Polysaccharide from Salviae miltiorrhizae Radix et Rhizoma Attenuates the Progress of Obesity-Induced Non-Alcoholic Fatty Liver Disease through Modulating Intestinal Microbiota-Related Gut–Liver Axis
Abstract
:1. Introduction
2. Results
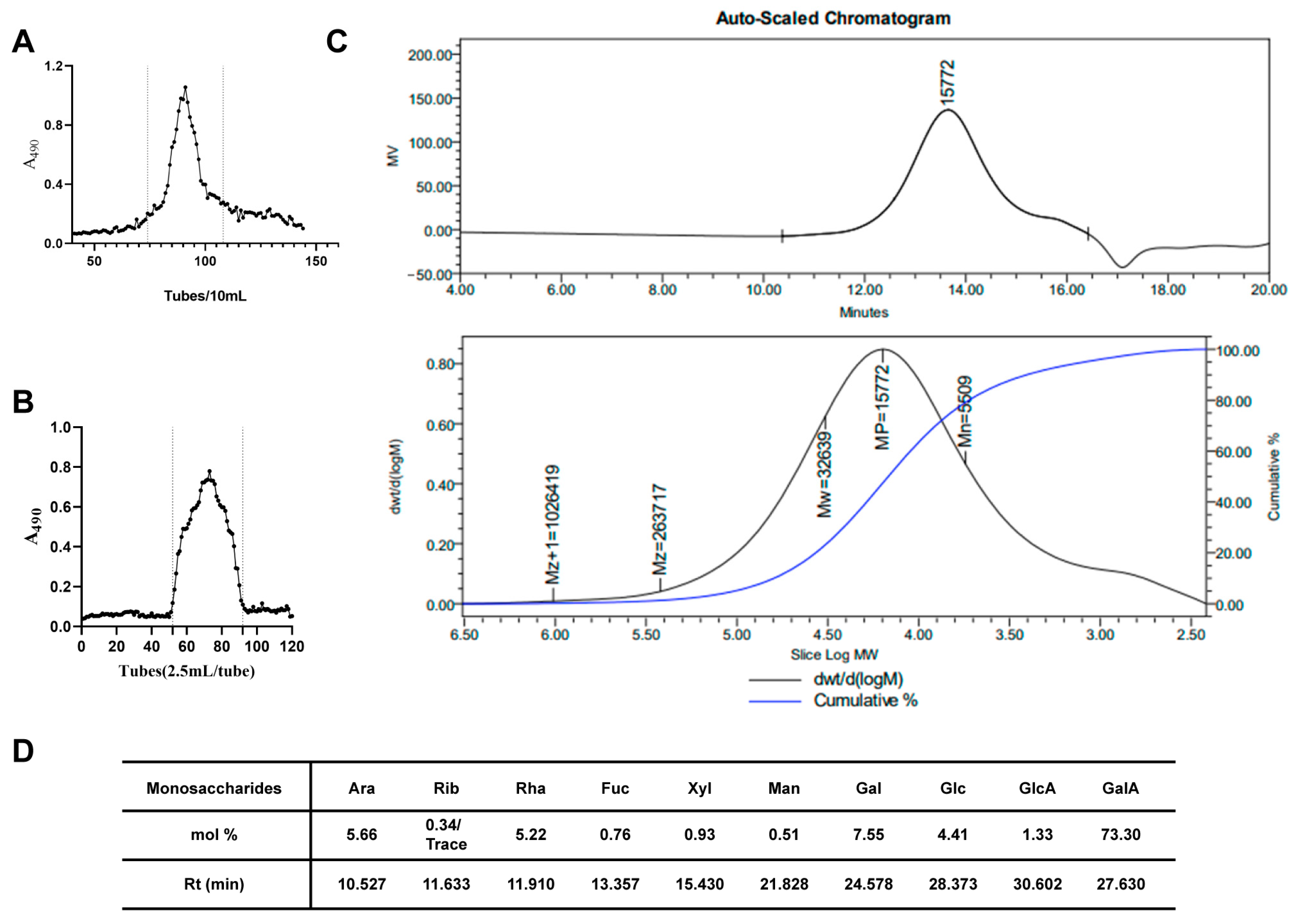
2.1. Isolation, Purification and Characterization of SMRR Polysaccharide
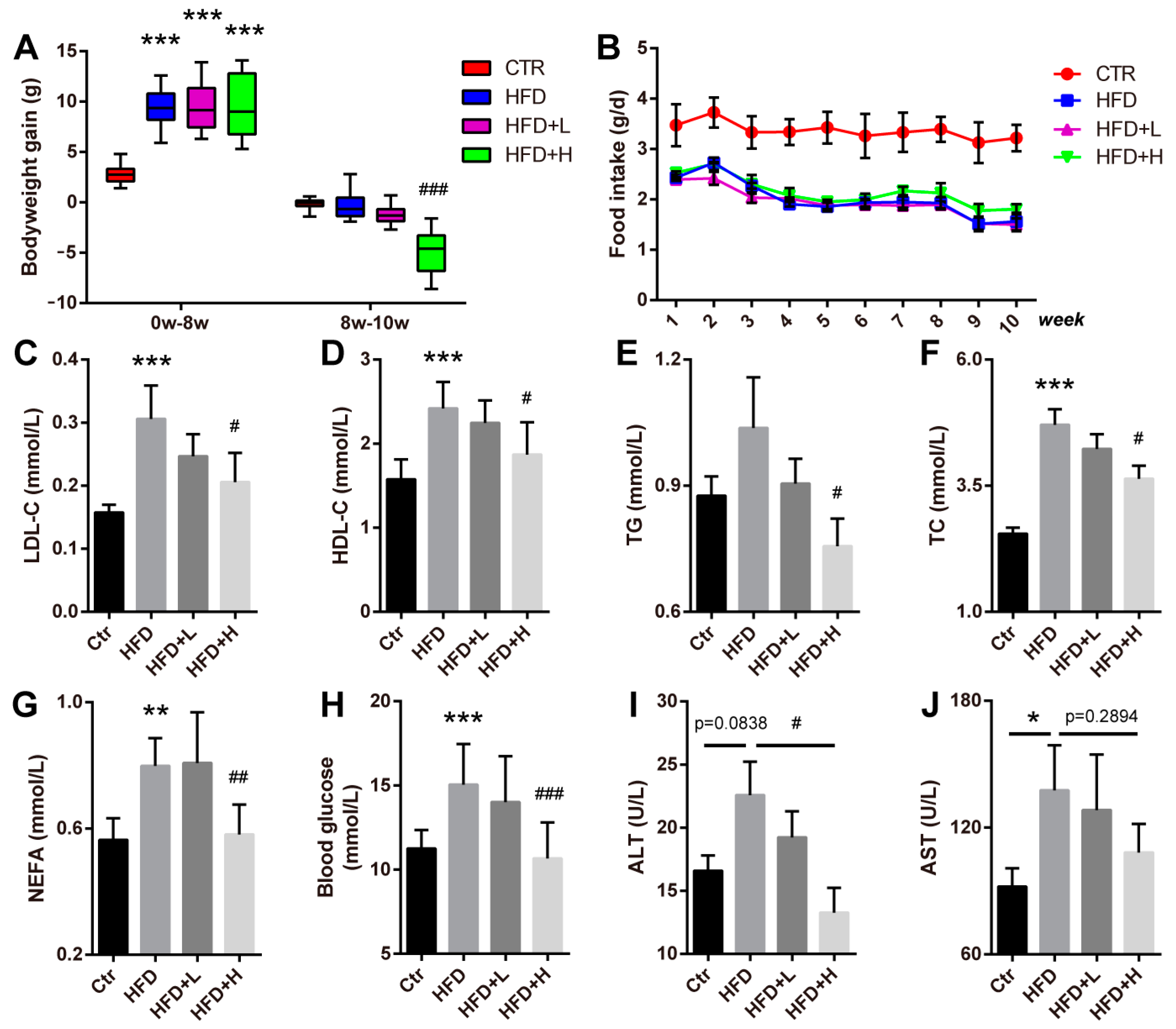
2.2. SMRR Polysaccharide Attenuates High-Fat-Diet-Induced Obesity
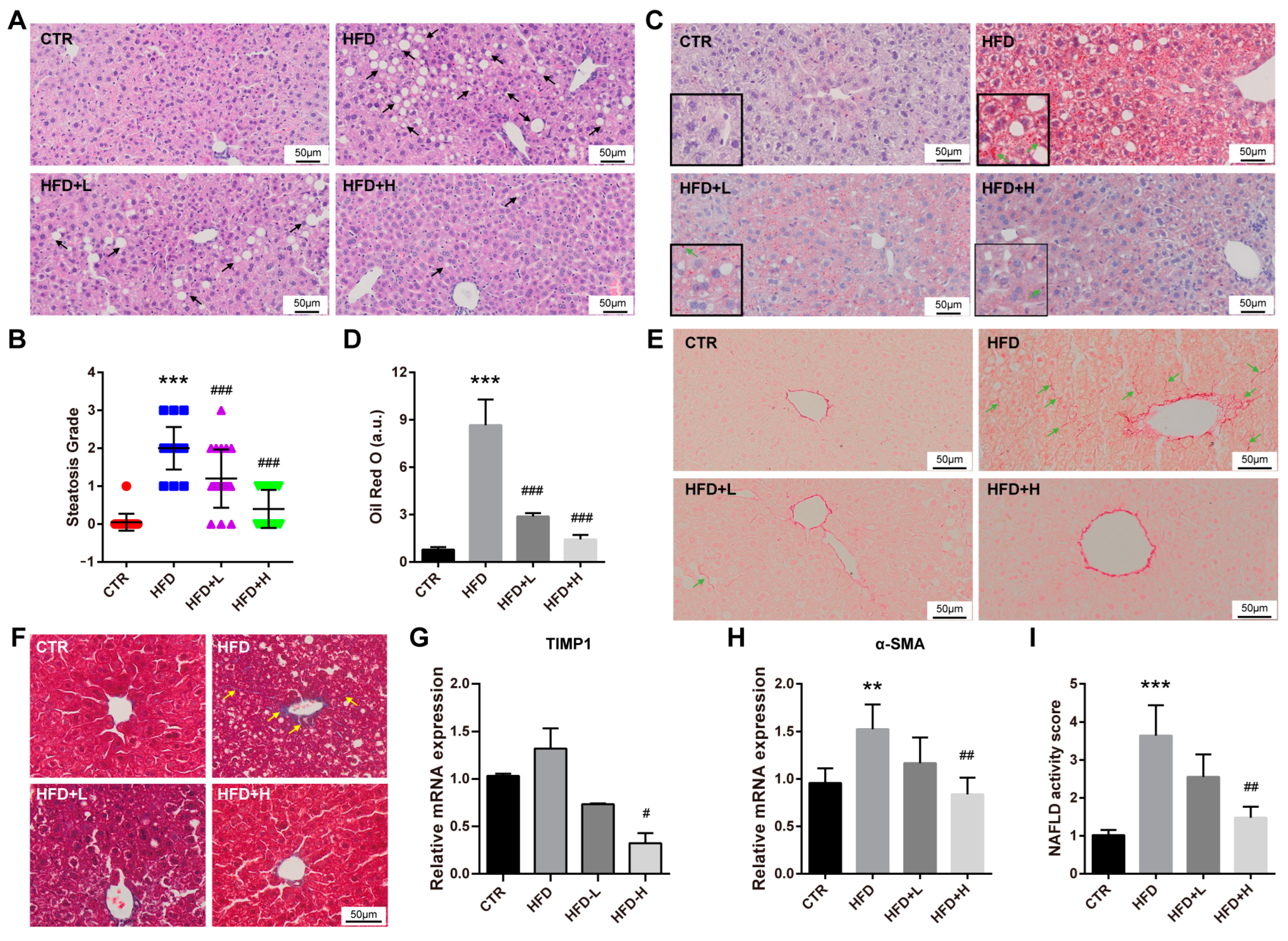
2.3. SMRR Polysaccharide Attenuates High-Fat-Diet-Induced NAFLD Progress
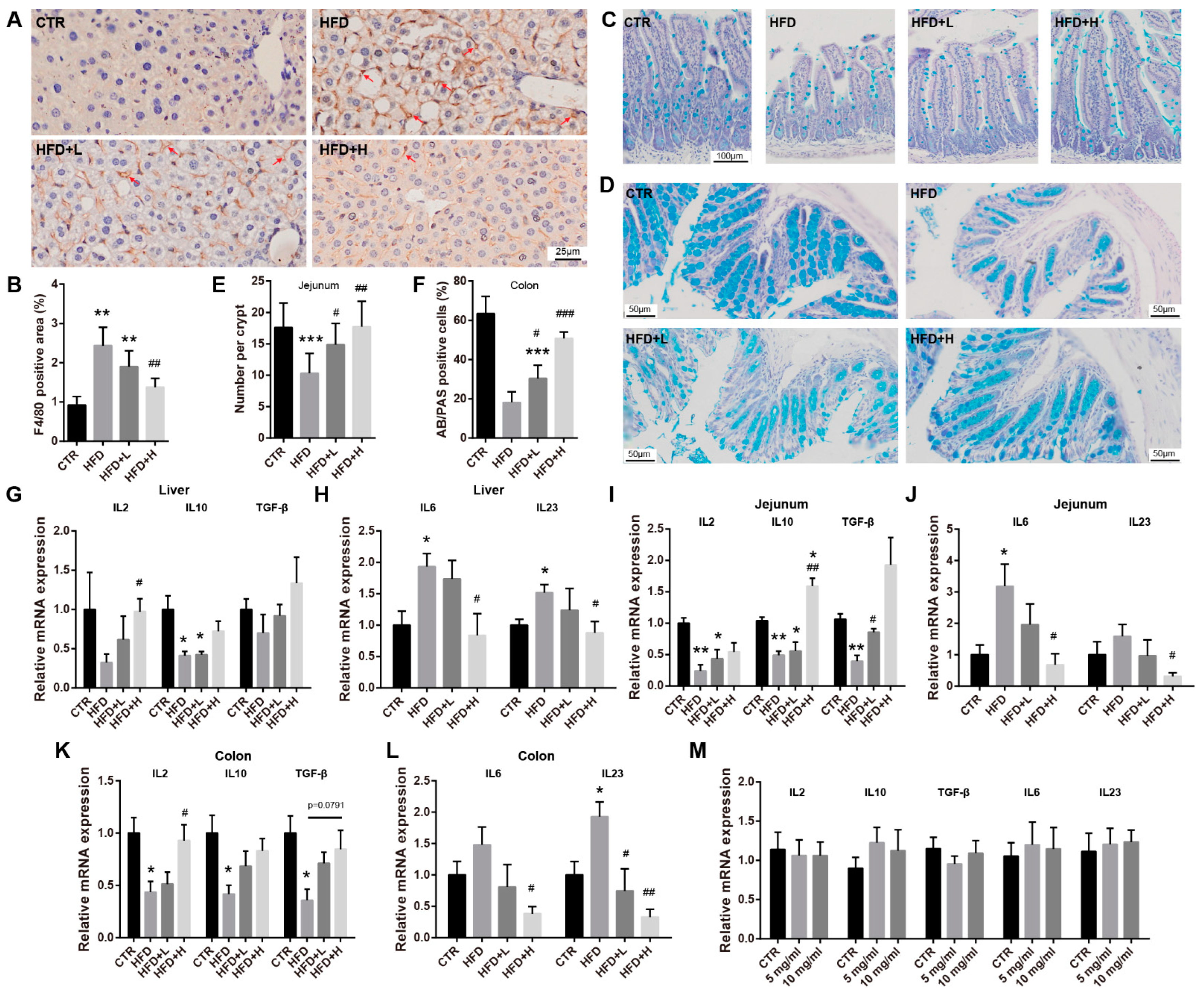
2.4. SMRR Polysaccharide Ameliorates Defects in Gut Structure and Permeability Induced by High-Fat Diet
2.5. SMRR Polysaccharide Attenuates Obesity-Induced Inflammation in Liver and Intestine
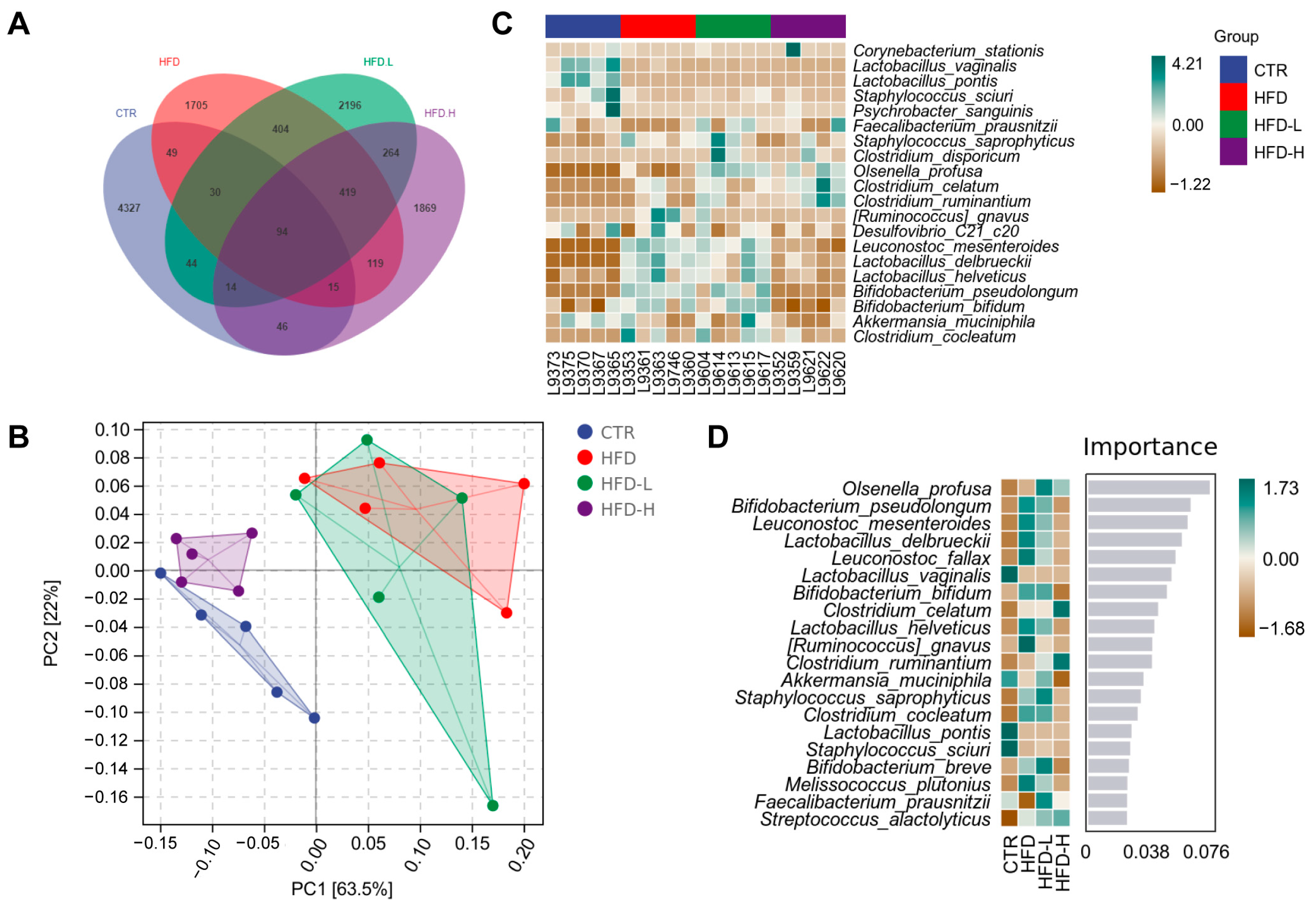
2.6. SMRR Polysaccharide Regulates Gut–Liver Axis through Modulating Gut Microbiota
3. Discussion
4. Materials and Methods
4.1. Isolation, Purification and Characterization of SMRR Polysaccharide
4.2. Animals and Treatments
4.3. Histological Staining
4.4. Immunohistochemical Staining
4.5. Quantitative Realtime PCR
4.6. Cell Culture
4.7. Enzyme-Linked Immunosorbent Assay (ELISA)
4.8. Gut Microbiota Analysis
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Albillos, A.; de Gottardi, A.; Rescigno, M. The gut-liver axis in liver disease: Pathophysiological basis for therapy. J. Hepatol. 2020, 72, 558–577. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.; Debelius, J.; Brenner, D.A.; Karin, M.; Loomba, R.; Schnabl, B.; Knight, R. The gut-liver axis and the intersection with the microbiome. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Compare, D.; Coccoli, P.; Rocco, A.; Nardone, O.M.; De Maria, S.; Carteni, M.; Nardone, G. Gut--liver axis: The impact of gut microbiota on non alcoholic fatty liver disease. Nutr. Metab. Cardiovasc. Dis. NMCD 2012, 22, 471–476. [Google Scholar] [CrossRef]
- Shepherd, H.A.; Selby, W.S.; Chapman, R.W.; Nolan, D.; Barbatis, C.; McGee, J.O.; Jewell, D.P. Ulcerative colitis and persistent liver dysfunction. Q. J. Med. 1983, 52, 503–513. [Google Scholar] [PubMed]
- Uko, V.; Thangada, S.; Radhakrishnan, K. Liver disorders in inflammatory bowel disease. Gastroenterol. Res. Pract. 2012, 2012, 642923. [Google Scholar] [CrossRef]
- Zou, Z.Y.; Shen, B.; Fan, J.G. Systematic Review With Meta-analysis: Epidemiology of Nonalcoholic Fatty Liver Disease in Patients With Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2019, 25, 1764–1772. [Google Scholar] [CrossRef]
- Ming, Z.; Sun, K.; Wu, Y.; Yang, Y.; Patrick, T.; Wu, Z. Interactions between Intestinal Microbiota and Host Immune Response in Inflammatory Bowel Disease. Front. Immunol. 2017, 8, 942. [Google Scholar]
