Identification of a Novel Renal Metastasis Associated CpG-Based DNA Methylation Signature (RMAMS)
Abstract
1. Introduction
2. Results
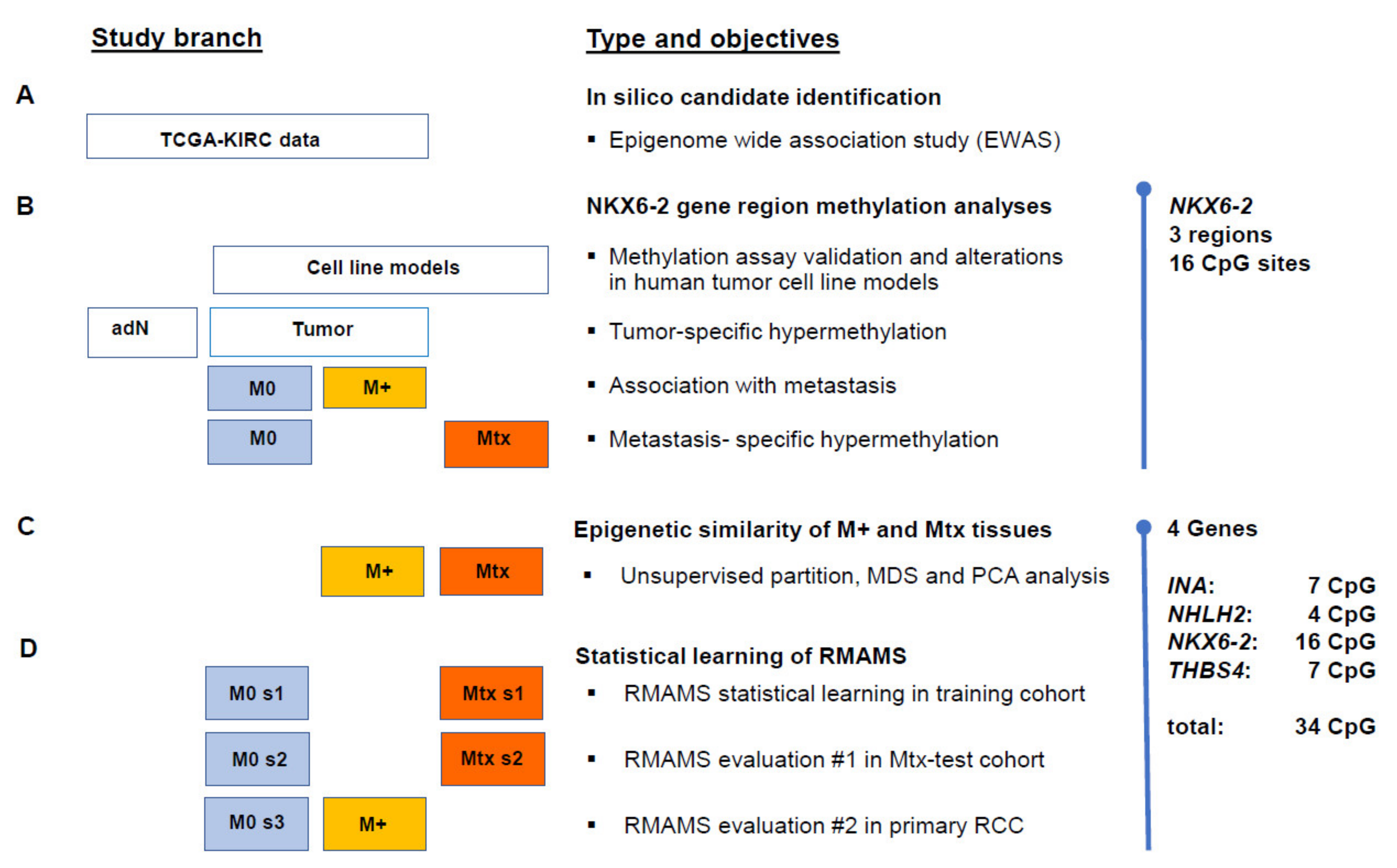
2.1. In Silico Identification of the Association of NKX6-2 Loci Methylation and State of Distant Metastasis
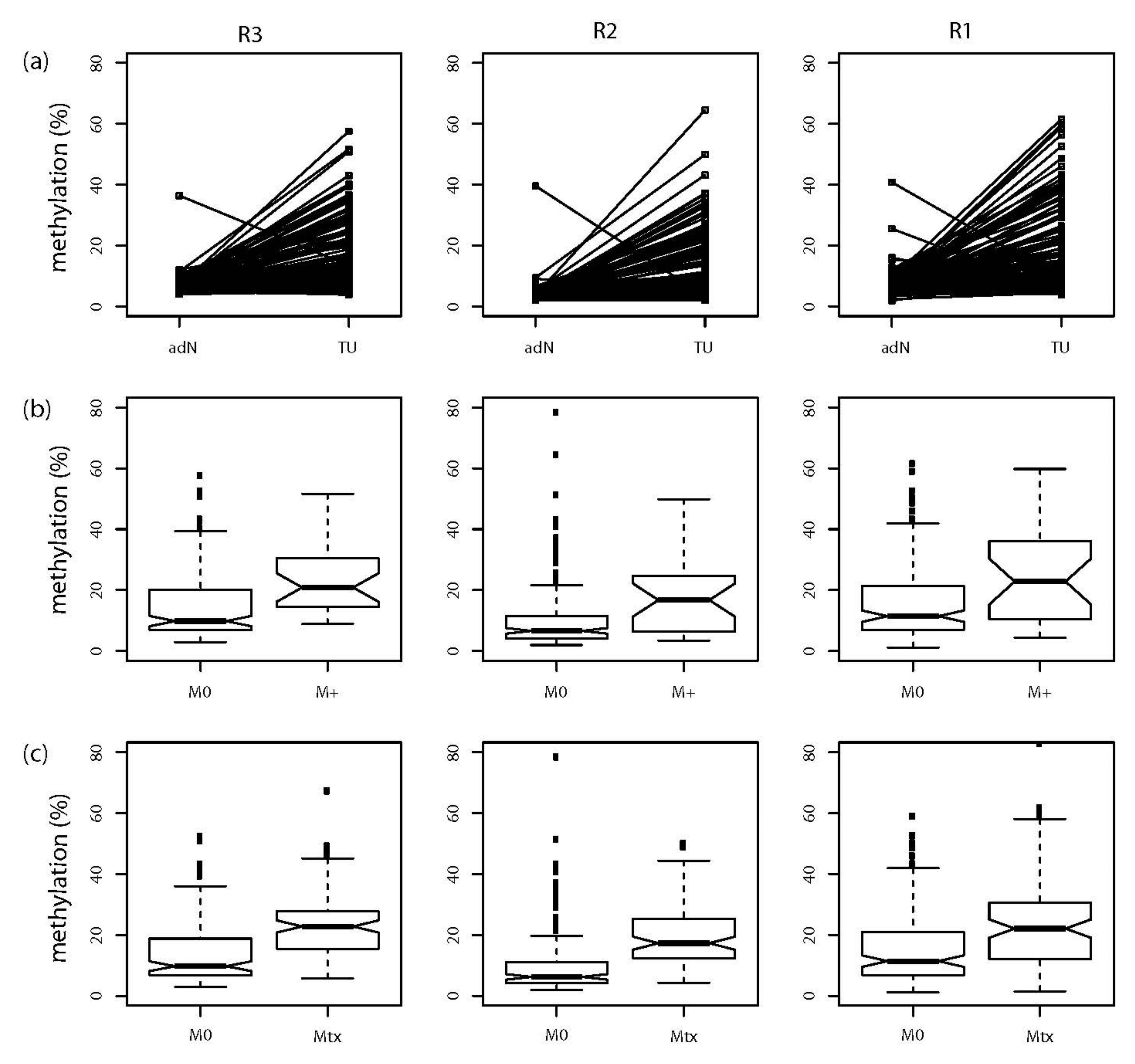
2.2. Evaluation of NKX6-2 Candidate Loci in Primary RCC, RCC-Associated Metastatic Tissues, and Cell Models
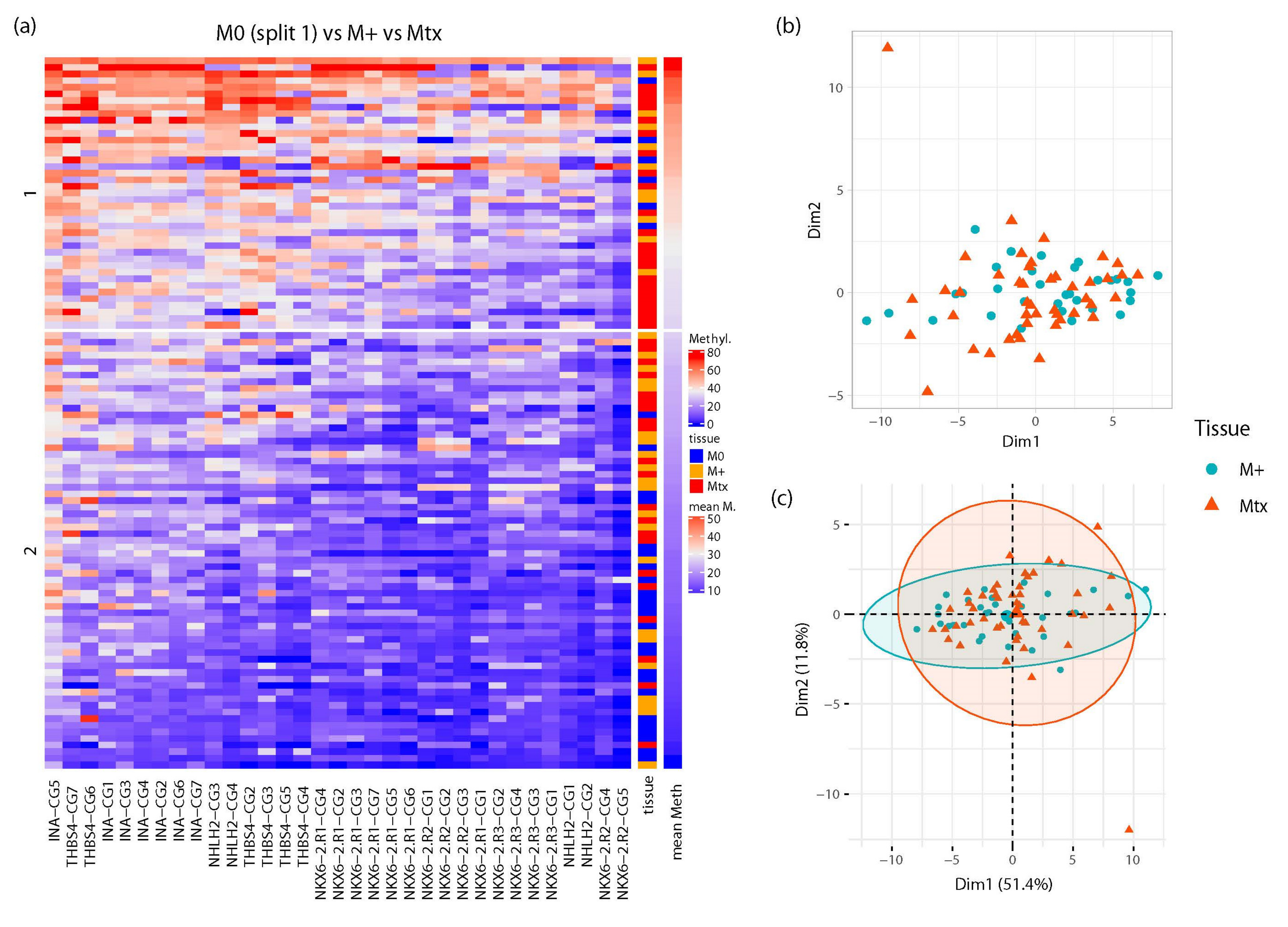
2.3. Similar CpG-Specific Methylation in Metastatic Primary Tumor Tissues and Metastatic Tissues
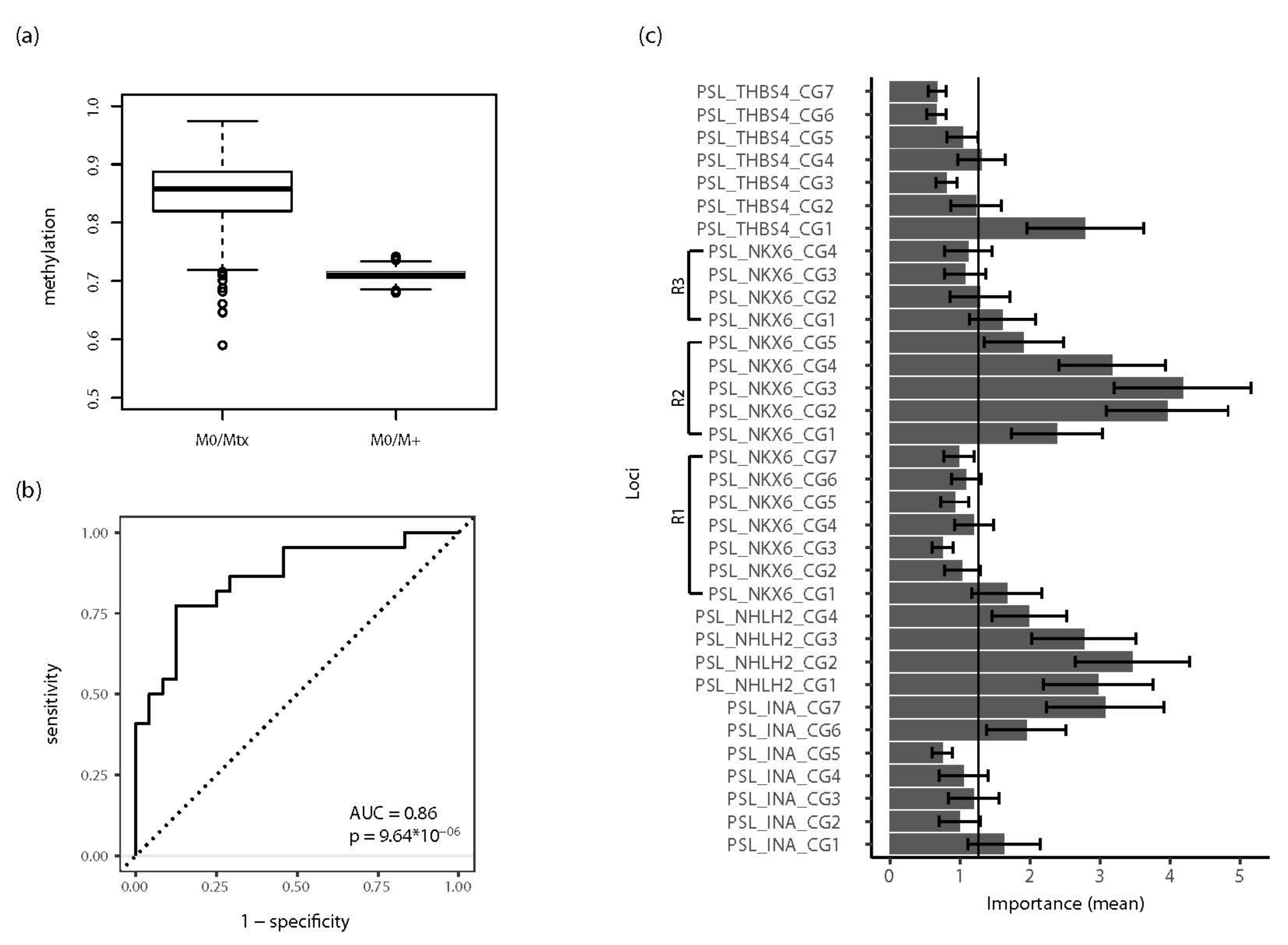
2.4. Development and Evaluation of a RMAMS
3. Discussion
4. Material and Methods
4.1. Study Design
4.2. Study Cohort
4.3. Nucleic Acid Extraction, DNA Bisulfite Conversion, and DNA Methylation Analysis
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Capitanio, U.; Bensalah, K.; Bex, A.; Boorjian, S.A.; Bray, F.; Coleman, J.; Gore, J.L.; Sun, M.; Wood, C.; Russo, P. Epidemiology of Renal Cell Carcinoma. Eur. Urol. 2019, 75, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Marconi, L.; Sun, M.; Beisland, C.; Klatte, T.; Ljungberg, B.; Stewart, G.D.; Dabestani, S.; Choueiri, T.K.; Bex, A. Prevalence, Disease-free, and Overall Survival of Contemporary Patients With Renal Cell Carcinoma Eligible for Adjuvant Checkpoint Inhibitor Trials. Clin. Genitourin. Cancer 2021, 19, e92–e99. [Google Scholar] [CrossRef] [PubMed]
- Dabestani, S.; Thorstenson, A.; Lindblad, P.; Harmenberg, U.; Ljungberg, B.; Lundstam, S. Renal cell carcinoma recurrences and metastases in primary non-metastatic patients: A population-based study. World J. Urol. 2016, 34, 1081–1086. [Google Scholar] [CrossRef] [PubMed]
- Patel, H.D.; Gupta, M.; Joice, G.A.; Srivastava, A.; Alam, R.; Allaf, M.E.; Pierorazio, P.M. Clinical Stage Migration and Survival for Renal Cell Carcinoma in the United States. Eur. Urol. Oncol. 2019, 2, 343–348. [Google Scholar] [CrossRef]
