Antisense Transcription in Plants: A Systematic Review and an Update on cis-NATs of Sugarcane
Abstract
1. Introduction
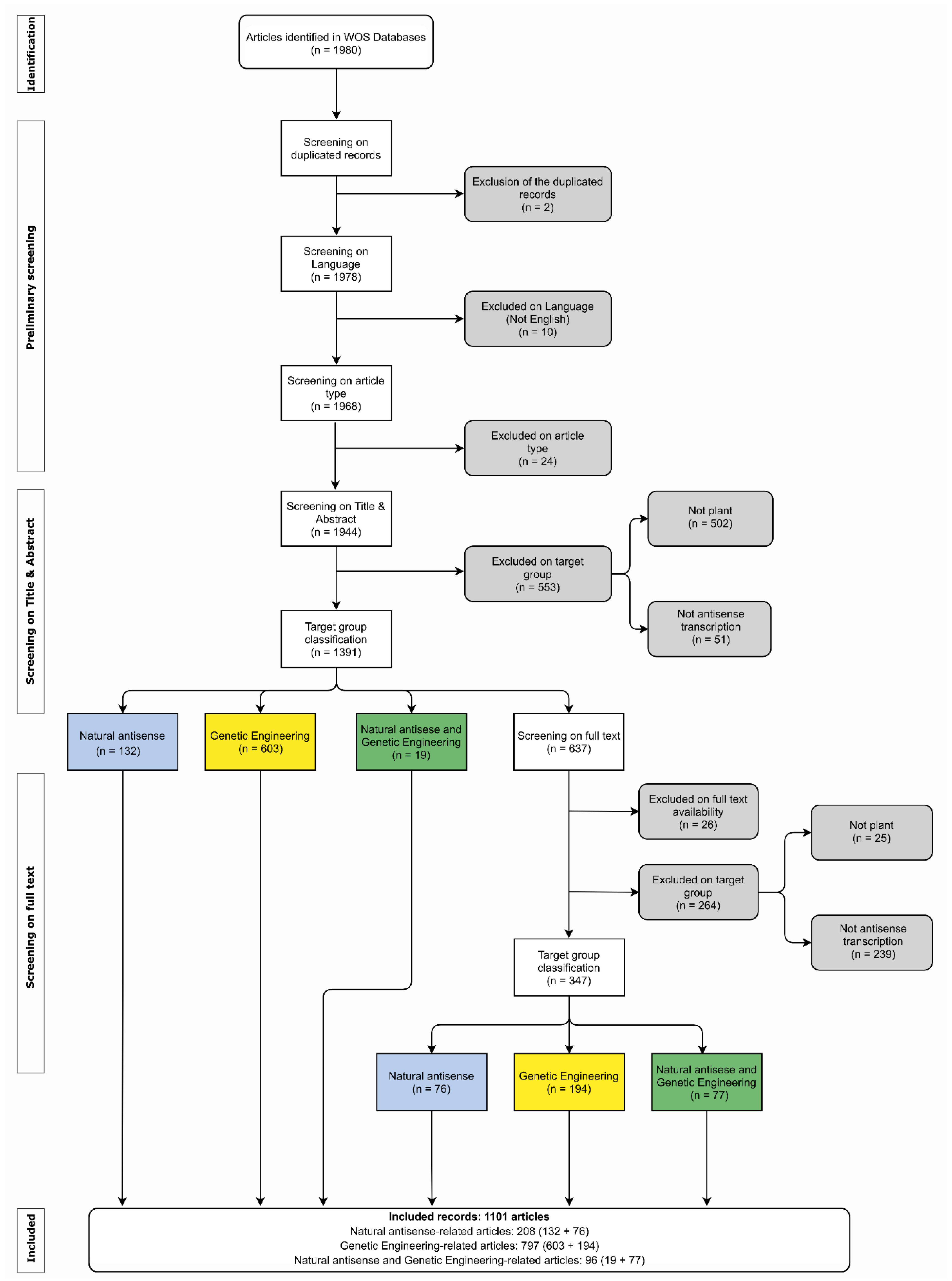
2. Methods
2.1. Plants asRNA Systematic Review
2.2. Sugarcane asRNA
3. Results and Discussion
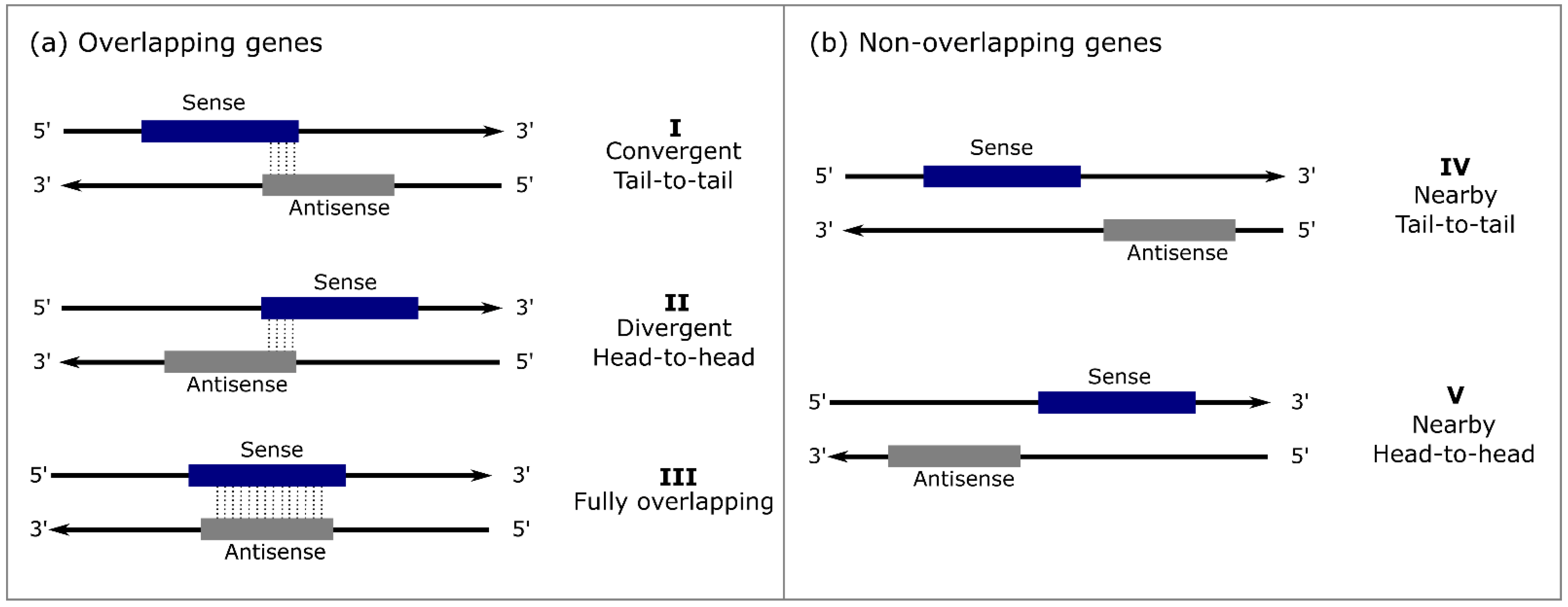
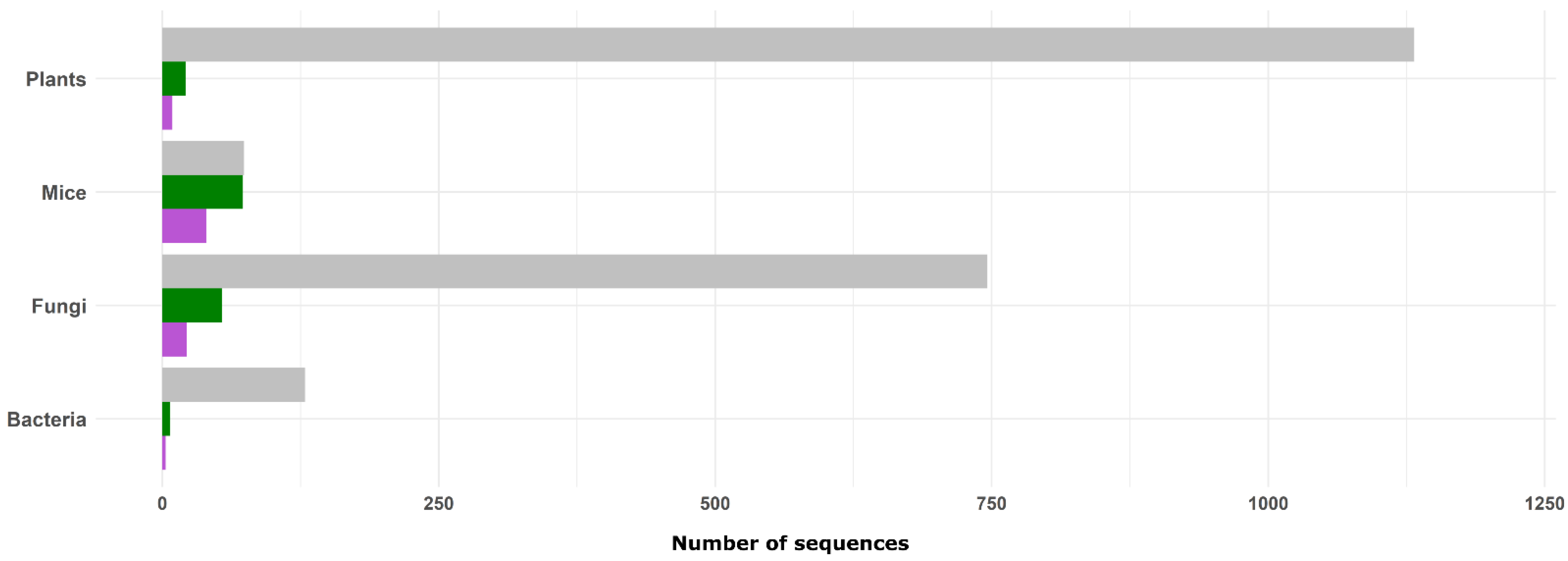
3.1. Plants as RNA Systematic Review
3.2. Sugarcane asRNA
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lee, H.; Zhang, Z.; Krause, H.M. Long Noncoding RNAs and Repetitive Elements: Junk or Intimate Evolutionary Partners? Trends Genet. 2019, 35, 892–902. [Google Scholar] [CrossRef]
- Fonouni-Farde, C.; Ariel, F.; Crespi, M. Plant Long Noncoding RNAs: New Players in the Field of Post-Transcriptional Regulations. Non-Coding RNA 2021, 7, 12. [Google Scholar] [CrossRef]
- Palazzo, A.F.; Koonin, E.V. Functional Long Non-Coding RNAs Evolve from Junk Transcripts. Cell 2020, 183, 1151–1161. [Google Scholar] [CrossRef]
- Weigel, D.; Mott, R. The 1001 Genomes Project for Arabidopsis thaliana. Genome Biol. 2009, 10, 107. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, E.J.; Matasci, N.; Ayyampalayam, S.; Wu, S.; Sun, J.; Yu, J.; Jimenez Vieira, F.R.; Bowler, C.; Dorrell, R.G.; Gitzendanner, M.A.; et al. Access to RNA-Sequencing Data from 1,173 Plant Species: The 1000 Plant Transcriptomes Initiative (1KP). GigaScience 2019, 8, giz126. [Google Scholar] [CrossRef] [PubMed]
- One Thousand Plant Transcriptomes Initiative. One Thousand Plant Transcriptomes and the Phylogenomics of Green Plants. Nature 2019, 574, 679–685. [Google Scholar] [CrossRef] [PubMed]
- NCBI Resource Coordinators; Agarwala, R.; Barrett, T.; Beck, J.; Benson, D.A.; Bollin, C.; Bolton, E.; Bourexis, D.; Brister, J.R.; Bryant, S.H.; et al. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018, 46, D8–D13. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A Comparative Platform for Green Plant Genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Berardini, T.Z.; Reiser, L.; Li, D.; Mezheritsky, Y.; Muller, R.; Strait, E.; Huala, E. The Arabidopsis Information Resource: Making and Mining the “Gold Standard” Annotated Reference Plant Genome: Tair: Making and Mining the “Gold Standard” Plant Genome. Genesis 2015, 53, 474–485. [Google Scholar] [CrossRef] [PubMed]
- Jabnoune, M.; Secco, D.; Lecampion, C.; Robaglia, C.; Shu, Q.; Poirier, Y. A Rice Cis-Natural Antisense RNA Acts as a Translational Enhancer for Its Cognate MRNA and Contributes to Phosphate Homeostasis and Plant Fitness. Plant Cell 2013, 25, 4166–4182. [Google Scholar] [CrossRef] [PubMed]
- Reis, R.S.; Deforges, J.; Schmidt, R.R.; Schippers, J.H.M.; Poirier, Y. An Antisense Noncoding RNA Enhances Translation via Localized Structural Rearrangements of Its Cognate MRNA. Plant Cell 2021, 33, 1381–1397. [Google Scholar] [CrossRef]
- Wan, Q.; Guan, X.; Yang, N.; Wu, H.; Pan, M.; Liu, B.; Fang, L.; Yang, S.; Hu, Y.; Ye, W.; et al. Small Interfering RNAs from Bidirectional Transcripts of GhMML3_A12 Regulate Cotton Fiber Development. New Phytol. 2016, 210, 1298–1310. [Google Scholar] [CrossRef] [PubMed]
- Reis, R.S.; Poirier, Y. Making Sense of the Natural Antisense Transcript Puzzle. Trends Plant Sci. 2021, 26, 1104–1115. [Google Scholar] [CrossRef]
- Boi, S.; Solda, G.; Tenchini, M. Shedding Light on the Dark Side of the Genome: Overlapping Genes in Higher Eukaryotes. Curr. Genom. 2004, 5, 509–524. [Google Scholar] [CrossRef]
- Jen, C.H.; Michalopoulos, I.; Westhead, D.R.; Meyer, P. Natural Antisense Transcripts with Coding Capacity in Arabidopsis May Have a Regulatory Role That Is Not Linked to Double-Stranded RNA Degradation. Genome Biol. 2005, 6, R51. [Google Scholar] [CrossRef]
- Wang, X. Genome-Wide Identification of R Genes and Exploitation of Candidate RGA Markers in Rice. Chin. Sci. Bull. 2005, 50, 1120. [Google Scholar] [CrossRef]
- Osato, N.; Suzuki, Y.; Ikeo, K.; Gojobori, T. Transcriptional Interferences in Cis Natural Antisense Transcripts of Humans and Mice. Genetics 2007, 176, 1299–1306. [Google Scholar] [CrossRef]
- Pelechano, V.; Steinmetz, L.M. Gene Regulation by Antisense Transcription. Nat. Rev. Genet. 2013, 14, 880–893. [Google Scholar] [CrossRef] [PubMed]
- Deforges, J.; Reis, R.S.; Jacquet, P.; Sheppard, S.; Gadekar, V.P.; Hart-Smith, G.; Tanzer, A.; Hofacker, I.L.; Iseli, C.; Xenarios, I.; et al. Control of Cognate Sense MRNA Translation by Cis-Natural Antisense RNAs. Plant Physiol. 2019, 180, 305–322. [Google Scholar] [CrossRef]
- Takayama, K.M.; Inouye, M. ANTISENSE RNA. Crit. Rev. Biochem. Mol. Biol. 1990, 25, 155–184. [Google Scholar] [CrossRef]
- Zhao, X.; Li, J.; Lian, B.; Gu, H.; Li, Y.; Qi, Y. Global Identification of Arabidopsis LncRNAs Reveals the Regulation of MAF4 by a Natural Antisense RNA. Nat. Commun. 2018, 9, 5056. [Google Scholar] [CrossRef] [PubMed]
- Lucero, L.; Fonouni-Farde, C.; Crespi, M.; Ariel, F. Long Noncoding RNAs Shape Transcription in Plants. Transcription 2020, 11, 160–171. [Google Scholar] [CrossRef] [PubMed]
- Csorba, T.; Questa, J.I.; Sun, Q.; Dean, C. Antisense COOLAIR Mediates the Coordinated Switching of Chromatin States at FLC during Vernalization. Proc. Natl. Acad. Sci. USA 2014, 111, 16160–16165. [Google Scholar] [CrossRef]