- Szabo, G.; Bala, S.; Petrasek, J.; Gattu, A. Gut-liver axis and sensing microbes. Dig. Dis. 2010, 28, 737–744. [Google Scholar] [CrossRef]
- Siebler, J.; Galle, P.R.; Weber, M.M. The gut-liver-axis: Endotoxemia, inflammation, insulin resistance and NASH. J. Hepatol. 2008, 48, 1032–1034. [Google Scholar] [CrossRef]
- Yu, L.X.; Schwabe, R.F. The gut microbiome and liver cancer: Mechanisms and clinical translation. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 527–539. [Google Scholar] [CrossRef]
- Wong, V.W.; Adams, L.A.; de Ledinghen, V.; Wong, G.L.; Sookoian, S. Noninvasive biomarkers in NAFLD and NASH—current progress and future promise. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 461–478. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.M.; Marchesini, G.; Pinto-Cortez, H.; Petta, S.J.T. Epidemiology of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis: Implications for liver transplantation. Transplantation 2019, 103, 22–27. [Google Scholar] [CrossRef]
- Hardy, T.; Oakley, F.; Anstee, Q.M.; Day, C.P. Nonalcoholic fatty liver disease: Pathogenesis and disease spectrum. Annu. Rev. Pathol. Mech. Dis. 2016, 11, 451–496. [Google Scholar] [CrossRef] [PubMed]
- Polyzos, S.A.; Kountouras, J.; Mantzoros, C.S. Obesity and nonalcoholic fatty liver disease: From pathophysiology to therapeutics. Metab. Clin. Exp. 2019, 92, 82–97. [Google Scholar] [CrossRef] [PubMed]
- Magri, S.; Paduano, D.; Chicco, F.; Cingolani, A.; Farris, C.; Delogu, G.; Tumbarello, F.; Lai, M.; Melis, A.; Casula, L.; et al. Nonalcoholic fatty liver disease in patients with inflammatory bowel disease: Beyond the natural history. World J. Gastroenterol. WJG 2019, 25, 5676–5686. [Google Scholar] [CrossRef] [PubMed]
- Rahman, K.; Desai, C.; Iyer, S.S.; Thorn, N.E.; Kumar, P.; Liu, Y.; Smith, T.; Neish, A.S.; Li, H.; Tan, S.; et al. Loss of Junctional Adhesion Molecule A Promotes Severe Steatohepatitis in Mice on a Diet High in Saturated Fat, Fructose, and Cholesterol. Gastroenterology 2016, 151, 733–746.e12. [Google Scholar] [CrossRef] [PubMed]
- Chassaing, B.; Etienne-Mesmin, L.; Gewirtz, A.T. Microbiota-liver axis in hepatic disease. Hepatology 2014, 59, 328–339. [Google Scholar] [CrossRef]
- Federico, A.; Dallio, M.; Godos, J.; Loguercio, C.; Salomone, F. Targeting gut-liver axis for the treatment of nonalcoholic steatohepatitis: Translational and clinical evidence. Transl. Res. J. Lab. Clin. Med. 2016, 167, 116–124. [Google Scholar] [CrossRef]
- Bawa, M.; Saraswat, V.A. Gut-liver axis: Role of inflammasomes. J. Clin. Exp. Hepatol. 2013, 3, 141–149. [Google Scholar] [CrossRef]
- Kirpich, I.A.; Marsano, L.S.; McClain, C.J. Gut-liver axis, nutrition, and non-alcoholic fatty liver disease. Clin. Biochem. 2015, 48, 923–930. [Google Scholar] [CrossRef]
