Transcriptome Analysis of the Responses of Rice Leaves to Chilling and Subsequent Recovery
Abstract
1. Introduction
2. Results
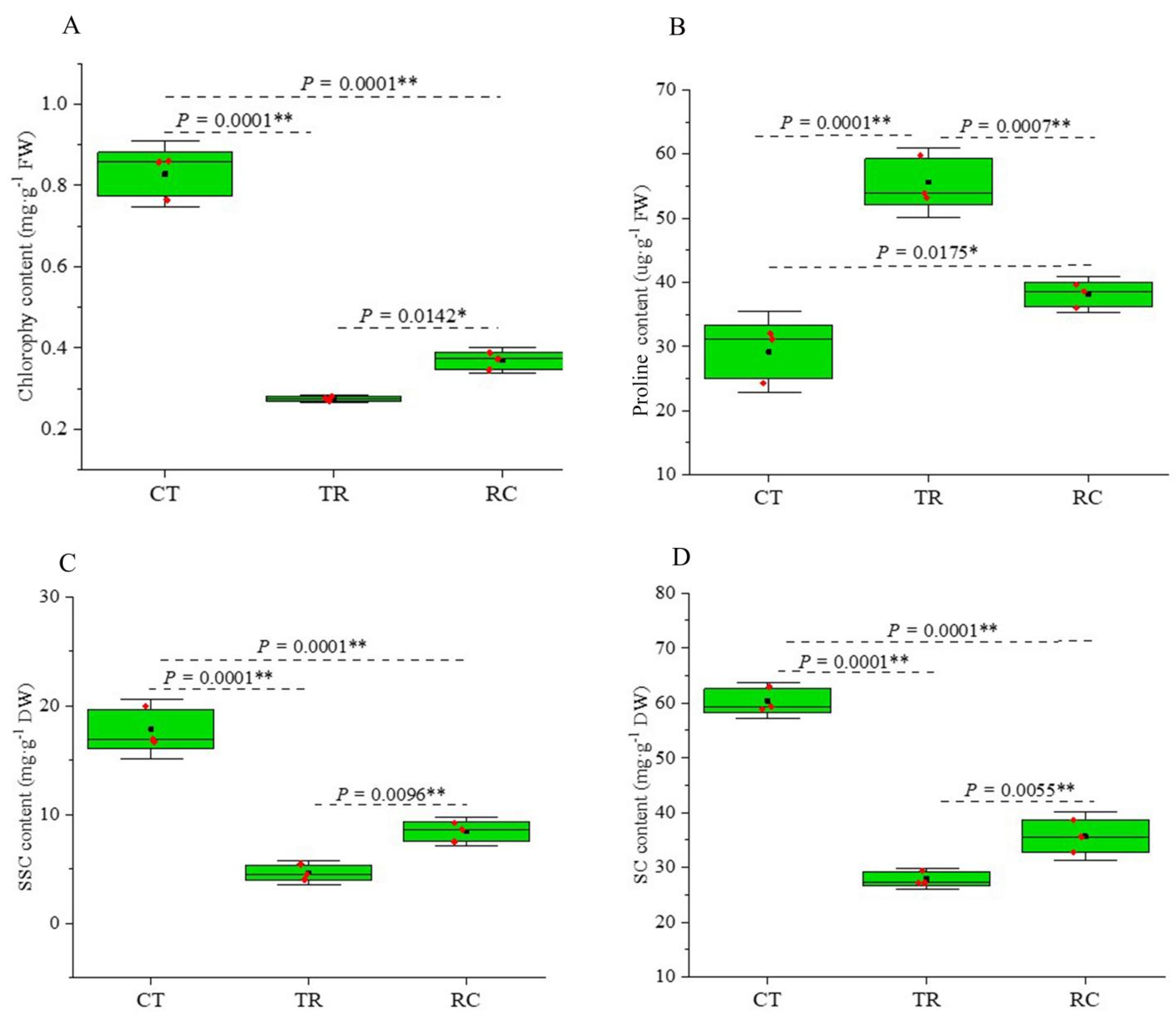
2.1. Changes in Chlorophyll and Proline and Non-Structure Carbohydrate (NSC) Contents of Rice Leaves
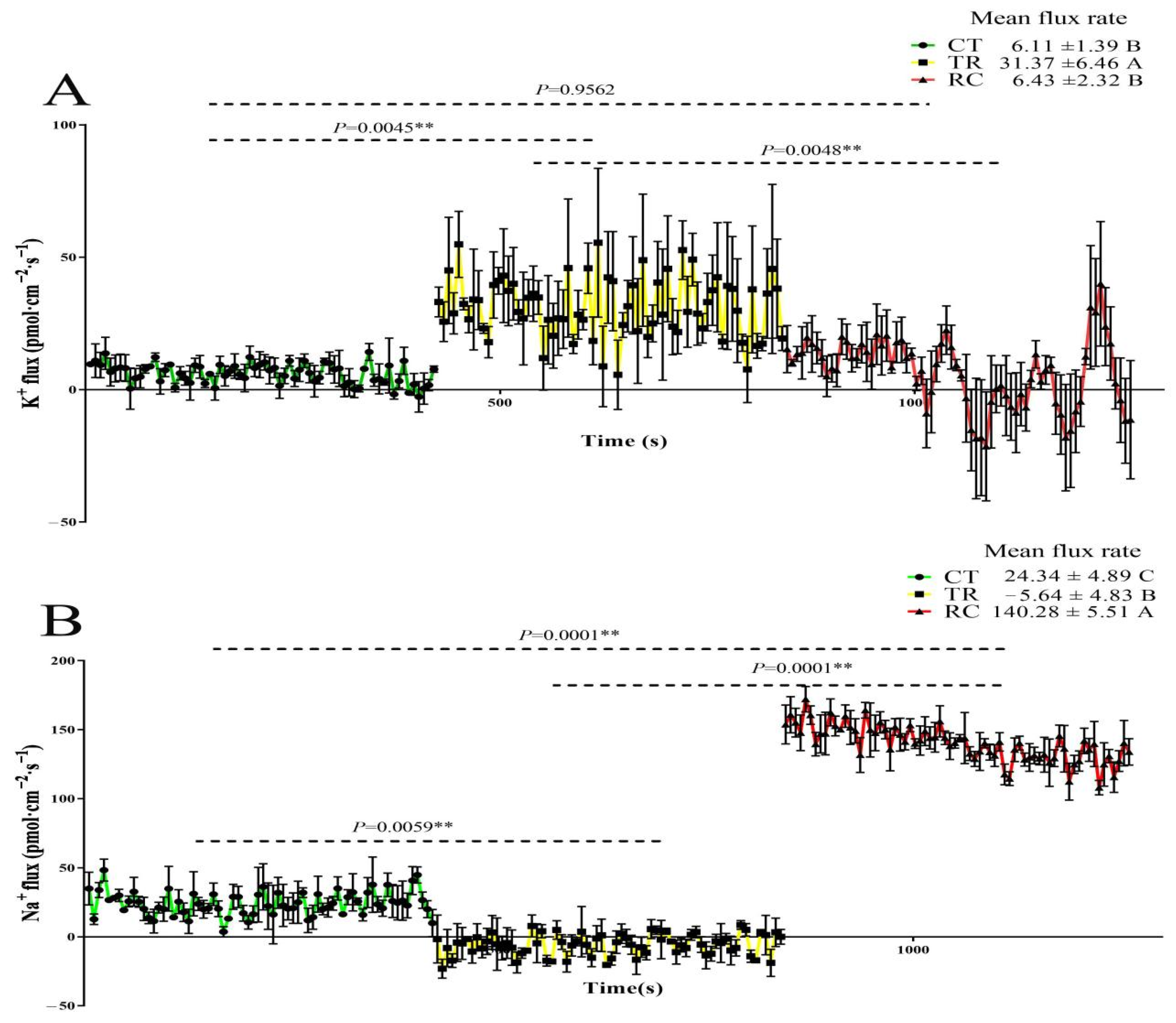
2.2. Changes in K+ and Na+ Flux Rate of Rice Roots
2.3. Gene Identification
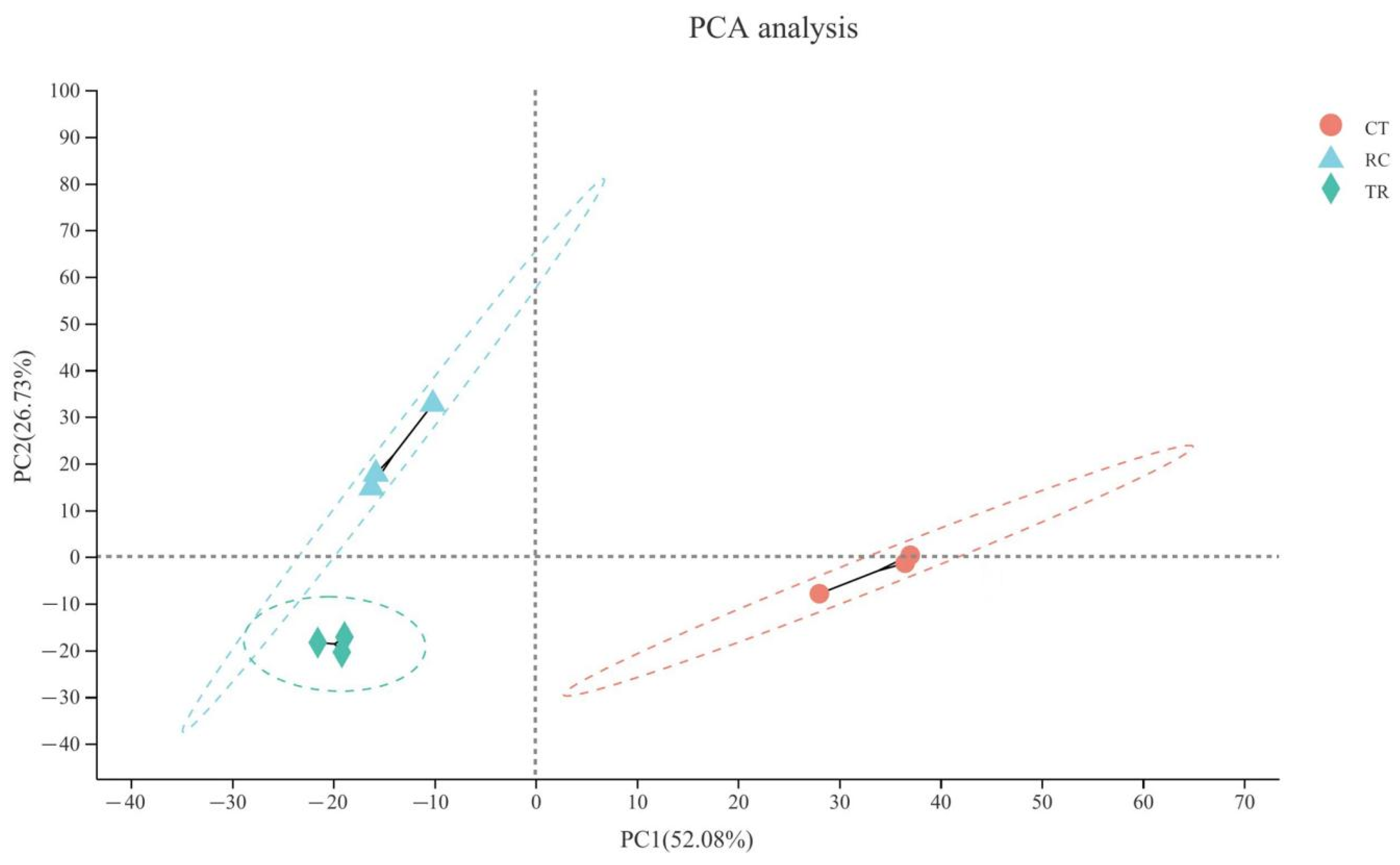
2.4. Correlation between Different Temperature Treatments in Dular Samples
2.5. Transcriptomic Response to Chilling and Normal Temperature Recovery in the Leaves of Dular
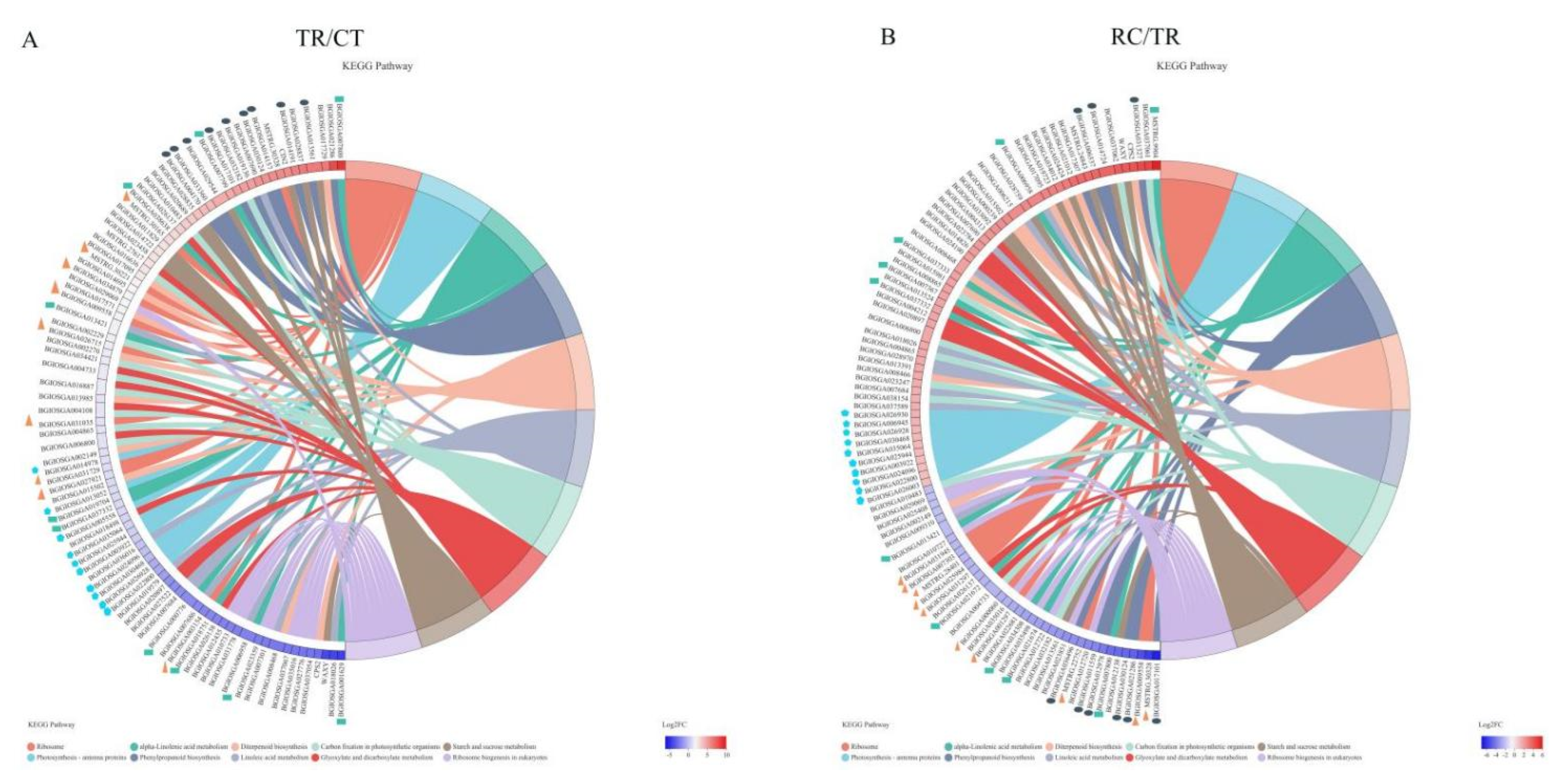
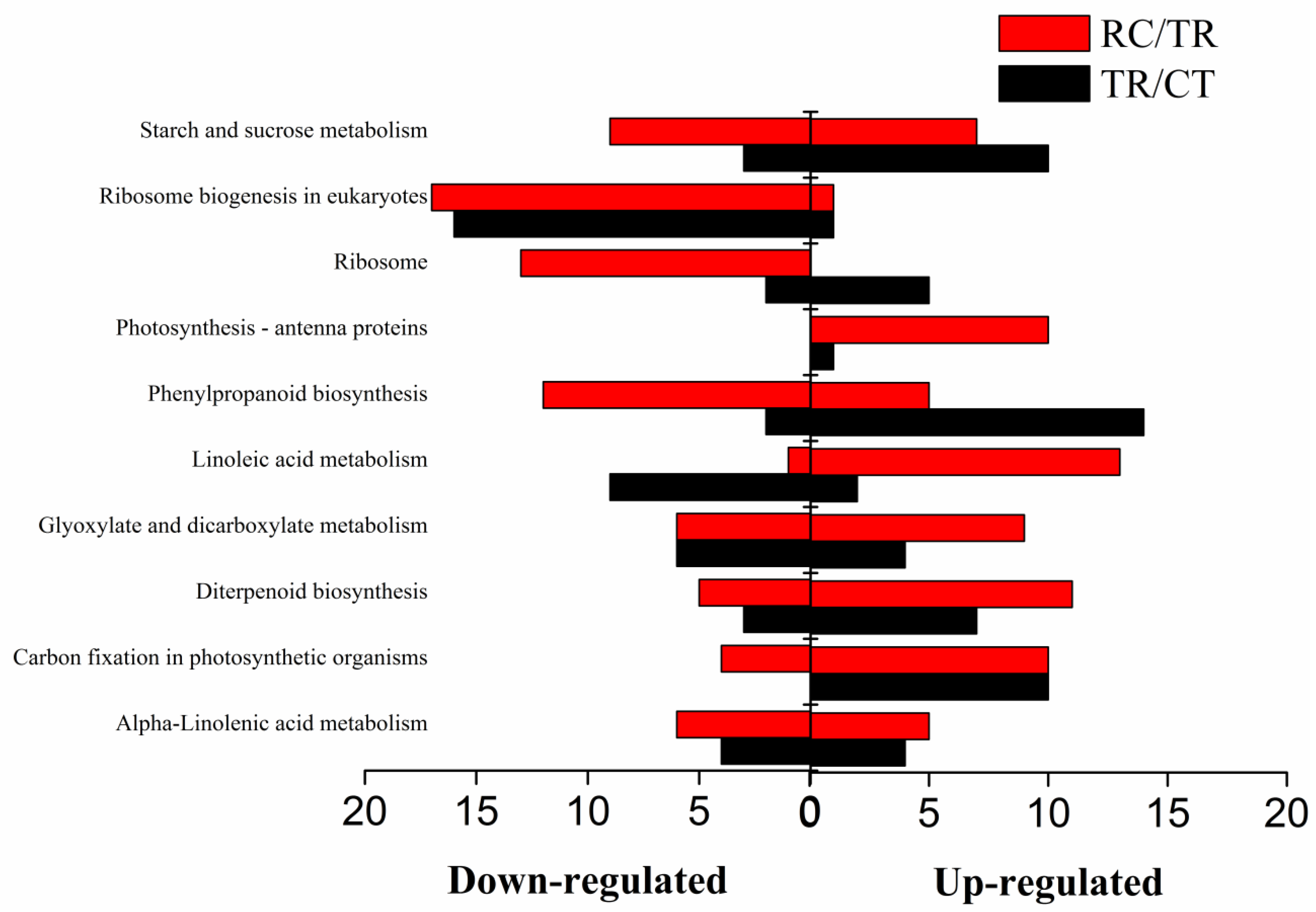
2.6. KEGG Pathway Enrichment under Chilling and Subsequent Recovery
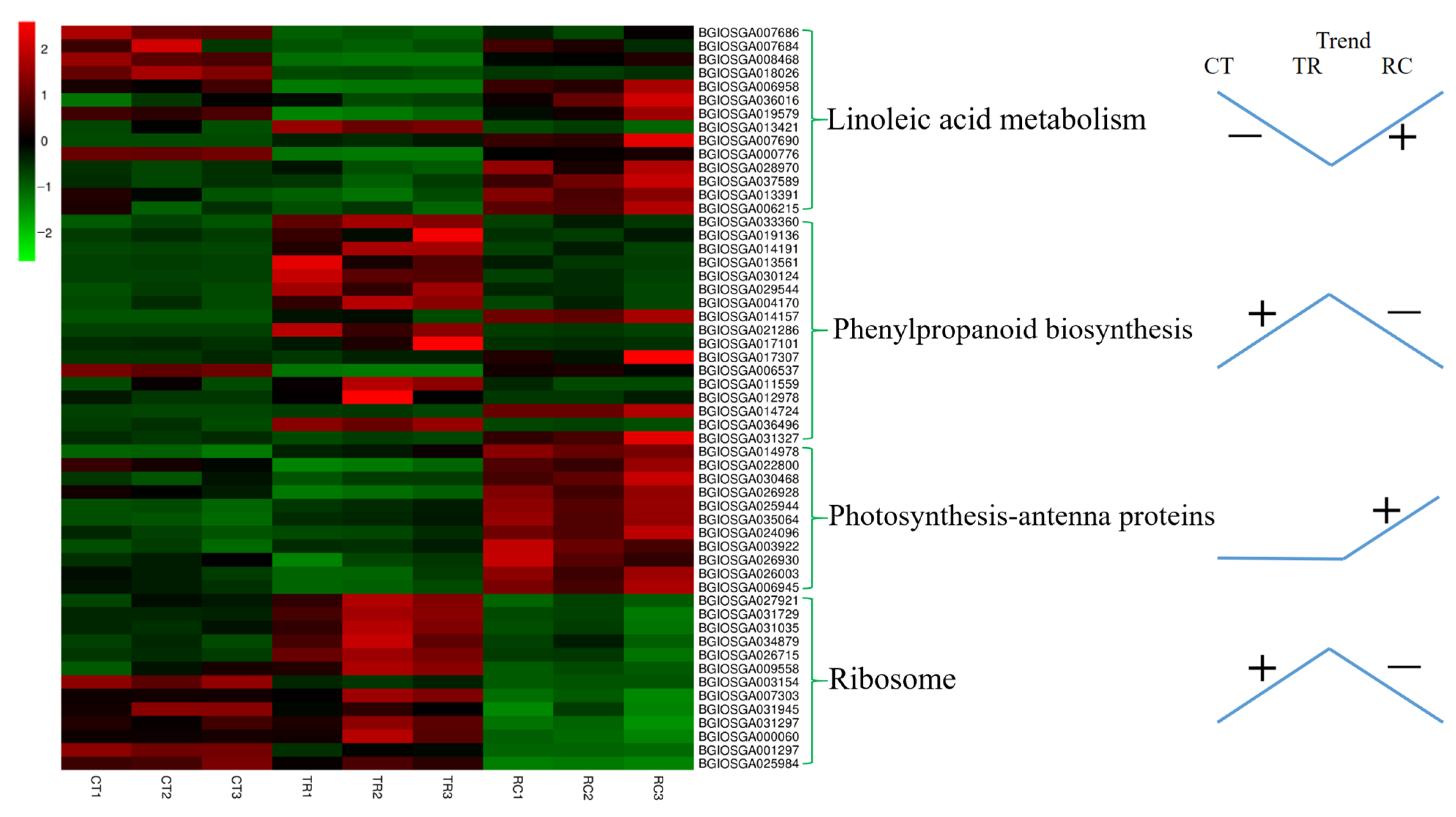
2.7. DEGs Enriched for KEGG under Chilling and Subsequent Recovery
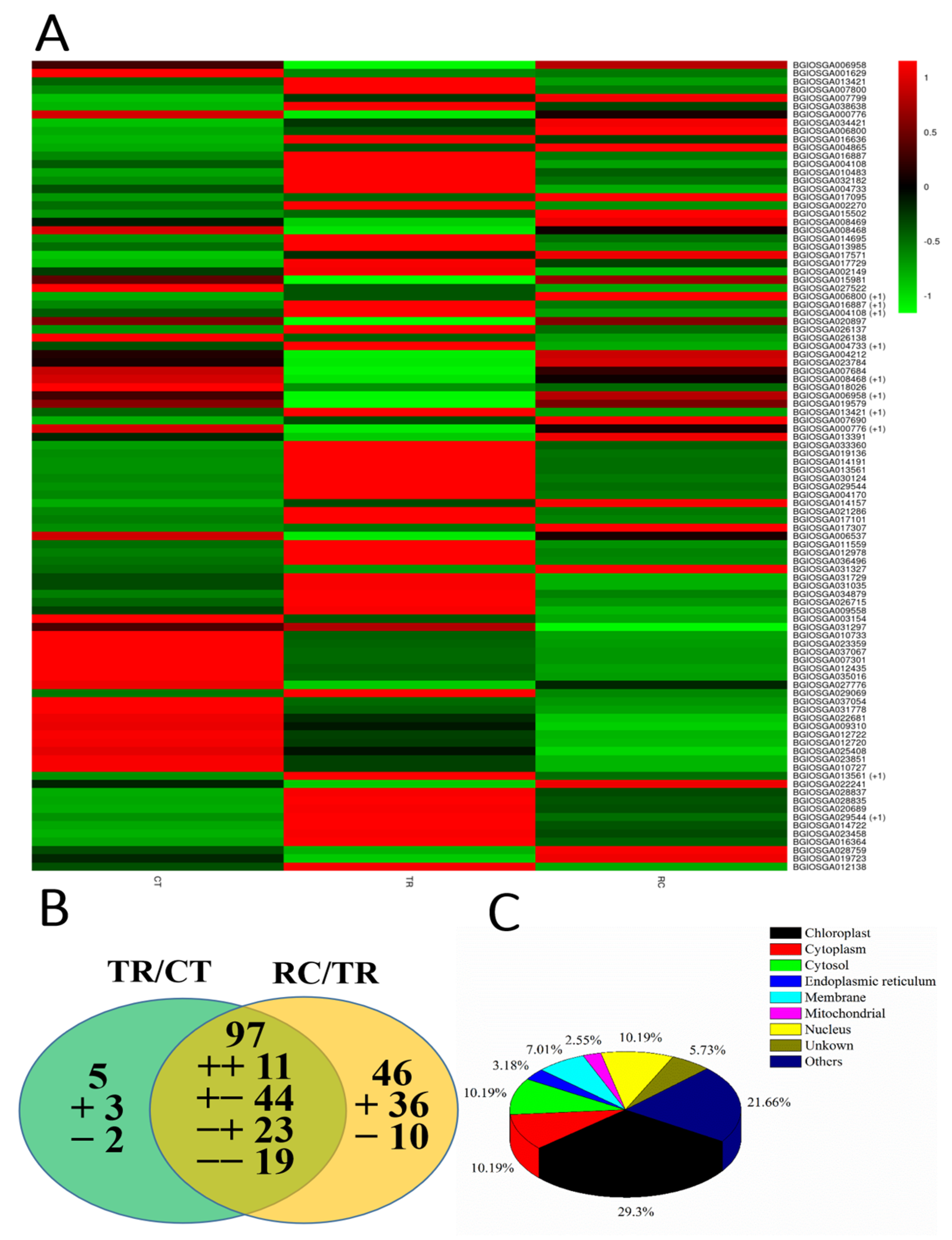
2.8. DEGs Analysis of Common Responsive Genes between TR/CT and RC/TR
2.9. Analysis of Genes Only Responsive in TR/CT or RC/TR
2.10. The Networking Analysis of Genes Responsive to Chilling and Subsequent Recovery
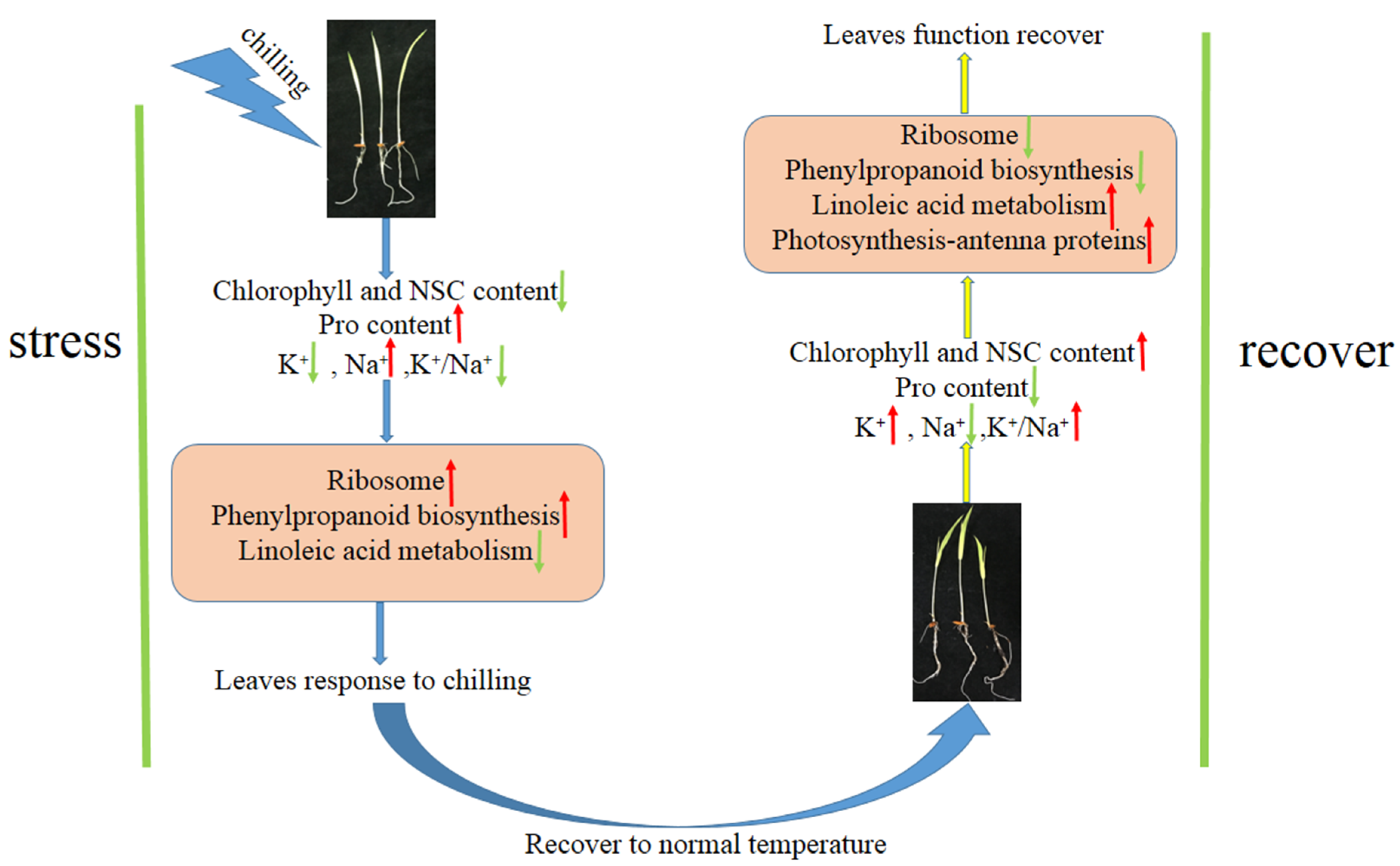
3. Discussion
3.1. Leaves Responses to Temperature Change
3.2. DEGs Involved in the Pathway of Photosynthesis
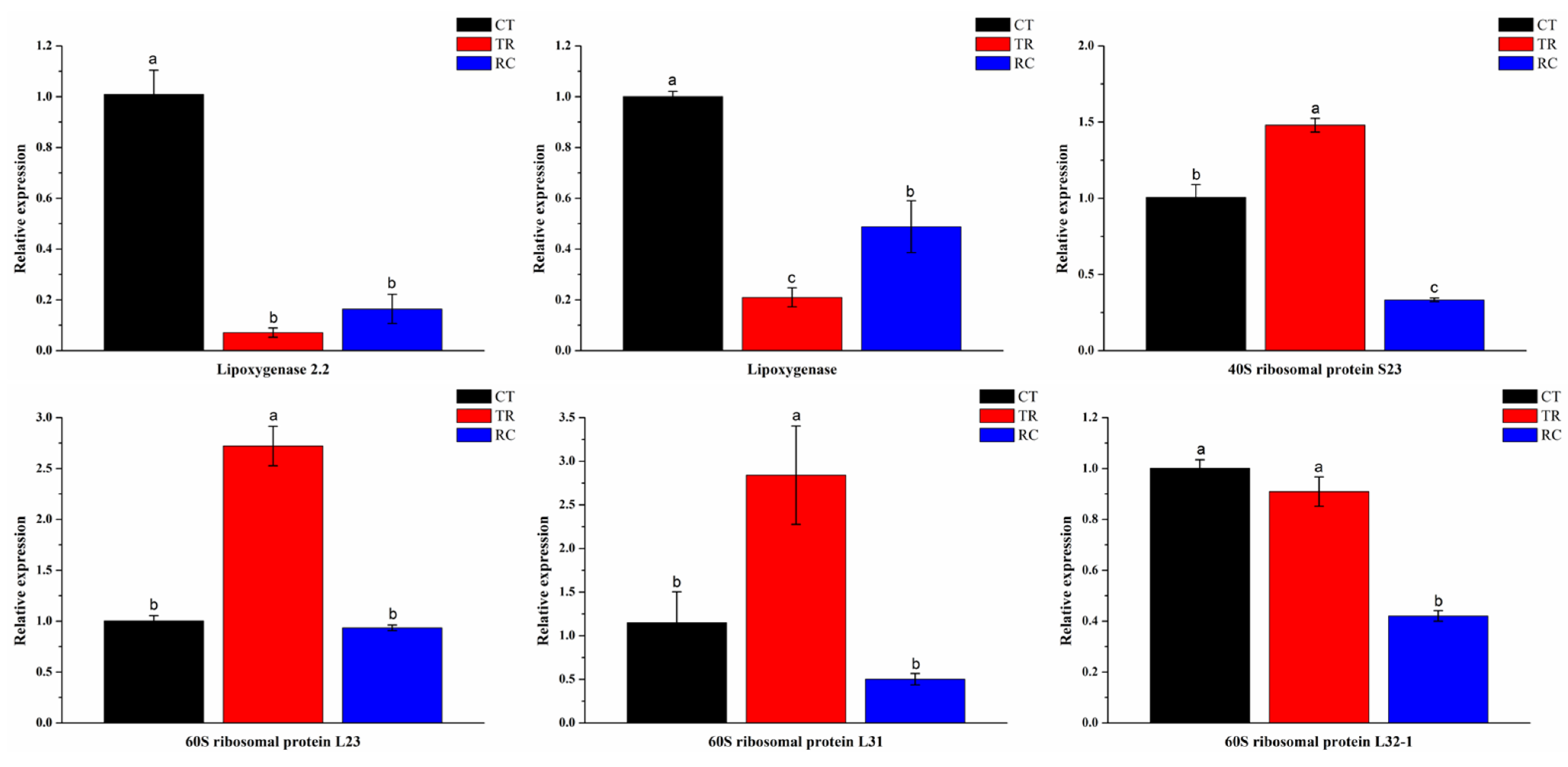
3.3. DEGs Involved in the Pathway of Linoleic Acid Metabolism
3.4. DEGs Involved in the Pathway of Phenylpropanoid Biosynthesis
3.5. DEGs Involved in the Pathway of Ribosome
4. Materials and Methods
4.1. Plant Growth and Treatment
4.2. Determination of Chlorophyll, Proline and NSC Content
4.2.1. Determination of Chlorophyll Content
4.2.2. Determination of Proline Content
4.2.3. Determination of NSC Content
4.3. The Measurement of K+ and Na+ Fluxes
4.4. RNA Extraction
4.5. Transcriptome Sequencing
4.6. Bioinformatics Analysis of Transcriptomic Data from Oryza sativa L.
4.7. qRT-PCR Validation
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, Z.; Li, J.; Li, F.; Liu, H.; Yang, W.; Chong, K.; Xu, Y. OsMAPK3 Phosphorylates OsbHLH002/OsICE1 and Inhibits Its Ubiquitination to Activate OsTPP1 and Enhances Rice Chilling Tolerance. Dev. Cell 2017, 43, 731–743. [Google Scholar] [CrossRef]
- Tartoura, K.A.H.; Youssef, S.A. Stimulation of ROS-scavenging systems in squash (Cucurbita pepo L.) plants by compost supplementation under normal and low temperature conditions. Sci. Hortic. 2011, 130, 862–868. [Google Scholar] [CrossRef]