- Kindgren, P.; Ard, R.; Ivanov, M.; Marquardt, S. Transcriptional Read-through of the Long Non-Coding RNA SVALKA Governs Plant Cold Acclimation. Nat. Commun. 2018, 9, 4561. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Luo, X.; Sun, F.; Hu, J.; Zha, X.; Su, W.; Yang, J. Overexpressing LncRNA LAIR Increases Grain Yield and Regulates Neighbouring Gene Cluster Expression in Rice. Nat. Commun. 2018, 9, 3516. [Google Scholar] [CrossRef] [PubMed]
- Borsani, O.; Zhu, J.; Verslues, P.E.; Sunkar, R.; Zhu, J.-K. Endogenous SiRNAs Derived from a Pair of Natural Cis-Antisense Transcripts Regulate Salt Tolerance in Arabidopsis. Cell 2005, 123, 1279–1291. [Google Scholar] [CrossRef] [PubMed]
- Bøvre, K.; Szybalski, W. Patterns of Convergent and Overlapping Transcription within the B2 Region of Coliphage λ. Virology 1969, 38, 614–626. [Google Scholar] [CrossRef]
- Rogers, J.C. RNA complementary to alpha-amylase messenger-RNA in barley. Plant Mol. Biol. 1988, 11, 125–138. [Google Scholar] [CrossRef]
- Rosikiewicz, W.; Makałowska, I. Biological Functions of Natural Antisense Transcripts. Acta Biochim. Pol. 2017, 63, 665–673. [Google Scholar] [CrossRef]
- Yu, X.; Yang, J.; Li, X.; Liu, X.; Sun, C.; Wu, F.; He, Y. Global Analysis of Cis-Natural Antisense Transcripts and Their Heat-Responsive Nat-SiRNAs in Brassica rapa. BMC Plant Biol. 2013, 13, 208. [Google Scholar] [CrossRef]
- Jiang, H.; Jia, Z.; Liu, S.; Zhao, B.; Li, W.; Jin, B.; Wang, L. Identification and Characterization of Long Non-Coding RNAs Involved in Embryo Development of Ginkgo biloba. Plant Signal. Behav. 2019, 14, 1674606. [Google Scholar] [CrossRef] [PubMed]
- Akter, M.A.; Mehraj, H.; Miyaji, N.; Takahashi, S.; Takasaki-Yasuda, T.; Seki, M.; Dennis, E.S.; Fujimoto, R.; Osabe, K. Transcriptional Association between MRNAs and Their Paired Natural Antisense Transcripts Following Fusarium oxysporum Inoculation in Brassica rapa L. Horticulturae 2021, 8, 17. [Google Scholar] [CrossRef]
- Wijma, M.; Lembke, C.G.; Diniz, A.L.; Santini, L.; Zambotti-Villela, L.; Colepicolo, P.; Carneiro, M.S.; Souza, G.M. Planting Season Impacts Sugarcane Stem Development, Secondary Metabolite Levels, and Natural Antisense Transcription. Cells 2021, 10, 3451. [Google Scholar] [CrossRef]
- Pandorf, C.E.; Haddad, F.; Roy, R.R.; Qin, A.X.; Edgerton, V.R.; Baldwin, K.M. Dynamics of Myosin Heavy Chain Gene Regulation in Slow Skeletal Muscle. J. Biol. Chem. 2006, 281, 38330–38342. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Li, W.; Sun, Y.; Yu, D.; Wen, X.; Wang, H.; Cui, J.; Wang, G.; Hoffman, A.R.; Hu, J.-F. A Novel Antisense Long Noncoding RNA within the IGF1R Gene Locus Is Imprinted in Hematopoietic Malignancies. Nucleic Acids Res. 2014, 42, 9588–9601. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, M.; Higo, K. Accumulation of Sense–Antisense Transcripts of the Rice Catalase Gene CatB under Dark Conditions Requires Signals from Shoots. Gene 2006, 377, 186–194. [Google Scholar] [CrossRef]
- Camblong, J.; Iglesias, N.; Fickentscher, C.; Dieppois, G.; Stutz, F. Antisense RNA Stabilization Induces Transcriptional Gene Silencing via Histone Deacetylation in S. cerevisiae. Cell 2007, 131, 706–717. [Google Scholar] [CrossRef]
- Richards, M.; Tan, S.-P.; Chan, W.-K.; Bongso, A. Reverse Serial Analysis of Gene Expression (SAGE) Characterization of Orphan SAGE Tags from Human Embryonic Stem Cells Identifies the Presence of Novel Transcripts and Antisense Transcription of Key Pluripotency Genes. Stem Cells 2006, 24, 1162–1173. [Google Scholar] [CrossRef]
- Poole, R.L.; Barker, G.L.; Werner, K.; Biggi, G.F.; Coghill, J.; Gibbings, J.G.; Berry, S.; Dunwell, J.M.; Edwards, K.J. Analysis of Wheat SAGE Tags Reveals Evidence for Widespread Antisense Transcription. BMC Genom. 2008, 9, 475. [Google Scholar] [CrossRef]
- Obermeier, C.; Hosseini, B.; Friedt, W.; Snowdon, R. Gene Expression Profiling via LongSAGE in a Non-Model Plant Species: A Case Study in Seeds of Brassica napus. BMC Genom. 2009, 10, 295. [Google Scholar] [CrossRef]
- Obermeier, C.; Hosseini, B.; Friedt, W.; Snowdon, R. Serial Analysis of Gene Expression (SAGE) during Brassica napus seed development. Acta Hortic. 2010, 867, 89–95. [Google Scholar] [CrossRef]
- Robinson, S.J.; Cram, D.J.; Lewis, C.T.; Parkin, I.A.P. Maximizing the Efficacy of SAGE Analysis Identifies Novel Transcripts in Arabidopsis. Plant Physiol. 2004, 136, 3223–3233. [Google Scholar] [CrossRef] [PubMed]
- Calsa, T., Jr.; Figueira, A. Serial Analysis of Gene Expression in Sugarcane (Saccharum Spp.) Leaves Revealed Alternative C4 Metabolism and Putative Antisense Transcripts. Plant Mol. Biol. 2007, 63, 745–762. [Google Scholar] [CrossRef]
- Lembke, C.G.; Nishiyama, M.Y., Jr.; Sato, P.M.; de Andrade, R.F.; Souza, G.M. Identification of Sense and Antisense Transcripts Regulated by Drought in Sugarcane. Plant Mol. Biol. 2012, 79, 461–477. [Google Scholar] [CrossRef]