- Choueiri, T.K.; Tomczak, P.; Park, S.H.; Venugopal, B.; Ferguson, T.; Chang, Y.-H.; Hajek, J.; Symeonides, S.N.; Lee, J.L.; Sarwar, N.; et al. Adjuvant Pembrolizumab after Nephrectomy in Renal-Cell Carcinoma. N. Engl. J. Med. 2021, 385, 683–694. [Google Scholar] [CrossRef]
- Klatte, T.; Rossi, S.H.; Stewart, G.D. Prognostic factors and prognostic models for renal cell carcinoma: A literature review. World J. Urol. 2018, 36, 1943–1952. [Google Scholar] [CrossRef]
- Correa, A.F.; Jegede, O.; Haas, N.B.; Flaherty, K.T.; Pins, M.R.; Messing, E.M.; Manola, J.; Wood, C.G.; Kane, C.J.; Jewett, M.A.S.; et al. Predicting Renal Cancer Recurrence: Defining Limitations of Existing Prognostic Models With Prospective Trial-Based Validation. J. Clin. Oncol. 2019, 37, 2062–2071. [Google Scholar] [CrossRef]
- Joosten, S.C.; Smits, K.M.; Aarts, M.J.; Melotte, V.; Koch, A.; Tjan-Heijnen, V.C.; van Engeland, M. Epigenetics in renal cell cancer: Mechanisms and clinical applications. Nat. Rev. Urol. 2018, 15, 430–451. [Google Scholar] [CrossRef]
- TCGA. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013, 499, 43–49. [Google Scholar] [CrossRef]
- Peters, I.; Merseburger, A.S.; Tezval, H.; Lafos, M.; Tabrizi, P.F.; Mazdak, M.; Wolters, M.; Kuczyk, M.A.; Serth, J.; von Klot, C.-A.J. The Prognostic Value of DNA Methylation Markers in Renal Cell Cancer: A Systematic Review. KCA 2020, 4, 3–13. [Google Scholar] [CrossRef]
- Morris, M.R.; Ricketts, C.J.; Gentle, D.; McRonald, F.; Carli, N.; Khalili, H.; Brown, M.; Kishida, T.; Yao, M.; Banks, R.E.; et al. Genome-wide methylation analysis identifies epigenetically inactivated candidate tumour suppressor genes in renal cell carcinoma. Oncogene 2011, 30, 1390–1401. [Google Scholar] [CrossRef]
- Van Vlodrop, I.J.; Niessen, H.E.; Derks, S.; Baldewijns, M.; Van Criekinge, W.; Herman, J.G.; van Engeland, M. Analysis of promoter CpG island hypermethylation in cancer: Location, location, location! Clin. Cancer Res. 2011, 17, 4225–4231. [Google Scholar] [CrossRef]