- Taghavi, T.; Aghaani, M. Effect of chilling duration on runner production and vegetative growth of ‘Pajaro’ strawberry. Acta Hortic. 2017, 1156, 505–508. [Google Scholar] [CrossRef]
- Zhang, X.D.; Liu, T.; Zhu, S.H.; Wang, D.; Sun, S.; Xin, L. Short-term hypobaric treatment alleviates chilling injury by regulating membrane fatty acids metabolism in peach fruit. J. Food Biochem. 2022, 46, e14113. [Google Scholar] [CrossRef]
- Mao, D.; Xin, Y.; Tan, Y.; Hu, X.; Bai, J.; Liu, Z.Y.; Yu, Y.; Li, L.; Peng, C.; Fan, T.; et al. Natural variation in the HAN1 gene confers chilling tolerance in rice and allowed adaptation to a temperate climate. Proc. Natl. Acad. Sci. USA 2019, 116, 3494–3501. [Google Scholar] [CrossRef]
- Ma, Y.; Dai, X.; Xu, Y.; Luo, W.; Zheng, X.; Zeng, D.; Pan, Y.; Lin, X.; Liu, H.; Zhang, D.; et al. COLD1 confers chilling tolerance in rice. Cell 2015, 160, 1209–1921. [Google Scholar] [CrossRef]
- Ma, Q.; Dai, X.; Xu, Y.; Guo, J.; Liu, Y.; Chen, N.; Xiao, J.; Zhang, D.; Xu, Z.; Zhang, X.; et al. Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol. 2009, 150, 244–256. [Google Scholar] [CrossRef]
- Kuk, Y.; Shin, J.; Burgos, N.; Hwang, T.; Han, O.; Cho, B.; Guh, J. Antioxidative enzymes offer protection from chilling damage in rice plants. Crop Sci. 2003, 43, 2109–2117. [Google Scholar] [CrossRef]
- Browse, J.; Xin, Z. Temperature sensing and cold acclimation. Curr. Opin. Plant Biol. 2001, 4, 241–246. [Google Scholar] [CrossRef]
- Thomashow, F. Plant Cold Acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef]
- Hughes, M.A.; Alison, D.M. The molecular biology of plant acclimation to low temperature. J. Exp. Bot. 1996, 47, 291–305. [Google Scholar] [CrossRef]
- Bohnert, J. Adaptations to Environmental Stresses. Plant Cell 1995, 7, 1099–1111. [Google Scholar] [CrossRef]
- Chen, T.; Murata, N. Enhancement of tolerance of abiotic stress by metabolic engineering of betaines and other compatible solutes. Curr. Opin. Plant Biol. 2002, 5, 250–257. [Google Scholar] [CrossRef]
- Prasad, T.K.; Anderson, M.D.; Martin, B.A.; Stewart, C.R. Evidence for chilling-induced oxidative stress in maize seedlings and a regulatory role for hydrogen peroxide. Plant Cell 1994, 6, 65–74. [Google Scholar] [CrossRef]
- Zhou, A.; Liu, E.; Li, H.; Li, Y.; Feng, S.; Gong, S.; Wang, J. PsCor413pm2, a plasma membrane-localized, cold-regulated protein from phlox subulata, confers low temperature tolerance in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2579. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.-K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef]