- Pang, H.; Wu, L.; Tang, Y.; Zhou, G.; Qu, C.; Duan, J.A. Chemical Analysis of the Herbal Medicine Salviae miltiorrhizae Radix et Rhizoma (Danshen). Molecules 2016, 21, 51. [Google Scholar] [CrossRef] [Green Version]
- Li, G.H.; Li, Y.R.; Jiao, P.; Zhao, Y.; Hu, H.X.; Lou, H.X.; Shen, T. Therapeutic Potential of Salviae Miltiorrhizae Radix et Rhizoma against Human Diseases Based on Activation of Nrf2-Mediated Antioxidant Defense System: Bioactive Constituents and Mechanism of Action. Oxidative Med. Cell. Longev. 2018, 2018, 7309073. [Google Scholar] [CrossRef] [PubMed]
- Su, C.Y.; Ming, Q.L.; Rahman, K.; Han, T.; Qin, L.P. Salvia miltiorrhiza: Traditional medicinal uses, chemistry, and pharmacology. Chin. J. Nat. Med. 2015, 13, 163–182. [Google Scholar] [CrossRef]
- Han, C.; Wei, Y.; Wang, X.; Cui, Y.; Shi, W. Salvia miltiorrhiza polysaccharides protect against lipopolysaccharide-induced liver injury by regulating NF-κb and Nrf2 pathway in mice. Food Agric. Immunol. 2019, 30, 979–994. [Google Scholar] [CrossRef]
- Wang, X.; Han, C.; Qin, J.; Wei, Y.; Qian, X. Pretreatment with Salvia miltiorrhiza Polysaccharides Protects from Lipopolysaccharides/d-Galactosamine-Induced Liver Injury in Mice Through Inhibiting TLR4/MyD88 Signaling Pathway. J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res. 2019, 39, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.G.; Fu, X.Q.; Cai, H.B.; Liu, Q.; Li, C.H.; Liu, Y.W.; Li, Y.J.; Liu, Z.F.; Song, Y.H.; Lv, Z.P. Proteomic Analysis of Protective Effects of Polysaccharides from Salvia miltiorrhiza Against Immunological Liver Injury in Mice. Phytother. Res. 2011, 25, 1087–1094. [Google Scholar] [CrossRef]
- Song, Y.H.; Liu, Q.; Lv, Z.P.; Chen, Y.Y.; Zhou, Y.C.; Sun, X.G. Protection of a polysaccharide from Salvia miltiorrhiza, a Chinese medicinal herb, against immunological liver injury in mice. Int. J. Biol. Macromol. 2008, 43, 170–175. [Google Scholar] [CrossRef]
- Brunt, E.M.; Wong, V.W.; Nobili, V.; Day, C.P.; Sookoian, S.; Maher, J.J.; Bugianesi, E.; Sirlin, C.B.; Neuschwander-Tetri, B.A.; Rinella, M.E. Nonalcoholic fatty liver disease. Nat. Rev. Dis. Primers 2015, 1, 15080. [Google Scholar] [CrossRef]
- Stal, P. Liver fibrosis in non-alcoholic fatty liver disease—diagnostic challenge with prognostic significance. World J. Gastroenterol. WJG 2015, 21, 11077–11087. [Google Scholar] [CrossRef]
- Huang, C.; Wei, X.; Luo, Q.; Xia, Y.; Pan, T.; He, J.; Jahangir, A.; Jia, L.; Liu, W.; Zou, Y.; et al. Loss of TRIM67 Attenuates the Progress of Obesity-Induced Non-Alcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2022, 23, 7475. [Google Scholar] [CrossRef]