- Joosten, S.C.; Deckers, I.A.G.; Aarts, M.J.; Hoeben, A.; van Roermund, J.G.; Smits, K.M.; Melotte, V.; van Engeland, M.; Tjan-Heijnen, V.C. Prognostic DNA methylation markers for renal cell carcinoma: A systematic review. Epigenomics 2017, 9, 1243–1257. [Google Scholar] [CrossRef]
- Ricketts, C.J.; De Cubas, A.A.; Fan, H.; Smith, C.C.; Lang, M.; Reznik, E.; Bowlby, R.; Gibb, E.A.; Akbani, R.; Beroukhim, R.; et al. The Cancer Genome Atlas Comprehensive Molecular Characterization of Renal Cell Carcinoma. Cell Rep. 2018, 23, 313–326.e5. [Google Scholar] [CrossRef]
- Peters, I.; Dubrowinskaja, N.; Hennenlotter, J.R.; Antonopoulos, W.; Von Klot, C.; Tezval, H.; Stenzl, A.; Kuczyk, M.; Serth, J.R. DNA methylation of neural EGFL like 1 (NELL1) is associated with advanced disease and the metastatic state of renal cell cancer patients. Oncol. Rep. 2018, 40, 3861–3868. [Google Scholar] [CrossRef]
- Peters, I.; Gebauer, K.; Dubrowinskaja, N.; Atschekzei, F.; Kramer, M.W.; Hennenlotter, J.; Tezval, H.; Abbas, M.; Scherer, R.; Merseburger, A.S.; et al. GATA5 CpG island hypermethylation is an independent predictor for poor clinical outcome in renal cell carcinoma. Oncol. Rep. 2014, 31, 1523–1530. [Google Scholar] [CrossRef]
- Tezval, H.; Dubrowinskaja, N.; Peters, I.; Reese, C.; Serth, K.; Atschekzei, F.; Hennenlotter, J.; Stenzl, A.; Kuczyk, M.A.; Serth, J. Tumor Specific Epigenetic Silencing of Corticotropin Releasing Hormone -Binding Protein in Renal Cell Carcinoma: Association of Hypermethylation and Metastasis. PLoS ONE 2016, 11, e0163873. [Google Scholar] [CrossRef]
- Katzendorn, O.; Peters, I.; Dubrowinskaja, N.; Tezval, H.; Tabrizi, P.F.; Von Klot, C.A.; Hennenlotter, J.; Lafos, M.; Kuczyk, M.A.; Serth, J. DNA methylation of tumor associated calcium signal transducer 2 (TACSTD2) loci shows association with clinically aggressive renal cell cancers. BMC Cancer 2021, 21, 444. [Google Scholar] [CrossRef]