- Wang, H.; Chung, P.J.; Liu, J.; Jang, I.-C.; Kean, M.J.; Xu, J.; Chua, N.-H. Genome-Wide Identification of Long Noncoding Natural Antisense Transcripts and Their Responses to Light in Arabidopsis. Genome Res. 2014, 24, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Żmieńko, A.; Guzowska-Nowowiejska, M.; Urbaniak, R.; Pląder, W.; Formanowicz, P.; Figlerowicz, M. A Tiling Microarray for Global Analysis of Chloroplast Genome Expression in Cucumber and Other Plants. Plant Methods 2011, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Matsui, A.; Ishida, J.; Morosawa, T.; Mochizuki, Y.; Kaminuma, E.; Endo, T.A.; Okamoto, M.; Nambara, E.; Nakajima, M.; Kawashima, M.; et al. Arabidopsis Transcriptome Analysis under Drought, Cold, High-Salinity and ABA Treatment Conditions Using a Tiling Array. Plant Cell Physiol. 2008, 49, 1135–1149. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, Y.; Matsui, A.; Hanada, K.; Kawashima, M.; Ishida, J.; Morosawa, T.; Tanaka, M.; Kaminuma, E.; Mochizuki, Y.; Matsushima, A.; et al. Genome-Wide Suppression of Aberrant mRNA-like Noncoding RNAs by NMD in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 2453–2458. [Google Scholar] [CrossRef] [PubMed]
- Hazen, S.P.; Naef, F.; Quisel, T.; Gendron, J.M.; Chen, H.; Ecker, J.R.; Borevitz, J.O.; Kay, S.A. Exploring the Transcriptional Landscape of Plant Circadian Rhythms Using Genome Tiling Arrays. Genome Biol. 2009, 10, R17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, Y.; Xie, X.; Xu, H.; Xu, Z.; Ma, J.; Li, B.; Lin, S.; Nie, Q.; Luo, Q.; et al. A Systematic Analysis on mRNA and MicroRNA Expression in Runting and Stunting Chickens. PLoS ONE 2015, 10, e0127342. [Google Scholar] [CrossRef]
- Zhu, B.; Yang, Y.; Li, R.; Fu, D.; Wen, L.; Luo, Y.; Zhu, H. RNA Sequencing and Functional Analysis Implicate the Regulatory Role of Long Non-Coding RNAs in Tomato Fruit Ripening. J. Exp. Bot. 2015, 66, 4483–4495. [Google Scholar] [CrossRef]
- Loh, S.C.; Othman, A.S.; Veera Singham, G. Identification and Characterization of Jasmonic Acid- and Linolenic Acid-Mediated Transcriptional Regulation of Secondary Laticifer Differentiation in Hevea brasiliensis. Sci. Rep. 2019, 9, 14296. [Google Scholar] [CrossRef]
- Giosa, D.; Felice, M.R.; Giuffrè, L.; Aiese Cigliano, R.; Paytuví-Gallart, A.; Lo Passo, C.; Barresi, C.; D’Alessandro, E.; Huang, H.; Criseo, G.; et al. Transcriptome-Wide Expression Profiling of Sporothrix schenckii Yeast and Mycelial Forms and the Establishment of the Sporothrix Genome DataBase. Microb. Genom. 2020, 6, mgen000445. [Google Scholar] [CrossRef] [PubMed]
- Hoolwerff, M.; Metselaar, P.I.; Tuerlings, M.; Suchiman, H.E.D.; Lakenberg, N.; Ramos, Y.F.M.; Cats, D.; Nelissen, R.G.H.H.; Broekhuis, D.; Mei, H.; et al. Elucidating Epigenetic Regulation by Identifying Functional Cis-Acting Long Noncoding RNAs and Their Targets in Osteoarthritic Articular Cartilage. Arthritis Rheumatol. 2020, 72, 1845–1854. [Google Scholar] [CrossRef]
- Jia, X.; He, Y.; Chen, S.-Y.; Wang, J.; Hu, S.; Lai, S.-J. Genome-Wide Identification and Characterisation of Long Non-Coding RNAs in Two Chinese Cattle Breeds. Ital. J. Anim. Sci. 2020, 19, 383–391. [Google Scholar] [CrossRef]
- Datson, N. Scaling Down SAGE: From MiniSAGE to MicroSAGE. Curr. Pharm. Biotechnol. 2008, 9, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Choudhuri, S. Genomic Technologies. In Bioinformatics for Beginners; Elsevier: Amsterdam, The Netherlands, 2014; pp. 55–72. ISBN 978-0-12-410471-6. [Google Scholar]
- Mills, J.D.; Chen, B.J.; Ueberham, U.; Arendt, T.; Janitz, M. The Antisense Transcriptome and the Human Brain. J. Mol. Neurosci. 2016, 58, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Halley, P.; Kadakkuzha, B.M.; Faghihi, M.A.; Magistri, M.; Zeier, Z.; Khorkova, O.; Coito, C.; Hsiao, J.; Lawrence, M.; Wahlestedt, C. Regulation of the Apolipoprotein Gene Cluster by a Long Noncoding RNA. Cell Rep. 2014, 6, 222–230. [Google Scholar] [CrossRef]
- Whitworth, D.E.; Swain, M.T. A Survey of Non-Coding RNAs in the Social and Predatory Myxobacterium Myxococcus xanthus DK1622. Mol. Omics 2020, 16, 492–502. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.-P.; Li, W. CPAT: Coding-Potential Assessment Tool Using an Alignment-Free Logistic Regression Model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef] [PubMed]
- Schwalb, B.; Michel, M.; Zacher, B.; Frühauf, K.; Demel, C.; Tresch, A.; Gagneur, J.; Cramer, P. TT-Seq Maps the Human Transient Transcriptome. Science 2016, 352, 1225–1228. [Google Scholar] [CrossRef]
- Hetzel, J.; Duttke, S.H.; Benner, C.; Chory, J. Nascent RNA Sequencing Reveals Distinct Features in Plant Transcription. Proc. Natl. Acad. Sci. USA 2016, 113, 12316–12321. [Google Scholar] [CrossRef] [PubMed]
- Kindgren, P.; Ivanov, M.; Marquardt, S. Native Elongation Transcript Sequencing Reveals Temperature Dependent Dynamics of Nascent RNAPII Transcription in Arabidopsis. Nucleic Acids Res. 2020, 48, 2332–2347. [Google Scholar] [CrossRef] [PubMed]
- Szabo, E.X.; Reichert, P.; Lehniger, M.-K.; Ohmer, M.; de Francisco Amorim, M.; Gowik, U.; Schmitz-Linneweber, C.; Laubinger, S. Metabolic Labeling of RNAs Uncovers Hidden Features and Dynamics of the Arabidopsis Transcriptome. Plant Cell 2020, 32, 871–887. [Google Scholar] [CrossRef]
- Barbieri, E.; Hill, C.; Quesnel-Vallières, M.; Zucco, A.J.; Barash, Y.; Gardini, A. Rapid and Scalable Profiling of Nascent RNA with FastGRO. Cell Rep. 2020, 33, 108373. [Google Scholar] [CrossRef]
- Zhu, J.; Liu, M.; Liu, X.; Dong, Z. RNA Polymerase II Activity Revealed by GRO-Seq and pNET-Seq in Arabidopsis. Nat. Plants 2018, 4, 1112–1123. [Google Scholar] [CrossRef]