- Zaldivar, M.M.; Pauels, K.; von Hundelshausen, P.; Berres, M.L.; Schmitz, P.; Bornemann, J.; Kowalska, M.A.; Gassler, N.; Streetz, K.L.; Weiskirchen, R.; et al. CXC chemokine ligand 4 (Cxcl4) is a platelet-derived mediator of experimental liver fibrosis. Hepatology 2010, 51, 1345–1353. [Google Scholar] [CrossRef] [PubMed]
- Brunt, E.M.; Kleiner, D.E.; Wilson, L.A.; Belt, P.; Neuschwander-Tetri, B.A. Nonalcoholic fatty liver disease (NAFLD) activity score and the histopathologic diagnosis in NAFLD: Distinct clinicopathologic meanings. Hepatology 2011, 53, 810–820. [Google Scholar] [CrossRef] [PubMed]
- Cockburn, D.W.; Koropatkin, N.M. Polysaccharide Degradation by the Intestinal Microbiota and Its Influence on Human Health and Disease. J. Mol. Biol. 2016, 428, 3230–3252. [Google Scholar] [CrossRef] [PubMed]
- Sherman, M.P. Intestinal microbes and obesity: A reality check. Commentary on f.B. Morel et al.: Can antibiotic treatment in preweaning rats alter body composition in adulthood? Neonatology 2013, 103, 190–192. [Google Scholar] [CrossRef]
- Winer, D.A.; Luck, H.; Tsai, S.; Winer, S. The Intestinal Immune System in Obesity and Insulin Resistance. Cell Metab. 2016, 23, 413–426. [Google Scholar] [CrossRef]
- Bradley, L.; Janet, S.; Amie, S. Association between increased intestinal permeability and disease: A systematic review. Adv. Integr. Med. 2018, 6, 23–24. [Google Scholar]
- Visser, J.; Rozing, J.; Sapone, A.; Lammers, K.; Fasano, A. Tight junctions, intestinal permeability, and autoimmunity: Celiac disease and type 1 diabetes paradigms. Ann. N. Y. Acad. Sci. 2009, 1165, 195–205. [Google Scholar] [CrossRef]
- Bischoff, S.C.; Barbara, G.; Buurman, W.; Ockhuizen, T.; Schulzke, J.D.; Serino, M.; Tilg, H.; Watson, A.; Wells, J.M. Intestinal permeability--a new target for disease prevention and therapy. BMC Gastroenterol. 2014, 14, 189. [Google Scholar] [CrossRef]
- Ferolla, S.M.; Armiliato, G.N.; Couto, C.A.; Ferrari, T.C. The role of intestinal bacteria overgrowth in obesity-related nonalcoholic fatty liver disease. Nutrients 2014, 6, 5583–5599. [Google Scholar] [CrossRef]
- Karczewski, J.; Sledzinska, E.; Baturo, A.; Jonczyk, I.; Maleszko, A.; Samborski, P.; Begier-Krasinska, B.; Dobrowolska, A. Obesity and inflammation. Eur. Cytokine Netw. 2018, 29, 83–94. [Google Scholar] [CrossRef]
- Peverill, W.; Powell, L.W.; Skoien, R. Evolving concepts in the pathogenesis of NASH: Beyond steatosis and inflammation. Int. J. Mol. Sci. 2014, 15, 8591–8638. [Google Scholar] [CrossRef]
- Dao, M.C.; Clement, K. Gut microbiota and obesity: Concepts relevant to clinical care. Eur. J. Intern. Med. 2018, 48, 18–24. [Google Scholar] [CrossRef] [Green Version]
- Al-Assal, K.; Martinez, A.C.; Torrinhas, R.S.; Cardinelli, C.; Dan, W. Gut Microbiota and obesity. Clin. Nutr. Exp. 2018, 20, 60–64. [Google Scholar] [CrossRef]