- Costa, V.L.; Henrique, R.; Ribeiro, F.R.; Pinto, M.; Oliveira, J.; Lobo, F.; Teixeira, M.R.; Jeronimo, C. Quantitative promoter methylation analysis of multiple cancer-related genes in renal cell tumors. BMC Cancer 2007, 7, 133. [Google Scholar] [CrossRef]
- Katzendorn, O.; Peters, I.; Dubrowinskaja, N.; Moog, J.M.; Reese, C.; Tezval, H.; Faraj Tabrizi, P.; Hennenlotter, J.; Lafos, M.; Kuczyk, M.A.; et al. DNA Methylation in INA, NHLH2, and THBS4 Is Associated with Metastatic Disease in Renal Cell Carcinoma. Cancers 2021, 14, 39. [Google Scholar] [CrossRef]
- Serth, J.; Peters, I.; Hill, B.; Hübscher, T.; Hennenlotter, J.; Klintschar, M.; Kuczyk, M.A. Age-Related DNA Methylation in Normal Kidney Tissue Identifies Epigenetic Cancer Risk Susceptibility Loci in the ANKRD34B and ZIC1 Genes. Int. J. Mol. Sci. 2022, 23, 5327. [Google Scholar] [CrossRef]
- Morris, M.R.; Ricketts, C.; Gentle, D.; Abdulrahman, M.; Clarke, N.; Brown, M.; Kishida, T.; Yao, M.; Latif, F.; Maher, E.R. Identification of candidate tumour suppressor genes frequently methylated in renal cell carcinoma. Oncogene 2010, 29, 2104–2117. [Google Scholar] [CrossRef]
- Atschekzei, F.; Hennenlotter, J.; Janisch, S.; Grosshennig, A.; Trankenschuh, W.; Waalkes, S.; Peters, I.; Dork, T.; Merseburger, A.S.; Stenzl, A.; et al. SFRP1 CpG island methylation locus is associated with renal cell cancer susceptibility and disease recurrence. Epigenetics 2012, 7, 447–457. [Google Scholar] [CrossRef]
- Gebauer, K.; Peters, I.; Dubrowinskaja, N.; Hennenlotter, J.; Abbas, M.; Scherer, R.; Tezval, H.; Merseburger, A.S.; Stenzl, A.; Kuczyk, M.A.; et al. Hsa-mir-124-3 CpG island methylation is associated with advanced tumours and disease recurrence of patients with clear cell renal cell carcinoma. Br. J. Cancer 2013, 108, 131–138. [Google Scholar] [CrossRef]