- Singh, K.B.; Foley, R.C.; Oñate-Sánchez, L. Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 2002, 5, 430–436. [Google Scholar] [CrossRef]
- Zhang, M.; Ye, J.; Xu, Q.; Feng, Y.; Yuan, X.; Yu, H.; Wang, Y.; Wei, X.; Yang, Y. Genome-wide association study of cold tolerance of Chinese indica rice varieties at the bud burst stage. Plant Cell Rep. 2018, 37, 529–539. [Google Scholar] [CrossRef]
- Kim, C.; Vo, K.; Nguyen, C.; Jeong, D.; Lee, S.; Kumar, M.; Kim, S.; Park, S.; Kim, J.; Jeon, J. Functional analysis of a cold-responsive rice WRKY gene, OsWRKY71. Plant Biotechnol. Rep. 2016, 10, 13–23. [Google Scholar] [CrossRef]
- Su, C.; Wang, Y.; Hsieh, T.; Lu, C.; Tseng, T.; Yu, S. A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant physiol. 2010, 153, 145–158. [Google Scholar] [CrossRef]
- Yokotani, N.; Sato, Y.; Tanabe, S.; Chujo, T.; Shimizu, T.; Okada, K.; Yamane, H.; Shimono, M.; Sugano, S.; Takatsuji, H.; et al. WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J. Exp. Bot. 2013, 64, 5085–5097. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, C.; Zhu, Y.; Zhang, L.; Chen, T.; Zhou, F.; Chen, H.; Lin, Y. The calcium-dependent kinase OsCPK24 functions in cold stress responses in rice. J. Integr. Plant Biol. 2018, 60, 173–188. [Google Scholar] [CrossRef]
- Saijo, Y.; Kinoshita, N.; Ishiyama, K.; Hata, S.; Izui, K. A Ca2+-dependent protein kinase that endows rice plants with cold- and salt-stress tolerancefunctions in Vascular Bundles. Plant Cell Physiol. 2001, 42, 1228–1233. [Google Scholar] [CrossRef] [PubMed]
- Bulgari, R.; Cocetta, G.; Trivellini, A.; Vernieri, P.; Ferrante, A. Biostimulants and crop responses: A review. Biol. Agric. Hortic. 2015, 31, 1–17. [Google Scholar] [CrossRef]
- Sharma, H.; Fleming, C.; Selby, C.; Rao, J.; Martin, T. Plant biostimulants: A review on the processing of macroalgae and use of extracts for crop management to reduce abiotic and biotic stresses. J. Appl. Phycol. 2014, 26, 465–490. [Google Scholar] [CrossRef]
- Light, M.; Daws, M.; Van Staden, J. Smoke-derived butenolide: Towards understanding its biological effects. S. Afr. J. Bot. 2009, 75, 1–7. [Google Scholar] [CrossRef]
- Kulkarni, M.; Light, M.; Van Staden, J. Plant-derived smoke: Old technology with possibilities for economic applications in agriculture and horticulture. S. Afr. J. Bot. 2011, 77, 972–979. [Google Scholar] [CrossRef]