- Murray, B.R.; Rice, B.L.; Keith, D.A.; Myerscough, P.J.; Howell, J.; Floyd, A.G.; Mills, K.; Westoby, M. Species in the tail of rank–abundance curves. Ecology 1999, 80, 1806–1816. [Google Scholar]
- Chao, A. Nonparametric estimation of the number of classes in a population. Scand. J. Statist. 1984, 11, 265–270. [Google Scholar]
- Wang, S.; Schuurmans, D.; Peng, F.; Zhao, Y. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Michie, M.G. Use of the Bray-Curtis similarity measure in cluster analysis of foraminiferal data. J. Int. Assoc. Math. Geol. 1982, 14, 661–667. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ckhed, F.B.; Fulton, L.; Gordon, J.I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 2008, 3, 213–223. [Google Scholar] [CrossRef]
- Hildebrandt, M.A.; Hoffmann, C.; Sherrill-Mix, S.A.; Keilbaugh, S.A.; Hamady, M.; Chen, Y.Y.; Knight, R.; Ahima, R.S.; Bushman, F.; Wu, G.D. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology 2009, 137, 1716–1724.e2. [Google Scholar] [CrossRef]
- Petriz, B.A.; Castro, A.P.; Almeida, J.A.; Gomes, C.P.; Fernandes, G.R.; Kruger, R.H.; Pereira, R.W.; Franco, O.L. Exercise induction of gut microbiota modifications in obese, non-obese and hypertensive rats. BMC Genom. 2014, 15, 511. [Google Scholar] [CrossRef]
- Henke, M.T.; Kenny, D.J.; Cassilly, C.D.; Vlamakis, H.; Xavier, R.J.; Clardy, J. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn’s disease, produces an inflammatory polysaccharide. Proc. Natl. Acad. Sci. USA 2019, 116, 12672–12677. [Google Scholar] [CrossRef]
- He, K.; Ma, H.; Xu, H.; Zou, Z.; Feng, M.; Li, X.; Ye, X. Anti-hyperlipidemic effects of Rhizoma Coptidis alkaloids are achieved through modulation of the enterohepatic circulation of bile acids and cross-talk between the gut microbiota and the liver. J. Funct. Foods 2017, 35, 205–215. [Google Scholar] [CrossRef]
- Wu, C.H.; Ko, J.L.; Liao, J.M.; Huang, S.S.; Lin, M.Y.; Lee, L.H.; Chang, L.Y.; Ou, C.C. D-methionine alleviates cisplatin-induced mucositis by restoring the gut microbiota structure and improving intestinal inflammation. Ther. Adv. Med. Oncol. 2019, 11, 1758835918821021. [Google Scholar] [CrossRef]
- Chan, Y.K.; Brar, M.S.; Kirjavainen, P.V.; Chen, Y.; Peng, J.; Li, D.; Leung, F.C.-C.; El-Nezami, H. High fat diet induced atherosclerosis is accompanied with low colonic bacterial diversity and altered abundances that correlates with plaque size, plasma A-FABP and cholesterol: A pilot study of high fat diet and its intervention with Lactobacillus rhamnosus GG (LGG) or telmisartan in ApoE−/− mice. BMC Microbiol. 2016, 16, 1–13. [Google Scholar]