- Deckers, I.A.G.; Schouten, L.J.; Van Neste, L.; van Vlodrop, I.J.H.; Soetekouw, P.M.M.B.; Baldewijns, M.M.L.L.; Jeschke, J.; Ahuja, N.; Herman, J.G.; van den Brandt, P.A.; et al. Promoter Methylation of CDO1 Identifies Clear-Cell Renal Cell Cancer Patients with Poor Survival Outcome. Clin. Cancer Res. 2015, 21, 3492–3500. [Google Scholar] [CrossRef]
- Vlodrop, I.J.H.v.; Joosten, S.C.; Meyer, T.D.; Smits, K.M.; Neste, L.V.; Melotte, V.; Baldewijns, M.M.L.L.; Schouten, L.J.; Brandt, P.A.v.d.; Jeschke, J.; et al. A Four-Gene Promoter Methylation Marker Panel Consisting of GREM1, NEURL, LAD1, and NEFH Predicts Survival of Clear Cell Renal Cell Cancer Patients. Clin. Cancer Res. 2017, 23, 2006–2018. [Google Scholar] [CrossRef]
- Dubrowinskaja, N.; Gebauer, K.; Peters, I.; Hennenlotter, J.; Abbas, M.; Scherer, R.; Tezval, H.; Merseburger, A.S.; Stenzl, A.; Grünwald, V.; et al. Neurofilament Heavy polypeptide CpG island methylation associates with prognosis of renal cell carcinoma and prediction of antivascular endothelial growth factor therapy response. Cancer Med. 2014, 3, 300–309. [Google Scholar] [CrossRef]
- Peters, I.; Dubrowinskaja, N.; Abbas, M.; Seidel, C.; Kogosov, M.; Scherer, R.; Gebauer, K.; Merseburger, A.S.; Kuczyk, M.A.; Grunwald, V.; et al. DNA methylation biomarkers predict progression-free and overall survival of metastatic renal cell cancer (mRCC) treated with antiangiogenic therapies. PLoS ONE 2014, 9, e91440. [Google Scholar] [CrossRef]
- Werner, R.J.; Kelly, A.D.; Issa, J.-P.J. Epigenetics and Precision Oncology. Cancer J. 2017, 23, 262–269. [Google Scholar] [CrossRef]
- Wei, J.-H.; Haddad, A.; Wu, K.-J.; Zhao, H.-W.; Kapur, P.; Zhang, Z.-L.; Zhao, L.-Y.; Chen, Z.-H.; Zhou, Y.-Y.; Zhou, J.-C.; et al. A CpG-methylation-based assay to predict survival in clear cell renal cell carcinoma. Nat. Commun. 2015, 6, 8699. [Google Scholar] [CrossRef]
- Chelban, V.; Patel, N.; Vandrovcova, J.; Zanetti, M.N.; Lynch, D.S.; Ryten, M.; Botía, J.A.; Bello, O.; Tribollet, E.; Efthymiou, S.; et al. Mutations in NKX6-2 Cause Progressive Spastic Ataxia and Hypomyelination. Am. J. Hum. Genet. 2017, 100, 969–977. [Google Scholar] [CrossRef]