- Nardi, S.; Pizzeghello, D.; Schiavon, M.; Ertani, A. Plant biostimulants: Physiological responses induced by protein hydrolyzed-based products and humic substances in plant metabolism. Sci. Agric. 2016, 73, 18–23. [Google Scholar] [CrossRef]
- Yakhin, O.; Lubyanov, A.; Yakhin, I.; Brown, P. Biostimulants in plant science: A global perspective. Front. Plant Sci. 2017, 7, 2049. [Google Scholar] [CrossRef]
- Kannan, R.R.; Aderogba, M.A.; Ndhlala, A.R.; Stirk, W.A.; Van Staden, J. Acetylcholinesterase inhibitory activity of phlorotannins isolated from the brown alga, Ecklonia maxima (Osbeck) Papenfuss. Food Res. Int. 2013, 54, 1250–1254. [Google Scholar] [CrossRef]
- Mi, J.; Li, G.; Huang, J.; Yu, H.; Zhou, F.; Zhang, Q.; Ouyang, Y.; Mou, T. Stacking S5-n and f5-n to overcome sterility in indica–japonica hybrid rice. Theor. Appl. Genet. 2016, 129, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, K.; Xu, C.; Li, H.; Zhang, Q. The high level of wide-compatibility of variety ‘Dular’ has a complex genetic basis. Theor. Appl. Genet. 1998, 97, 407–412. [Google Scholar] [CrossRef]
- Azeem, S.; Li, Z.; Zheng, H.; Lin, W.; Arafat, Y.; Zhang, Z.; Lin, X.; Lin, W. Quantitative proteomics study onLsi1in regulation of rice (Oryza sativa L.) cold resistance. Plant Growth Regul. 2016, 78, 307–323. [Google Scholar] [CrossRef]
- Li, Z.; Feng, S.; Zhan, W.; Xu, L.; Fang, C.; Zhang, Z.; Lin, W. Lsi1 plays an active role in enhancing the chilling tolerance of rice roots. Plant Growth Regul. 2020, 90, 529–543. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Pan, Y.; Li, J.; Zhou, L.; Shi, H.; Zeng, Y.; Guo, H.; Yang, S.; Zheng, W. Natural variation in CTB4a enhances rice adaptation to cold habitats. Nat. Commun. 2017, 8, 14788. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, S.; Yang, T.; Zeng, Z.; Huang, Z.; Liu, Q.; Wang, X.; Leach, J.; Leung, H.; Liu, B. Global transcriptional profiling of a cold-tolerant rice variety under moderate cold stress reveals different cold stress response mechanisms. Physiol. Plant. 2015, 154, 381–394. [Google Scholar] [CrossRef]
- Shimono, H.; Hasegawa, T.; Fujimura, S.; Iwama, K. Responses of leaf photosynthesis and plant water status in rice to low water temperature at different growth stages. Field Crop. Res. 2004, 89, 71–83. [Google Scholar] [CrossRef]
- Ariizumi, T.; Kishitani, S.; Inatsugi, R.; Nishida, I.; Murata, N.; Toriyama, K. An increase in unsaturation of fatty acids in phosphatidylglycerol from leaves improves the rates of photosynthesis and growth at low temperatures in transgenic rice seedlings. Plant Cell Physiol. 2002, 43, 751–758. [Google Scholar] [CrossRef]