- Yeom, Y.; Kim, B.S.; Kim, S.J.; Kim, Y. Sasa quelpaertensis leaf extract regulates microbial dysbiosis by modulating the composition and diversity of the microbiota in dextran sulfate sodium-induced colitis mice. BMC Complement. Altern. Med. 2016, 16, 481. [Google Scholar] [CrossRef] [Green Version]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Li, Z.M.; Xu, S.W.; Liu, P.Q. Salvia miltiorrhizaBurge (Danshen): A golden herbal medicine in cardiovascular therapeutics. Acta Pharmacol. Sin. 2018, 39, 802–824. [Google Scholar] [CrossRef]
- Chen, W.; Wang, N.; Li, R.C.; Xu, G.F.; Bao, G.; Jiang, H.T.; Wang, M.D. Salvianolic acid B renders glioma cells more sensitive to radiation via Fis-1-mediated mitochondrial dysfunction. Biomed. Pharmacother. Biomed. Pharmacother. 2018, 107, 1230–1236. [Google Scholar] [CrossRef]
- Li, W.; Zhao, F.; Pan, J.; Qu, H. Influence of ethanol concentration of extraction solvent on metabolite profiling for Salviae Miltiorrhizae Radix et Rhizoma extract by 1H NMR spectroscopy and multivariate data analysis. Process Biochem. 2020, 97, 158–167. [Google Scholar] [CrossRef]
- Wang, R.N.; Zhao, H.C.; Huang, J.Y.; Wang, H.L.; Di, L.Q. Challenges and strategies in progress of drug delivery system for traditional Chinese medicine Salviae Miltiorrhizae Radix et Rhizoma (Danshen). Chin. Herb. Med. 2020, 13, 78–79. [Google Scholar] [CrossRef]
- Zhang, W.; Zheng, L.; Zhang, Z.; Hai, C.X. Protective effect of a water-soluble polysaccharide from Salvia miltiorrhiza Bunge on insulin resistance in rats. Carbohydr. Polym. 2012, 89, 890–898. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Willför, R.S.; Xu, C. A review of bioactive plant polysaccharides: Biological activities, functionalization, and biomedical applications. Bioact. Carbohydr. Diet. Fibre 2015, 5, 31–61. [Google Scholar] [CrossRef]
- Foster, J.A.; Rinaman, L.; Cryan, J.F. Stress & the gut-brain axis: Regulation by the microbiome. Neurobiol. Stress 2017, 7, 124–136. [Google Scholar]
- Marsland, B.J.; Trompette, A.; Gollwitzer, E.S. The Gut-Lung Axis in Respiratory Disease. Ann. Am. Thorac. Soc. 2015, 12 (Suppl. S2), S150–S156. [Google Scholar] [CrossRef]
- Salem, I.; Ramser, A.; Isham, N.; Ghannoum, M.A. The gut microbiome as a major regulator of the gut-skin axis. Front. Microbiol. 2018, 9, 1459. [Google Scholar] [CrossRef]
- Li, H.; Xie, Z.; Zhang, Y.; Liu, Y.; Niu, A.; Liu, Y.; Zhang, L.; Guan, L. Rosa rugosa polysaccharide attenuates alcoholic liver disease in mice through the gut-liver axis. Food Biosci. 2021, 44, 101385. [Google Scholar] [CrossRef]
- Jiang, W.; Zhu, H.; Liu, C.; Hu, B.; Guo, Y.; Cheng, Y.; Qian, H. In-depth investigation of the mechanisms of Echinacea purpurea polysaccharide mitigating alcoholic liver injury in mice via gut microbiota informatics and liver metabolomics. Int. J. Biol. Macromol. 2022, 209, 1327–1338. [Google Scholar] [CrossRef]