- Nelson, S.B.; Schaffer, A.E.; Sander, M. The transcription factors Nkx6.1 and Nkx6.2 possess equivalent activities in promoting beta-cell fate specification in Pdx1+ pancreatic progenitor cells. Development 2007, 134, 2491–2500. [Google Scholar] [CrossRef] [PubMed]
- Chung, W.; Bondaruk, J.; Jelinek, J.; Lotan, Y.; Liang, S.; Czerniak, B.; Issa, J.-P.J. Detection of Bladder Cancer Using Novel DNA Methylation Biomarkers in Urine Sediments. Cancer Epidemiol. Biomark. Prev. 2011, 20, 1483–1491. [Google Scholar] [CrossRef] [PubMed]
- Ashktorab, H.; Shakoori, A.; Zarnogi, S.; Sun, X.; Varma, S.; Lee, E.; Shokrani, B.; Laiyemo, A.O.; Washington, K.; Brim, H. Reduced Representation Bisulfite Sequencing Determination of Distinctive DNA Hypermethylated Genes in the Progression to Colon Cancer in African Americans. Gastroenterol. Res. Pract. 2016, 2016, 2102674. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, M.P.; Desai, A.; Palakal, M.J. Systems biology approach to stage-wise characterization of epigenetic genes in lung adenocarcinoma. BMC Syst Biol 2013, 7, 141. [Google Scholar] [CrossRef] [PubMed]
- Arai, E.; Chiku, S.; Mori, T.; Gotoh, M.; Nakagawa, T.; Fujimoto, H.; Kanai, Y. Single-CpG-resolution methylome analysis identifies clinicopathologically aggressive CpG island methylator phenotype clear cell renal cell carcinomas. Carcinogenesis 2012, 33, 1487–1493. [Google Scholar] [CrossRef]
- Power, M.; Fell, G.; Wright, M. Principles for high-quality, high-value testing. BMJ Evid.-Based Med. 2013, 18, 5–10. [Google Scholar] [CrossRef]
- Joosten, S.C.; Odeh, S.N.O.; Koch, A.; Buekers, N.; Aarts, M.J.B.; Baldewijns, M.M.L.L.; Van Neste, L.; van Kuijk, S.; Schouten, L.J.; van den Brandt, P.A.; et al. Development of a prognostic risk model for clear cell renal cell carcinoma by systematic evaluation of DNA methylation markers. Clin. Epigenet. 2021, 13, 103. [Google Scholar] [CrossRef]