- Yahmed, J.; Oliveira, T.; Novillo, P.; Quinones, A.; Forner, M.; Salvador, A.; Froelicher, Y.; Mimoun, M.; Talon, M.; Ollitrault, P. A simple, fast and inexpensive method to assess salt stress tolerance of aerial plant part: Investigations in the mandarin group. J. Plant Physiol. 2016, 190, 36–43. [Google Scholar] [CrossRef]
- Jeong, S.; Choi, S.; Lee, D.; Ahn, S.; Hur, Y.; Chow, W.; Park, Y. Differential susceptibility of photosynthesis to light-chilling stress in rice (Oryza sativa L.) depends on the capacity for photochemical dissipation of light. Mol. Cells 2002, 13, 419–428. [Google Scholar] [PubMed]
- Zhang, K.; Tian, G.; Li, X.; Zhang, Z.; Liu, J.; Li, Y.; Xie, J.; Wang, P. ROS produced via BsRBOHD plays an important role in low temperature-induced anthocyanin biosynthesis in begonia semperflorens. Russ. J. Plant Physiol. 2020, 67, 250–258. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Wang, L.; Xu, Y.; Chen, N.; Ma, Q.; Li, F.; Chong, K. Overexpression of OsCOIN, a putative cold inducible zinc finger protein, increased tolerance to chilling, salt and drought, and enhanced proline level in rice. Planta 2007, 226, 1007–1101. [Google Scholar] [CrossRef] [PubMed]
- Trotel-Aziz, P.; Niogret, M.F.; Deleu, C.; Bouchereau, A.; Aziz, A.; Larher, F.R. The control of proline consumption by abscisic acid during osmotic stress recovery of canola leaf discs. Physiol. Plant. 2003, 117, 213–221. [Google Scholar] [CrossRef]
- Sun, J.; Li, L.; Liu, M.; Wang, M.; Ding, M.; Deng, S.; Lu, C.; Zhou, X.; Shen, X.; Zheng, X. Hydrogen peroxide and nitric oxide mediate K+/Na+ homeostasis and antioxidant defense in NaCl-stressed callus cells of two contrasting poplars. Plant Cell Tissue Organ Cult. 2010, 103, 205–215. [Google Scholar] [CrossRef]
- Sun, J.; Wang, M.; Ding, M.; Deng, S.; Liu, M.; Lu, C.; Zhou, X.; Shen, X.; Zheng, X.; Zhang, Z. H2O2 and cytosolic Ca2+ signals triggered by the PM H+-coupled transport system mediate K+/Na+ homeostasis in NaCl-stressed Populus euphratica cells. Plant Cell Environ. 2010, 33, 943–958. [Google Scholar] [CrossRef]
- Tyerman, S.; Skerrett, I. Root ion channels and salinity. Sci. Hortic. 1998, 78, 175–235. [Google Scholar] [CrossRef]
- Cuin, T.; Betts, S.; Chalmandrier, R.; Shabala, S. A root’s ability to retain K+ correlates with salt tolerance in wheat. J. Exp. Bot. 2008, 59, 2697–2706. [Google Scholar] [CrossRef]