- Wu, H.-Y.L.; Song, G.; Walley, J.W.; Hsu, P.Y. The Tomato Translational Landscape Revealed by Transcriptome Assembly and Ribosome Profiling. Plant Physiol. 2019, 181, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, B.; Han, J.; Wang, T.; Han, L. Competing Endogenous RNA Network Analysis for Screening Inflammation-related Long Non-coding RNAs for Acute Ischemic Stroke. Mol. Med. Rep. 2020, 22, 3081–3094. [Google Scholar] [CrossRef] [PubMed]
- Maamar, H.; Cabili, M.N.; Rinn, J.; Raj, A. Linc-HOXA1 Is a Noncoding RNA That Represses Hoxa1 Transcription in Cis. Genes Dev. 2013, 27, 1260–1271. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhang, G.; Li, J.; Zhang, X.; Huang, S.; Xiang, S.; Hu, X.; Liu, C. CRISPRlnc: A Manually Curated Database of Validated sgRNAs for lncRNAs. Nucleic Acids Res. 2019, 47, D63–D68. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Fu, D.; Zhu, B.; Luo, Y.; Zhu, H. CRISPR/Cas9-mediated Mutagenesis of LncRNA1459 Alters Tomato Fruit Ripening. Plant J. 2018, 94, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Esposito, R.; Bosch, N.; Lanzós, A.; Polidori, T.; Pulido-Quetglas, C.; Johnson, R. Hacking the Cancer Genome: Profiling Therapeutically Actionable Long Non-Coding RNAs Using CRISPR-Cas9 Screening. Cancer Cell 2019, 35, 545–557. [Google Scholar] [CrossRef]
- Ietswaart, R.; Rosa, S.; Wu, Z.; Dean, C.; Howard, M. Cell-Size-Dependent Transcription of FLC and Its Antisense Long Non-Coding RNA COOLAIR Explain Cell-to-Cell Expression Variation. Cell Syst. 2017, 4, 622–635.e9. [Google Scholar] [CrossRef] [PubMed]
- Wibowo, A.; Becker, C.; Marconi, G.; Durr, J.; Price, J.; Hagmann, J.; Papareddy, R.; Putra, H.; Kageyama, J.; Becker, J.; et al. Hyperosmotic Stress Memory in Arabidopsis Is Mediated by Distinct Epigenetically Labile Sites in the Genome and Is Restricted in the Male Germline by DNA Glycosylase Activity. eLife 2016, 5, e13546. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.; Brunton, J.; Graziosi, S. EPPI-Reviewer 4: Software for Research Synthesis; EPPI-Centre Software; Social Science Research Unit, UCL Institute of Education: London, UK, 2010. [Google Scholar]
- Thomas, J.; Graziosi, S.; Brunton, J.; Ghouze, Z.; O’Driscoll, P.; Bond, M. EPPI-Reviewer: Advanced Software for Systematic Reviews, Maps and Evidence Synthesis; UCL Social Research Institute: London, UK, 2020. [Google Scholar]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Hotta, C.T.; Nishiyama, M.Y.; Souza, G.M. Circadian Rhythms of Sense and Antisense Transcription in Sugarcane, a Highly Polyploid Crop. PLoS ONE 2013, 8, e71847. [Google Scholar] [CrossRef]
- Diniz, A.L.; da Silva, D.I.R.; Lembke, C.G.; Costa, M.D.-B.L.; ten-Caten, F.; Li, F.; Vilela, R.D.; Menossi, M.; Ware, D.; Endres, L.; et al. Amino Acid and Carbohydrate Metabolism Are Coordinated to Maintain Energetic Balance during Drought in Sugarcane. Int. J. Mol. Sci. 2020, 21, 9124. [Google Scholar] [CrossRef]
- Cunha, C.P.; Roberto, G.G.; Vicentini, R.; Lembke, C.G.; Souza, G.M.; Ribeiro, R.V.; Machado, E.C.; Lagôa, A.M.M.A.; Menossi, M. Ethylene-Induced Transcriptional and Hormonal Responses at the Onset of Sugarcane Ripening. Sci. Rep. 2017, 7, 43364. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, S.S.; Hotta, C.T.; Poelking, V.G.d.C.; Leite, D.C.C.; Buckeridge, M.S.; Loureiro, M.E.; Barbosa, M.H.P.; Carneiro, M.S.; Souza, G.M. Co-Expression Network Analysis Reveals Transcription Factors Associated to Cell Wall Biosynthesis in Sugarcane. Plant Mol. Biol. 2016, 91, 15–35. [Google Scholar] [CrossRef]
- Dantas, L.L.d.B.; Almeida-Jesus, F.M.; de Lima, N.O.; Alves-Lima, C.; Nishiyama-Jr, M.Y.; Carneiro, M.S.; Souza, G.M.; Hotta, C.T. Rhythms of Transcription in Field-Grown Sugarcane Are Highly Organ Specific. Sci. Rep. 2020, 10, 6565. [Google Scholar] [CrossRef] [PubMed]
- Souza, G.M.; Van Sluys, M.-A.; Lembke, C.G.; Lee, H.; Margarido, G.R.A.; Hotta, C.T.; Gaiarsa, J.W.; Diniz, A.L.; Oliveira, M.d.M.; Ferreira, S. de S.; et al. Assembly of the 373k Gene Space of the Polyploid Sugarcane Genome Reveals Reservoirs of Functional Diversity in the World’s Leading Biomass Crop. GigaScience 2019, 8, giz129. [Google Scholar] [CrossRef]
- Vettore, A.L.; da Silva, F.R.; Kemper, E.L.; Souza, G.M.; da Silva, A.M.; Ferro, M.I.T.; Henrique-Silva, F.; Giglioti, É.A.; Lemos, M.V.F.; Coutinho, L.L.; et al. Analysis and Functional Annotation of an Expressed Sequence Tag Collection for Tropical Crop Sugarcane. Genome Res. 2003, 13, 2725–2735. [Google Scholar] [CrossRef] [PubMed]
- Ecker, J.R.; Davis, R.W. Inhibition of Gene-Expression in Plant-Cells by Expression of Antisense RNA. Proc. Natl. Acad. Sci. USA 1986, 83, 5372–5376. [Google Scholar] [CrossRef] [PubMed]
- Rothstein, S.J.; DiMaio, J.; Strand, M.; Rice, D. Stable and Heritable Inhibition of the Expression of Nopaline Synthase in Tobacco Expressing Antisense RNA. Proc. Natl. Acad. Sci. USA 1987, 84, 8439–8443. [Google Scholar] [CrossRef]
- Hemenway, C.; Fang, R.X.; Kaniewski, W.K.; Chua, N.H.; Tumer, N.E. Analysis of the mechanism of protection in transgenic plants expressing the potato virus-X coat protein or its antisense RNA. Embo J. 1988, 7, 1273–1280. [Google Scholar] [CrossRef] [PubMed]
- Sandler, S.J.; Stayton, M.; Townsend, J.A.; Ralston, M.L.; Bedbrook, J.R.; Dunsmuir, P. Inhibition of Gene-Expression in Transformed Plants by Antisense RNA. Plant Mol. Biol. 1988, 11, 301–310. [Google Scholar] [CrossRef]