- Wang, X.; Shi, L.; Wang, X.; Feng, Y.; Wang, Y. MDG-1, an Ophiopogon polysaccharide, restrains process of non-alcoholic fatty liver disease via modulating the gut-liver axis. Int. J. Biol. Macromol. 2019, 141, 1013–1021. [Google Scholar] [CrossRef]
- Mushref, M.A.; Srinivasan, S. Effect of high fat-diet and obesity on gastrointestinal motility. Ann. Transl. Med. 2013, 1, 14. [Google Scholar] [PubMed]
- Ahmad, R.; Rah, B.; Bastola, D.; Dhawan, P.; Singh, A.B. Obesity-induces Organ and Tissue Specific Tight Junction Restructuring and Barrier Deregulation by Claudin Switching. Sci. Rep. 2017, 7, 5125. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Moon, K.M.; Kim, C.Y. Tight Junction in the Intestinal Epithelium: Its Association with Diseases and Regulation by Phytochemicals. J. Immunol. Res. 2018, 2018, 2645465. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.R.; Lee, S.H.; Kim, B.; Nam, M.H.; Ahn, Y.K.; Park, Y.M.; Jeong, S.M.; Park, M.J.; Song, K.B.; Lee, S.Y.; et al. Ruminococcus gnavus ameliorates atopic dermatitis by enhancing Treg cell and metabolites in BALB/c mice. Pediatric Allergy Immunol. Off. Publ. Eur. Soc. Pediatric Allergy Immunol. 2022, 33, e13678. [Google Scholar] [CrossRef]
- Jie, Z.; Yu, X.; Liu, Y.; Sun, L.; Chen, P.; Ding, Q.; Gao, Y.; Zhang, X.; Yu, M.; Liu, Y.; et al. The Baseline Gut Microbiota Directs Dieting-Induced Weight Loss Trajectories. Gastroenterology 2021, 160, 2029–2042.e16. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Wang, Y.; Kang, W.; Zhou, J.; Dong, R.; Feng, Q. Chitosan attenuates obesity by modifying the intestinal microbiota and increasing serum leptin levels in mice. J. Funct. Foods 2020, 64, 103659. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, S.; Tang, D.; Dong, R.; Feng, Q. Chitosan Oligosaccharide Ameliorates Metabolic Syndrome Induced by Overnutrition via Altering Intestinal Microbiota. Front. Nutr. 2021, 725. [Google Scholar] [CrossRef]
- Liu, S.; Li, F.; Zhang, X. Structural modulation of gut microbiota reveals Coix seed contributes to weight loss in mice. Appl. Microbiol. Biotechnol. 2019, 103, 5311–5321. [Google Scholar] [CrossRef]
- Liu, S.; Qin, P.; Wang, J. High-Fat Diet Alters the Intestinal Microbiota in Streptozotocin-Induced Type 2 Diabetic Mice. Microorganisms 2019, 7, 176. [Google Scholar] [CrossRef]
- Dao, M.C.; Everard, A.; Aron-Wisnewsky, J.; Sokolovska, N.; Prifti, E.; Verger, E.O.; Kayser, B.D.; Levenez, F.; Chilloux, J.; Hoyles, L.; et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: Relationship with gut microbiome richness and ecology. Gut 2016, 65, 426–436. [Google Scholar] [CrossRef]
- Minami, J.-I.; Kondo, S.; Yanagisawa, N.; Odamaki, T.; Xiao, J.-Z.; Abe, F.; Nakajima, S.; Hamamoto, Y.; Saitoh, S.; Shimoda, T. Oral administration of Bifidobacterium breve B-3 modifies metabolic functions in adults with obese tendencies in a randomised controlled trial. J. Nutr. Sci. 2015, 4, 2015. [Google Scholar] [CrossRef]