- Peters, I.; Eggers, H.; Atschekzei, F.; Hennenlotter, J.; Waalkes, S.; Trankenschuh, W.; Grosshennig, A.; Merseburger, A.S.; Stenzl, A.; Kuczyk, M.A.; et al. GATA5 CpG island methylation in renal cell cancer: A potential biomarker for metastasis and disease progression. BJU Int. 2012, 110, E144-52. [Google Scholar] [CrossRef]
- R Core Team R. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. 2021. Available online: https://www.R-project.org/ (accessed on 15 July 2022).
- Team, R. RStudio: Integrated Development for R. RStudio, PBC, Boston, MA. Available online: http://www.rstudio.com/ (accessed on 5 January 2022).
- Van Buuren, S.; Groothuis-Oudshoorn, K. mice: Multivariate Imputation by Chained Equations in R. J. Stat. Softw. 2011, 45, 1–67. [Google Scholar] [CrossRef]
- R ClusterTools Package. Available online: https://github.com/vzoche-golob/clustertools/blob/master/description (accessed on 15 July 2022).
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Kuhn, M.; Wickham, H. Tidymodels: A Collection of Packages for Modeling and Machine Learning Using Tidyverse Principles. Available online: https://www.tidymodels.org (accessed on 1 April 2022).
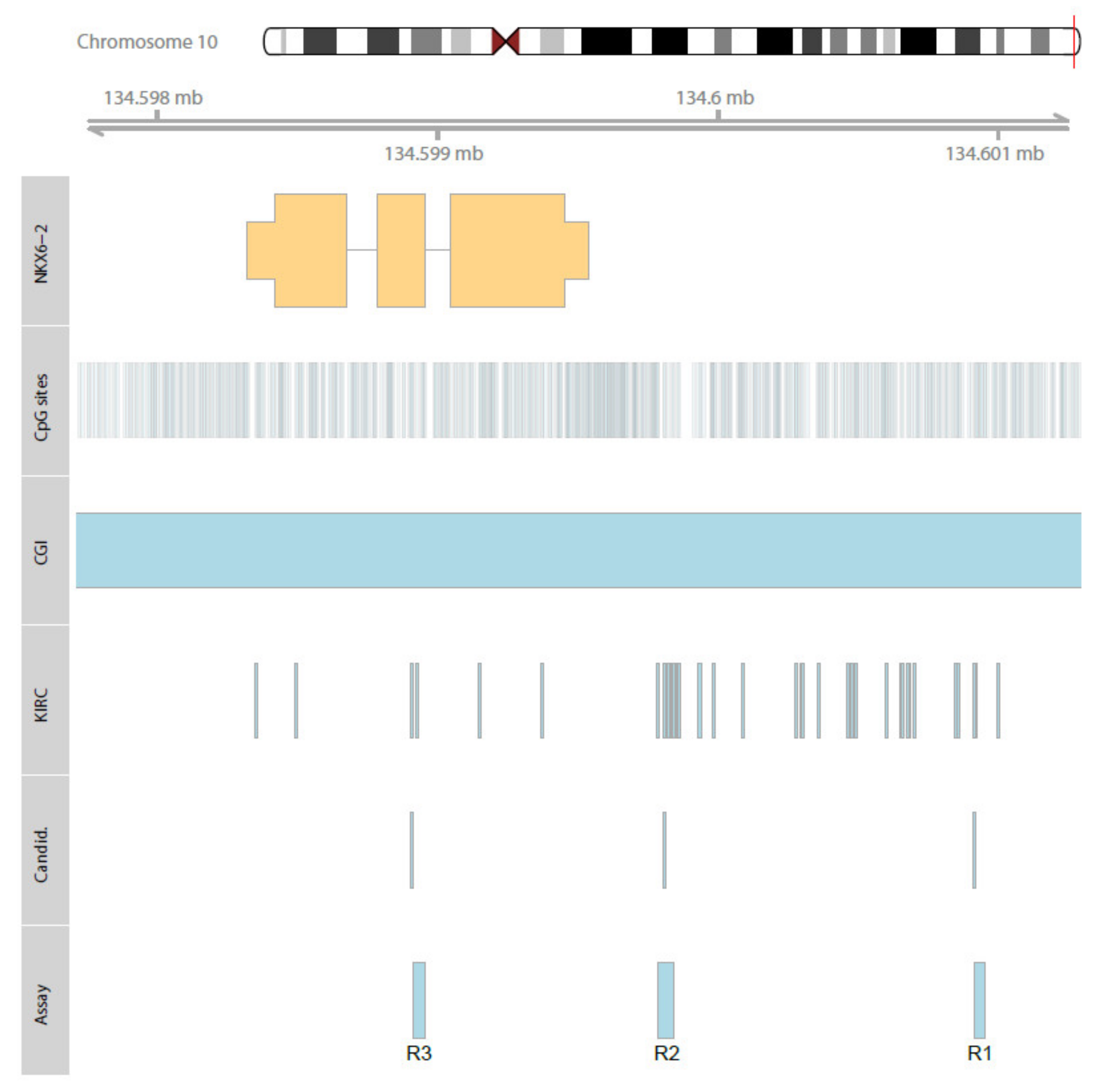

| Pyroassay | Gene | Chromosom | TCGA/KIRC Candidate | Genomic Position | AssayCG |
|---|---|---|---|---|---|
| NKX6-2 | 10 | cg06082548 | 134,598,909 | ||
| R3 | NKX6-2 | 10 | 134,598,942 | CG4 | |
| 134,598,945 | CG3 | ||||
| 134,598,948 | CG2 | ||||
| 134,598,952 | CG1 | ||||
| R2 | NKX6-2 | 10 | 134,599,807 | CG5 | |
| cg01384488 | 134,599,809 | CG4 | |||
| 134,599,823 | CG3 | ||||
| 134,599,836 | CG2 | ||||
| 134,599,841 | CG1 | ||||
| R1 | NKX6-2 | 10 | cg19701540 | 134,600,915 | CG1 |
| 134,600,919 | CG2 | ||||
| 134,600,922 | CG3 | ||||
| 134,600,932 | CG4 | ||||
| 134,600,934 | CG5 | ||||
| 134,600,938 | CG6 | ||||
| 134,600,949 | CG7 |
| Assay | Tumor Specific Hypermethylation | Metastatic Primary Cancer Hypermethylation | Metastatic Tissue Specific Hypermethylation | ||||||