- Green, B.; Pichersky, E.; Kloppstech, K. Chlorophyll a/b-binding proteins: An extended family. Trends Biochem. Sci. 1991, 16, 181–186. [Google Scholar] [CrossRef]
- Ruban, A.V. Nonphotochemical chlorophyll fluorescence quenching: Mechanism and effectiveness in protecting plants from photodamage. Plant Physiol. 2016, 170, 1903–1916. [Google Scholar] [CrossRef] [PubMed]
- Belgio, E.; Kapitonova, E.; Chmeliov, J.; Duffy, C.; Ungerer, P.; Valkunas, L.; Ruban, A. Economic photoprotection in photosystem II that retains a complete light-harvesting system with slow energy traps. Nat. Commun. 2014, 5, 4433. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Tan, Q.; Nie, Z.; Hu, C.; An, Y. Differential expression of proteins in response to molybdenum deficiency in winter wheatleaves under low-temperature stress. Plant Mol. Biol. Rep. 2014, 32, 1057–1069. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, H.; Huang, R. Expression analysis of low temperature responsive genes in eupatorium adenophorum spreng using cDNA-AFLP. Plant Mol. Biol. Rep. 2007, 25, 37–44. [Google Scholar] [CrossRef]
- Whelan, J.; Fritsche, K. Linoleic acid. Adv. Nutr. 2013, 4, 311–312. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wu, D.; Shi, J.; He, Y.; Pinot, F.; Grausem, B.; Yin, C.; Zhu, L.; Chen, M.; Luo, Z. Rice CYP703A3, a cytochrome P450 hydroxylase, is essential for development of anther cuticle and pollen exine. J. Integr. Plant Biol. 2014, 56, 979–994. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Pinot, F.; Sauveplane, V.; Werck-Reichhart, D.; Diehl, P.; Schreiber, L.; Franke, R.; Zhang, P.; Chen, L.; Gao, Y. Cytochrome P450 family member CYP704B2 catalyzes the ω-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 2010, 22, 173–190. [Google Scholar] [CrossRef]
- Wang, Z.; Benning, C. Chloroplast lipid synthesis and lipid trafficking through ER–plastid membrane contact sites. Biochem. Soc. Trans. 2012, 40, 457–463. [Google Scholar] [CrossRef]
- Yang, Y.; Benning, C. Functions of triacylglycerols during plant development and stress. Curr. Opin. Biotechnol. 2018, 49, 191–198. [Google Scholar] [CrossRef]
- Penfield, S.; Pinfield-Wells, H.; Graham, I. Storage reserve mobilisation and seedling establishment in Arabidopsis. Arab. Book/Am. Soc. Plant Biol. 2006, 4, e0100. [Google Scholar] [CrossRef][Green Version]
- Liu, H.; Wang, Z.G.; Xu, W.H.; Zeng, J.; Li, L.X.; Li, S.L.; Gao, Z. Bacillus pumilus LZP02 promotes rice root growth by improving carbohydrate metabolism and phenylpropanoid biosynthesis. Mol. Plant-Microbe Interact. 2020, 33, 1222–1231. [Google Scholar] [CrossRef] [PubMed]