- Smith, C.J.S.; Watson, C.F.; Ray, J.; Bird, C.R.; Morris, P.C.; Schuch, W.; Grierson, D. Antisense RNA Inhibition of Polygalacturonase Gene-Expression in Transgenic Tomatoes. Nature 1988, 334, 724–726. [Google Scholar] [CrossRef]
- Cuozzo, M.; Oconnell, K.M.; Kaniewski, W.; Fang, R.X.; Chua, N.H.; Tumer, N.E. Viral Protection in Transgenic Tobacco Plants Expressing the Cucumber Mosaic-Virus Coat Protein or its Antisense RNA. Bio-Technology 1988, 6, 549. [Google Scholar] [CrossRef]
- Delauney, A.J.; Tabaeizadeh, Z.; Verma, D.P.S. A Stable Bifunctional Antisense Transcript Inhibiting Gene-Expression in Transgenic Plants. Proc. Natl. Acad. Sci. USA 1988, 85, 4300–4304. [Google Scholar] [CrossRef]
- Sheehy, R.E.; Kramer, M.; Hiatt, W.R. Reduction of Polygalacturonase Activity in Tomato Fruit by Antisense RNA. Proc. Natl. Acad. Sci. USA. 1988, 85, 8805–8809. [Google Scholar] [CrossRef]
- Suwabe, K.; Yano, K. Omics Databases in Plant Science: Key to Systems Biology. Plant Biotechnol. 2008, 25, 413–422. [Google Scholar] [CrossRef]
- Xia, J.; Zeng, C.; Chen, Z.; Zhang, K.; Chen, X.; Zhou, Y.; Song, S.; Lu, C.; Yang, R.; Yang, Z.; et al. Endogenous Small-Noncoding RNAs and Their Roles in Chilling Response and Stress Acclimation in Cassava. BMC Genom. 2014, 15, 634. [Google Scholar] [CrossRef]
- Zuo, J.; Wang, Q.; Han, C.; Ju, Z.; Cao, D.; Zhu, B.; Luo, Y.; Gao, L. SRNAome and Degradome Sequencing Analysis Reveals Specific Regulation of SRNA in Response to Chilling Injury in Tomato Fruit. Physiol. Plantarum. 2017, 160, 142–154. [Google Scholar] [CrossRef]
- Shea, D.J.; Nishida, N.; Takada, S.; Itabashi, E.; Takahashi, S.; Akter, A.; Miyaji, N.; Osabe, K.; Mehraj, H.; Shimizu, M.; et al. Long Noncoding RNAs in Brassica rapa L. Following Vernalization. Sci. Rep. 2019, 9, 9302. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, B.; Habermann, K.; Arif, M.A.; Weil, H.L.; Garcia-Molina, A.; Kleine, T.; Mühlhaus, T.; Frank, W. Identification of Small RNAs during Cold Acclimation in Arabidopsis thaliana. BMC Plant Biol. 2020, 20, 298. [Google Scholar] [CrossRef] [PubMed]
- Calixto, C.P.G.; Tzioutziou, N.A.; James, A.B.; Hornyik, C.; Guo, W.; Zhang, R.; Nimmo, H.G.; Brown, J.W.S. Cold-Dependent Expression and Alternative Splicing of Arabidopsis Long Non-Coding RNAs. Front. Plant Sci. 2019, 10, 235. [Google Scholar] [CrossRef]
- Tiwari, B.; Habermann, K.; Arif, M.A.; Top, O.; Frank, W. Identification of Small RNAs During High Light Acclimation in Arabidopsis thaliana. Front. Plant Sci. 2021, 12, 656657. [Google Scholar] [CrossRef] [PubMed]
- Muthusamy, M.; Uma, S.; Backiyarani, S.; Saraswathi, M.S. Genome-Wide Screening for Novel, Drought Stress-Responsive Long Non-Coding RNAs in Drought-Stressed Leaf Transcriptome of Drought-Tolerant and -Susceptible Banana (Musa spp) Cultivars Using Illumina High-Throughput Sequencing. Plant Biotechnol. Rep. 2015, 9, 279–286. [Google Scholar] [CrossRef]
- Lu, X.; Chen, X.; Mu, M.; Wang, J.; Wang, X.; Wang, D.; Yin, Z.; Fan, W.; Wang, S.; Guo, L.; et al. Genome-Wide Analysis of Long Noncoding RNAs and Their Responses to Drought Stress in Cotton (Gossypium hirsutum L.). PLoS ONE 2016, 11, e0156723. [Google Scholar] [CrossRef]
- Li, S.; Yu, X.; Lei, N.; Cheng, Z.; Zhao, P.; He, Y.; Wang, W.; Peng, M. Genome-Wide Identification and Functional Prediction of Cold and/or Drought-Responsive LncRNAs in Cassava. Sci. Rep. 2017, 7, 45981. [Google Scholar] [CrossRef]
- Xu, J.; Wang, Q.; Freeling, M.; Zhang, X.; Xu, Y.; Mao, Y.; Tang, X.; Wu, F.; Lan, H.; Cao, M.; et al. Natural Antisense Transcripts Are Significantly Involved in Regulation of Drought Stress in Maize. Nucleic Acids Res. 2017, 45, 5126–5141. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Chen, S. Widespread Antisense Transcription of Populus Genome under Drought. Mol. Genet. Genom. 2018, 293, 1017–1033. [Google Scholar] [CrossRef]
- Eom, S.H.; Lee, H.J.; Lee, J.H.; Wi, S.H.; Kim, S.K.; Hyun, T.K. Identification and Functional Prediction of Drought-Responsive Long Non-Coding RNA in Tomato. Agronomy 2019, 9, 629. [Google Scholar] [CrossRef]
- Xu, Y.-C.; Zhang, J.; Zhang, D.-Y.; Nan, Y.-H.; Ge, S.; Guo, Y.-L. Identification of Long Noncoding Natural Antisense Transcripts (lncNATs) Correlated with Drought Stress Response in Wild Rice (Oryza nivara). BMC Genom. 2021, 22, 424. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.S.; Yang, S.N.; Li, H.; Zhu, C.C.; Liu, Z.P.; Yang, Z.M. Molecular Dissection of Mercury-Responsive Transcriptome and Sense/Antisense Genes in Medicago truncatula. J. Hazard. Mater. 2013, 252–253, 123–131. [Google Scholar] [CrossRef]
- Oono, Y.; Yazawa, T.; Kanamori, H.; Sasaki, H.; Mori, S.; Matsumoto, T. Genome-Wide Analysis of Rice Cis-Natural Antisense Transcription under Cadmium Exposure Using Strand-Specific RNA-Seq. BMC Genom. 2017, 18, 761. [Google Scholar] [CrossRef]
- Huanca-Mamani, W.; Arias-Carrasco, R.; Cárdenas-Ninasivincha, S.; Rojas-Herrera, M.; Sepúlveda-Hermosilla, G.; Caris-Maldonado, J.; Bastías, E.; Maracaja-Coutinho, V. Long Non-Coding RNAs Responsive to Salt and Boron Stress in the Hyper-Arid Lluteño Maize from Atacama Desert. Genes 2018, 9, 170. [Google Scholar] [CrossRef]
