping and Whole Genome Sequencing Identifies Candidate Genes for Alkalinity Tolerance in Rice (Oryza sativa). Int. J. Mol. Sci. 2022, 23, 11791. https://doi.org/10.3390/ijms231911791
Singh L, Coronejo S, Pruthi R, Chapagain S, Subudhi PK. Integration of QTL Mapping and Whole Genome Sequencing Identifies Candidate Genes for Alkalinity Tolerance in Rice (Oryza sativa). International Journal of Molecular Sciences. 2022; 23(19):11791. https://doi.org/10.3390/ijms231911791
Chicago/Turabian StyleSingh, Lovepreet, Sapphire Coronejo, Rajat Pruthi, Sandeep Chapagain, and Prasanta K. Subudhi. 2022. "Integration of QTL Mapping and Whole Genome Sequencing Identifies Candidate Genes for Alkalinity Tolerance in Rice (Oryza sativa)" International Journal of Molecular Sciences 23, no. 19: 11791. https://doi.org/10.3390/ijms231911791
APA StyleSingh, L., Coronejo, S., Pruthi, R., Chapagain, S., & Subudhi, P. K. (2022). Integration of QTL Mapping and Whole Genome Sequencing Identifies Candidate Genes for Alkalinity Tolerance in Rice (Oryza sativa). International Journal of Molecular Sciences, 23(19), 11791. https://doi.org/10.3390/ijms231911791