- Rattanawong, K.; Koiso, N.; Toda, E.; Kinoshita, A.; Tanaka, M.; Tsuji, H.; Okamoto, T. Regulatory functions of ROS dynamics via glutathione metabolism and glutathione peroxidase activity in developing rice zygote. Plant J. 2021, 108, 1097–1115. [Google Scholar] [CrossRef] [PubMed]
- Susumu, H.; Katsutomo, S.; Hiroyuki, I.; Yuko, O.; Hirokazu, M. A large family of class III plant peroxidases. Plant Cell Physiol. 2001, 42, 462–468. [Google Scholar]
- Almagro, L.; Gómez, R.; Belchi-Navarro, S.; Bru, R.; Ros, B.; Pedreño, M. Class III peroxidases in plant defence reactions. J. Exp. Bot. 2009, 60, 377–390. [Google Scholar] [CrossRef]
- Ma, Y.; Ren, X.; Liang, C. Exogenous Ca2+ enhances antioxidant defense in rice to simulated acid rain by regulating ascorbate peroxidase and glutathione reductase. Planta 2021, 254, 41. [Google Scholar] [CrossRef]
- Saloheimo, M.; Kuja-Panula, J.; Ylosmaki, E.; Ward, M.; Penttila, M. Enzymatic Properties and Intracellular Localization of the Novel Trichoderma reesei β-Glucosidase BGLII (Cel1A). Appl. Environ. Microbiol. 2002, 68, 4546–4553. [Google Scholar] [CrossRef]
- Lee, L. Enrichment of bioactive isoflavones in soymilk fermented with β-glucosidase-producing lactic acid bacteria. Food Res. Int. 2005, 38, 551–559. [Google Scholar]
- Hang, R.; Wang, Z.; Deng, X.; Liu, C.; Yan, B.; Yang, C.; Song, X.; Mo, B.; Cao, X. Ribosomal RNA biogenesis and its response to chilling stress in Oryza sativa. Plant Physiol. 2018, 177, 381–397. [Google Scholar] [CrossRef]
- Moin, M.; Saha, A.; Bakshi, A.; Madhav, M.S.; Kirti, P.B. Constitutive expression of Ribosomal Protein L6 modulates salt tolerance in rice transgenic plants. Gene 2021, 789, 145670. [Google Scholar] [CrossRef]
- Barakat, A.; Szick-Miranda, K.; Chang, I.-F.; Guyot, R.; Blanc, G.; Cooke, R.; Delseny, M.; Bailey-Serres, J. The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol. 2001, 127, 398–415. [Google Scholar] [CrossRef]
- Uzair, M.; Long, H.; Zafar, S.A.; Patil, S.B.; Chun, Y.; Li, L.; Fang, J.; Zhao, J.; Peng, L.; Yuan, S. Narrow leaf21, encoding ribosomal protein RPS3A, controls leaf development in rice. Plant Physiol. 2021, 186, 497–518. [Google Scholar] [CrossRef]
- Zhou, K.; Zhang, C.; Xia, J.; Yun, P.; Wang, Y.; Ma, T.; Li, Z. Albino seedling lethality 4; Chloroplast 30S Ri-bosomal protein S1 is required for chloroplast ribosome biogenesis and early chloroplast development in rice. Rice 2021, 14, 47. [Google Scholar] [CrossRef] [PubMed]