- Yao, Y.; Bilichak, A.; Golubov, A.; Blevins, T.; Kovalchuk, I. Differential Sensitivity of Arabidopsis siRNA Biogenesis Mutants to Genotoxic Stress. Plant Cell Rep. 2010, 29, 1401–1410. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.-Z.; Liu, M.; Zhao, M.-G.; Chen, R.; Zhang, W.-H. Identification and Characterization of Long Non-Coding RNAs Involved in Osmotic and Salt Stress in Medicago truncatula Using Genome-Wide High-Throughput Sequencing. BMC Plant Biol. 2015, 15, 131. [Google Scholar] [CrossRef]
- Gowda, M.; Venu, R.-C.; Li, H.; Jantasuriyarat, C.; Chen, S.; Bellizzi, M.; Pampanwar, V.; Kim, H.; Dean, R.A.; Stahlberg, E.; et al. Magnaporthe Grisea Infection Triggers RNA Variation and Antisense Transcript Expression in Rice. Plant Physiol. 2007, 144, 524–533. [Google Scholar] [CrossRef] [PubMed]
- Jiao, C.; Gao, M.; Wang, X.; Fei, Z. Transcriptome Characterization of Three Wild Chinese Vitis Uncovers a Large Number of Distinct Disease Related Genes. BMC Genom. 2015, 16, 223. [Google Scholar] [CrossRef]
- Summanwar, A.; Basu, U.; Rahman, H.; Kav, N. Identification of lncRNAs Responsive to Infection by Plasmodiophora brassicae in Clubroot-Susceptible and -Resistant Brassica napus Lines Carrying Resistance Introgressed from Rutabaga. Mol. Plant-Microbe Interact. 2019, 32, 1360–1377. [Google Scholar] [CrossRef] [PubMed]
- Xing, Q.; Zhang, W.; Liu, M.; Li, L.; Li, X.; Yan, J. Genome-Wide Identification of Long Non-Coding RNAs Responsive to Lasiodiplodia theobromae Infection in Grapevine. Evol. Bioinform. Online 2019, 15, 117693431984136. [Google Scholar] [CrossRef]
- Muthusamy, M.; Uma, S.; Suthanthiram, B.; Saraswathi, M.S.; Chandrasekar, A. Genome-Wide Identification of Novel, Long Non-Coding RNAs Responsive to Mycosphaerella eumusae and Pratylenchus coffeae Infections and Their Differential Expression Patterns in Disease-Resistant and Sensitive Banana Cultivars. Plant Biotechnol. Rep. 2019, 13, 73–83. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, X.; Wang, C.; Xu, Z.; Wang, Y.; Liu, X.; Kang, Z.; Ji, W. Long Non-Coding Genes Implicated in Response to Stripe Rust Pathogen Stress in Wheat (Triticum aestivum L.). Mol. Biol. Rep. 2013, 40, 6245–6253. [Google Scholar] [CrossRef] [PubMed]
- Sharma, Y.; Sharma, A.; Madhu; Shumayla; Singh, K.; Upadhyay, S.K. Long Non-Coding RNAs as Emerging Regulators of Pathogen Response in Plants. ncRNA 2022, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Visser, M.; Maree, H.J.; Rees, D.J.G.; Burger, J.T. High-Throughput Sequencing Reveals Small RNAs Involved in ASGV Infection. BMC Genom. 2014, 15, 568. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wu, S.; Jin, J.; Li, R. Identification of Herbivore-Elicited Long Non-Coding RNAs in Rice. Plant Signal. Behav. 2021, 16, 1916702. [Google Scholar] [CrossRef]
- Ansaldi, R.; Chaboud, A.; Dumas, C. Multiple S Gene Family Members Including Natural Antisense Transcripts Are Differentially Expressed during Development of Maize Flowers. J. Biol. Chem. 2000, 275, 24146–24155. [Google Scholar] [CrossRef] [PubMed]
- Podio, M.; Colono, C.; Siena, L.; Ortiz, J.P.A.; Pessino, S.C. A Study of the Heterochronic Sense/Antisense RNA Representation in Florets of Sexual and Apomictic Paspalum notatum. BMC Genom. 2021, 22, 185. [Google Scholar] [CrossRef] [PubMed]
- Akagi, H.; Sakamoto, M.; Shinjyo, C.; Shimada, H.; Fujimura, T. A Unique Sequence Located Downstream from the Rice Mitochondrial Atp6 May Cause Male Sterility. Curr. Genet 1994, 25, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Ruwe, H.; Wang, G.; Gusewski, S.; Schmitz-Linneweber, C. Systematic Analysis of Plant Mitochondrial and Chloroplast Small RNAs Suggests Organelle-Specific mRNA Stabilization Mechanisms. Nucleic Acids Res. 2016, 44, 7406–7417. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, J.; Yuan, G.; Liu, C. Complex Interplay among DNA Modification, Noncoding RNA Expression and Protein-Coding RNA Expression in Salvia miltiorrhiza Chloroplast Genome. PLoS ONE 2014, 9, e99314. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shimizu, K.; Hozumi, A.; Aoki, K. Organization of Vascular Cells in the Haustorium of the Parasitic Flowering Plant Cuscuta japonica. Plant Cell Physiol. 2018, 59, 720–728. [Google Scholar] [CrossRef] [PubMed]
- Gou, X.; Zhong, C.; Zhang, P.; Mi, L.; Li, Y.; Lu, W.; Zheng, J.; Xu, J.; Meng, Y.; Shan, W. MiR398b and AtC2GnT Form a Negative Feedback Loop to Regulate Arabidopsis thaliana Resistance against Phytophthora parasitica. Plant J. 2022, 111, 360–373. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhu, P.; Hepworth, J.; Bloomer, R.; Antoniou-Kourounioti, R.L.; Doughty, J.; Heckmann, A.; Xu, C.; Yang, H.; Dean, C. Natural temperature fluctuations promote COOLAIR regulation of FLC. Genes Dev. 2021, 35, 888–898. [Google Scholar] [CrossRef]
- Swiezewski, S.; Liu, F.; Magusin, A.; Dean, C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 2009, 462, 799–802. [Google Scholar] [CrossRef]
- Tian, Y.; Zheng, H.; Zhang, F.; Wang, S.; Ji, X.; Xu, C.; He, Y.; Ding, Y. PRC2 recruitment and H3K27me3 deposition at FLC require FCA binding of COOLAIR. Sci. Adv. 2019, 5, eaau7246. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Hu, Y.; Zhou, Y.; Zhu, Z.; Sun, Y.; Li, J.; Wu, R.; Miao, Y.; Sun, X. Profiling of the Receptor for Activated C Kinase 1a (RACK1a) Interaction Network in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2019, 520, 366–372. [Google Scholar] [CrossRef] [PubMed]