|---|---|---|---|---|---|---|---|---|---|
| p-Value 1 | Mean Meth. (%) | OR (95% CI) | p-Value 2 | Mean Meth. (%) | OR (95% CI) | p-Value 2 | |||
| M0 | M+ | M0 | Mtx | ||||||
| R3 | 6.60 × 10−17 | 14.70 | 23.08 | 1.06 (1.03–1.10) | 3.37 × 10−4 | 14.14 | 23.05 | 1.08 (1.05–1.11) | 1.14 × 10−7 |
| R2 | 1.25 × 10−14 | 11.19 | 17.23 | 1.04 (1.01–1.07) | 0.008 | 10.46 | 19.81 | 1.09 (1.06–1.12) | 7.20 × 10−8 |
| R1 | 1.16 × 10−14 | 16.31 | 23.35 | 1.04 (1.01–1.07) | 0.006 | 15.82 | 24.81 | 1.05 (1.03–1.07) | 1.04 × 10−5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Serth, J.; Peters, I.; Katzendorn, O.; Dang, T.N.; Moog, J.; Balli, Z.; Reese, C.; Hennenlotter, J.; Grote, A.; Lafos, M.; et al. Identification of a Novel Renal Metastasis Associated CpG-Based DNA Methylation Signature (RMAMS). Int. J. Mol. Sci. 2022, 23, 11190. https://doi.org/10.3390/ijms231911190
Serth J, Peters I, Katzendorn O, Dang TN, Moog J, Balli Z, Reese C, Hennenlotter J, Grote A, Lafos M, et al. Identification of a Novel Renal Metastasis Associated CpG-Based DNA Methylation Signature (RMAMS). International Journal of Molecular Sciences. 2022; 23(19):11190. https://doi.org/10.3390/ijms231911190
Chicago/Turabian StyleSerth, Jürgen, Inga Peters, Olga Katzendorn, Tu N. Dang, Joana Moog, Zarife Balli, Christel Reese, Jörg Hennenlotter, Alexander Grote, Marcel Lafos, and et al. 2022. "Identification of a Novel Renal Metastasis Associated CpG-Based DNA Methylation Signature (RMAMS)" International Journal of Molecular Sciences 23, no. 19: 11190. https://doi.org/10.3390/ijms231911190
APA StyleSerth, J., Peters, I., Katzendorn, O., Dang, T. N., Moog, J., Balli, Z., Reese, C., Hennenlotter, J., Grote, A., Lafos, M., Tezval, H., & Kuczyk, M. A. (2022). Identification of a Novel Renal Metastasis Associated CpG-Based DNA Methylation Signature (RMAMS). International Journal of Molecular Sciences, 23(19), 11190. https://doi.org/10.3390/ijms231911190