- Mazahar, M.; Anusree, S.; Achala, B.; Divya, D.; Madhav, M.S.; Kirti, P.B. Study on transcriptional responses and identification of Ri-bosomal protein genes for potential resistance against brown planthopper and gall midge pests in rice. Curr. Genom. 2021, 22, 98–110. [Google Scholar]
- Lin, J.; Lai, S.; Jia, R.; Xu, A.; Zhang, L.; Lu, J.; Ye, K. Structural basis for site-specific ribose methylation by box C/D RNA protein complexes. Nature 2011, 469, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Woolford, J.; Baserga, S. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 2013, 195, 643–681. [Google Scholar] [CrossRef]
- Wang, X. Principles and Techniques of Plant Physiological and Biochemical Experiments; Higher Education Press: Beijing, China, 2006. [Google Scholar]
- Zhao, R.; Sun, H.; Zhao, N.; Jing, X.; Shen, X.; Chen, S. The Arabidopsis Ca2+-dependent protein kinase CPK27 is required for plant response to salt-stress. Gene 2015, 563, 203–214. [Google Scholar] [CrossRef]
- Li, Q.; Li, B.; Kronzucker, H.; Shi, W. Root growth inhibition by NH4+ in Arabidopsis is mediated by the root tip and is linked to NH4+ efflux and GMPase activity. Plant Cell Environ. 2010, 33, 1529–1542. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Khan, M.U.; Letuma, P.; Xie, Y.; Zhan, W.; Wang, W.; Jiang, Y.; Lin, W.; Zhang, Z. Transcriptome Analysis of the Responses of Rice Leaves to Chilling and Subsequent Recovery. Int. J. Mol. Sci. 2022, 23, 10739. https://doi.org/10.3390/ijms231810739
Li Z, Khan MU, Letuma P, Xie Y, Zhan W, Wang W, Jiang Y, Lin W, Zhang Z. Transcriptome Analysis of the Responses of Rice Leaves to Chilling and Subsequent Recovery. International Journal of Molecular Sciences. 2022; 23(18):10739. https://doi.org/10.3390/ijms231810739
Chicago/Turabian StyleLi, Zhong, Muhammad Umar Khan, Puleng Letuma, Yuebin Xie, Wenshan Zhan, Wei Wang, Yuhang Jiang, Wenxiong Lin, and Zhixing Zhang. 2022. "Transcriptome Analysis of the Responses of Rice Leaves to Chilling and Subsequent Recovery" International Journal of Molecular Sciences 23, no. 18: 10739. https://doi.org/10.3390/ijms231810739
APA StyleLi, Z., Khan, M. U., Letuma, P., Xie, Y., Zhan, W., Wang, W., Jiang, Y., Lin, W., & Zhang, Z. (2022). Transcriptome Analysis of the Responses of Rice Leaves to Chilling and Subsequent Recovery. International Journal of Molecular Sciences, 23(18), 10739. https://doi.org/10.3390/ijms231810739