- Henriques, R.; Wang, H.; Liu, J.; Boix, M.; Huang, L.; Chua, N. The Antiphasic Regulatory Module Comprising CDF5 and Its Antisense RNA FLORE Links the Circadian Clock to Photoperiodic Flowering. New. Phytol. 2017, 216, 854–867. [Google Scholar] [CrossRef]
- Cui, J.; Luan, Y.; Jiang, N.; Bao, H.; Meng, J. Comparative Transcriptome Analysis between Resistant and Susceptible Tomato Allows the Identification of LncRNA16397 Conferring Resistance to Phytophthora Infestans by Co-Expressing Glutaredoxin. Plant J. 2017, 89, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Vega-Arreguín, J.C.; Ibarra-Laclette, E.; Jiménez-Moraila, B.; Martínez, O.; Vielle-Calzada, J.P.; Herrera-Estrella, L.; Herrera-Estrella, A. Deep Sampling of the Palomero Maize Transcriptome by a High Throughput Strategy of Pyrosequencing. BMC Genom. 2009, 10, 299. [Google Scholar] [CrossRef]
- Chu, Z.; Lu, Y.; Qin, R.; Dong, Y. LncRNA KCNQ1OT1 Promotes the Apoptosis and Inflammatory Response of Microglia by Regulating the MiR-589-5p/NPTN Axis after Spinal Cord Injury. An. Acad. Bras. Ciências 2022, 94, e20210188. [Google Scholar] [CrossRef]
- Ren, Y.; Gao, X.-P.; Liang, H.; Zhang, H.; Hu, C.-Y. LncRNA KCNQ1OT1 Contributes to Oxygen-Glucose-Deprivation/Reoxygenation-Induced Injury via Sponging MiR-9 in Cultured Neurons to Regulate MMP8. Exp. Mol. Pathol. 2020, 112, 104356. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Qin, Y.; Lv, J.; Wang, Y.; Che, H.; Chen, X.; Jiang, Y.; Li, A.; Sun, X.; Yue, E.; et al. Silencing Long Non-Coding RNA Kcnq1ot1 Alleviates Pyroptosis and Fibrosis in Diabetic Cardiomyopathy. Cell Death Dis. 2018, 9, 1000. [Google Scholar] [CrossRef] [PubMed]
- Navarro, F.J.; Chakravarty, P.; Nurse, P. Phosphorylation of the RNA binding protein Zfs1 modulates sexual differentiation in fission yeast. J. Cell Sci. 2017, 130, 4144–4154. [Google Scholar] [CrossRef]
- Coornaert, A.; Lu, A.; Mandin, P.; Springer, M.; Gottesman, S.; Guillier, M. MicA sRNA Links the PhoP Regulon to Cell Envelope Stress. Mol. Microbiol. 2010, 76, 467–479. [Google Scholar] [CrossRef]
- Udekwu, K.I.; Darfeuille, F.; Vogel, J.; Reimegård, J.; Holmqvist, E.; Wagner, E.G.H. Hfq-Dependent Regulation of OmpA Synthesis Is Mediated by an Antisense RNA. Genes Dev. 2005, 19, 2355–2366. [Google Scholar] [CrossRef] [PubMed]
- Moores, A.; Chipper-Keating, S.; Sun, L.; McVicker, G.; Wales, L.; Gashi, K.; Blomfield, I.C. RfaH Suppresses Small RNA MicA Inhibition of fimB Expression in Escherichia Coli K-12. J. Bacteriol. 2014, 196, 148–156. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Choi, H.; Kim, M.; Jeon, J.; Han, J.K.; Kim, K. Overexpression of MicA Induces Production of OmpC-Enriched Outer Membrane Vesicles That Protect against Salmonella Challenge. Biochem. Biophys. Res. Commun. 2017, 490, 991–996. [Google Scholar] [CrossRef]
- D’Hont, A. Unraveling the genome structure of polyploids using FISH and GISH; examples of sugarcane and banana. Cytogenet. Genome Res. 2005, 109, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Cheavegatti-Gianotto, A.; de Abreu, H.M.C.; Arruda, P.; Bespalhok Filho, J.C.; Burnquist, W.L.; Creste, S.; di Ciero, L.; Ferro, J.A.; de Oliveira Figueira, A.V.; de Sousa Filgueiras, T.; et al. Sugarcane (Saccharum × officinarum): A Reference Study for the Regulation of Genetically Modified Cultivars in Brazil. Trop. Plant Biol. 2011, 4, 62–89. [Google Scholar] [CrossRef] [PubMed]
- Cursi, D.E.; Hoffmann, H.P.; Barbosa, G.V.S.; Bressiani, J.A.; Gazaffi, R.; Chapola, R.G.; Fernandes Junior, A.R.; Balsalobre, T.W.A.; Diniz, C.A.; Santos, J.M.; et al. History and Current Status of Sugarcane Breeding, Germplasm Development and Molecular Genetics in Brazil. Sugar Tech 2022, 24, 112–133. [Google Scholar] [CrossRef]
- Piperidis, N.; D’Hont, A. Sugarcane Genome Architecture Decrypted with Chromosome-specific Oligo Probes. Plant J. 2020, 103, 2039–2051. [Google Scholar] [CrossRef] [PubMed]
- Garcia, A.A.F.; Mollinari, M.; Marconi, T.G.; Serang, O.R.; Silva, R.R.; Vieira, M.L.C.; Vicentini, R.; Costa, E.A.; Mancini, M.C.; Garcia, M.O.S.; et al. SNP Genotyping Allows an In-Depth Characterisation of the Genome of Sugarcane and Other Complex Autopolyploids. Sci. Rep. 2013, 3, 3399. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, X.; Tang, H.; Zhang, Q.; Hua, X.; Ma, X.; Zhu, F.; Jones, T.; Zhu, X.; Bowers, J.; et al. Allele-Defined Genome of the Autopolyploid Sugarcane Saccharum spontaneum L. Nat. Genet. 2018, 50, 1565–1573. [Google Scholar] [CrossRef] [PubMed]
- Garsmeur, O.; Droc, G.; Antonise, R.; Grimwood, J.; Potier, B.; Aitken, K.; Jenkins, J.; Martin, G.; Charron, C.; Hervouet, C.; et al. A Mosaic Monoploid Reference Sequence for the Highly Complex Genome of Sugarcane. Nat. Commun. 2018, 9, 2638. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, M.Y.; Ferreira, S.S.; Tang, P.-Z.; Becker, S.; Pörtner-Taliana, A.; Souza, G.M. Full-Length Enriched cDNA Libraries and ORFeome Analysis of Sugarcane Hybrid and Ancestor Genotypes. PLoS ONE 2014, 9, e107351. [Google Scholar] [CrossRef] [PubMed]
- Manimekalai, R.; Narayanan, J.; Ranjini, R.; Gokul, M.; Selvi, A.; Kumar, P.; Gomathi, R. Hydrogen Peroxide-Induced Oxidative Stress in Sugarcane and Response Expression Pattern of Stress-Responsive Genes Through Quantitative RT-PCR. Sugar Tech 2018, 20, 681–691. [Google Scholar] [CrossRef]
- Narayanan, J.; Manimekalai, R.; Selvi, A.; Arun Kumar, R. Physiological, Biochemical and Molecular Responses to Oxidative Stress in Saccharum spontaneum. Sugar Tech 2022, 1–12. [Google Scholar] [CrossRef]
- Tavares, E.Q.P.; Grandis, A.; Lembke, C.G.; Souza, G.M.; Purgatto, E.; De Souza, A.P.; Buckeridge, M.S. Roles of Auxin and Ethylene in Aerenchyma Formation in Sugarcane Roots. Plant Signal. Behav. 2018, 13, e1422464. [Google Scholar] [CrossRef] [PubMed]
- Cia, M.C.; de Carvalho, G.; Azevedo, R.A.; Monteiro-Vitorello, C.B.; Souza, G.M.; Nishiyama-Junior, M.Y.; Lembke, C.G.; Antunes de Faria, R.S.d.C.; Marques, J.P.R.; Melotto, M.; et al. Novel Insights into the Early Stages of Ratoon Stunting Disease of Sugarcane Inferred from Transcript and Protein Analysis. Phytopathology 2018, 108, 1455–1466. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yuan, D.; Tu, L.; Gao, W.; He, Y.; Hu, H.; Wang, P.; Liu, N.; Lindsey, K.; Zhang, X. Long Noncoding RNA s and Their Proposed Functions in Fibre Development of Cotton (Gossypium spp.). New Phytol. 2015, 207, 1181–1197. [Google Scholar] [CrossRef] [PubMed]
- Shen, E.; Zhu, X.; Hua, S.; Chen, H.; Ye, C.; Zhou, L.; Liu, Q.; Zhu, Q.-H.; Fan, L.; Chen, X. Genome-Wide Identification of Oil Biosynthesis-Related Long Non-Coding RNAs in Allopolyploid Brassica napus. BMC Genom. 2018, 19, 745. [Google Scholar] [CrossRef] [PubMed]


| GEO Series | Experiment ID | Experiment Description | Number of Hybridizations | Reference |
|---|---|---|---|---|
| Ancestral | Sugarcane ancestral genotypes: Saccharum officinarum, S. spontaneum, and S. robustum | 36 | Ferreira et al. [82] | |
| GSE42725 | Circadian I | Circadian rhythms of sense and antisense transcriptions | 22 | Hotta et al. [79] |
| GSE129543 | Circadian II | Vast differences in organ-specific rhythms of transcription in field-grown sugarcane | 84 | Dantas et al. [83] |
| GSE33574 | Drought I | Sugarcane expression data from stress time series | 6 | Lembke et al. [44] |
| GSE125070 | Drought II | Sugarcane drought experiment under field conditions | 12 | Diniz et al. [80] |
| GSE125069 | Drought III | SP80-3280 drought and rewatering | 12 | Diniz et al. [80] |
| GPL22278 | Ethylene | Ethephon- and AVG-induced transcriptional changes | 48 | Cunha et al. [81] |
| GSE124990 | G and M 1 | SP80-3280 growth and maturation under field conditions | 30 | Souza et al. [84]; Wijma et al. [33] |
| Antisense-Related Gene | Cognate Sense Gene | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene ID | Gene Symbol | Gene Type | Citations 1 | Gene ID | Gene Symbol | Gene Type | Citations 1 | Species |
| 5008140 | AT4G13505 | ncRNA | 1 | 826983 | AMT1;1 | protein-coding | 18 | Arabidopsis thaliana |
| 5008149 | AT4G20362 | ncRNA | 1 | 827784 | RABE1b | protein-coding | 30 | Arabidopsis thaliana |
| 6241025 | AT1G69572 | ncRNA | 1 | 843293 | AT1G69570 | protein-coding | 7 | Arabidopsis thaliana |
| 28720403 | AT5G01675 | ncRNA | 3 | 830878 | FLC | protein-coding | 142 | Arabidopsis thaliana |
| 109115966 | lncRNA16397 | ncRNA | 1 | 101250592 | GRX22 | protein-coding | 2 | Solanum lycopersicum |
| 109461486 | LOC109461486 | ncRNA | 1 | 100284505 | LOC100284505 | protein-coding | 4 | Zea mays |
| 5008210 | MIR398b | ncRNA | 7 | 831306 | AT5G14550 | protein-coding | 3 | Arabidopsis thaliana |
| 28721157 | MIR398c | miscRNA | 4 | 831308 | NRT2.7 | protein-coding | 5 | Arabidopsis thaliana |
| 104795691 | MIR398 | ncRNA | 3 | 103856086 | LOC103856086 | protein-coding | 0 | Brassica rapa |
| SS/AS Expression | Ancestral | Circadian I | Circadian II | Drought I | Drought II | Drought III | Ethylene | G and M 1 |
|---|---|---|---|---|---|---|---|---|
| Concordant | 0 | 41 | 21 | 3 | 21 | 1 | 4 | 1 |
| Discordant | 1 | 37 | 12 | 5 | 2 | 0 | 9 | 9 |
| Neutral | 55 | 651 | 225 | 295 | 186 | 47 | 136 | 549 |
| Total | 56 | 729 | 258 | 303 | 209 | 48 | 149 | 559 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santini, L.; Yoshida, L.; de Oliveira, K.D.; Lembke, C.G.; Diniz, A.L.; Cantelli, G.C.; Nishiyama-Junior, M.Y.; Souza, G.M. Antisense Transcription in Plants: A Systematic Review and an Update on cis-NATs of Sugarcane. Int. J. Mol. Sci. 2022, 23, 11603. https://doi.org/10.3390/ijms231911603
Santini L, Yoshida L, de Oliveira KD, Lembke CG, Diniz AL, Cantelli GC, Nishiyama-Junior MY, Souza GM. Antisense Transcription in Plants: A Systematic Review and an Update on cis-NATs of Sugarcane. International Journal of Molecular Sciences. 2022; 23(19):11603. https://doi.org/10.3390/ijms231911603
Chicago/Turabian StyleSantini, Luciane, Leonardo Yoshida, Kaique Dias de Oliveira, Carolina Gimiliani Lembke, Augusto Lima Diniz, Geraldo Cesar Cantelli, Milton Yutaka Nishiyama-Junior, and Glaucia Mendes Souza. 2022. "Antisense Transcription in Plants: A Systematic Review and an Update on cis-NATs of Sugarcane" International Journal of Molecular Sciences 23, no. 19: 11603. https://doi.org/10.3390/ijms231911603
APA StyleSantini, L., Yoshida, L., de Oliveira, K. D., Lembke, C. G., Diniz, A. L., Cantelli, G. C., Nishiyama-Junior, M. Y., & Souza, G. M. (2022). Antisense Transcription in Plants: A Systematic Review and an Update on cis-NATs of Sugarcane. International Journal of Molecular Sciences, 23(19), 11603. https://doi.org/10.3390/ijms231911603