- Choi, S.Y.; Ryu, S.H.; Park, J.I.; Jeong, E.S.; Park, J.H.; Ham, S.H.; Jeon, H.Y.; Kim, J.Y.; Kyeong, I.G.; Kim, D.G. Anti-obesity effect of robusta fermented with Leuconostoc mesenteroides in high-fat diet-induced obese mice. Exp. Ther. Med. 2017, 14, 3761–3767. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhao, Y.; Liu, L.; Li, M. Anti-hyperlipidemia efficacy of Lactobacillus delbrueckii on blood lipids and gut microbiota in high-fat diet-fed mice. Int. J. Clin. Exp. Med. 2018, 11, 8453–8459. [Google Scholar]
- Moya-Pérez, A.; Neef, A.; Sanz, Y. Bifidobacterium pseudocatenulatum CECT 7765 reduces obesity-associated inflammation by restoring the lymphocyte-macrophage balance and gut microbiota structure in high-fat diet-fed mice. PLoS ONE 2015, 10, e0126976. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Jin, H.; Oh, S.Y.; Ji, G.E. Anti-obese effects of two Lactobacilli and two Bifidobacteria on ICR mice fed on a high fat diet. Biochem. Biophys. Res. Commun. 2016, 480, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Peng, X.; Pang, D.-J.; Li, J.; Paulsen, B.-S.; Rise, F.; Chen, Y.-L.; Chen, Z.-L.; Jia, R.-Y.; Li, L.-X.; et al. Pectic polysaccharide from Nelumbo nucifera leaves promotes intestinal antioxidant defense in vitro and in vivo. Food Funct. 2021, 12, 10828–10841. [Google Scholar] [CrossRef]
- DuBois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.T.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Huang, C.; Cao, X.; Chen, X.; Fu, Y.; Zhu, Y.; Chen, Z.; Luo, Q.; Li, L.; Song, X.; Jia, R.; et al. A pectic polysaccharide from Ligusticum chuanxiong promotes intestine antioxidant defense in aged mice. Carbohydr. Polym. 2017, 174, 915–922. [Google Scholar] [CrossRef]
- Chambers, R.E.; Clamp, J.R. An assessment of methanolysis and other factors used in the analysis of carbohydrate-containing materials. Biochem. J. 1971, 125, 1009–1018. [Google Scholar] [CrossRef] [Green Version]
- Austarheim, I.; Christensen, B.E.; Hegna, I.K.; Petersen, B.O.; Duus, J.O.; Bye, R.; Michaelsen, T.E.; Diallo, D.; Inngjerdingen, M.; Paulsen, B.S. Chemical and biological characterization of pectin-like polysaccharides from the bark of the Malian medicinal tree Cola cordifolia. Carbohydr. Polym. 2012, 89, 259–268. [Google Scholar] [CrossRef]
- Mehlem, A.; Hagberg, C.E.; Muhl, L.; Eriksson, U.; Falkevall, A. Imaging of neutral lipids by oil red O for analyzing the metabolic status in health and disease. Nat. Protoc. 2013, 8, 1149–1154. [Google Scholar] [CrossRef]
- Tandra, S.; Yeh, M.M.; Brunt, E.M.; Vuppalanchi, R.; Cummings, O.W.; Unalp-Arida, A.; Wilson, L.A.; Chalasani, N. Presence and significance of microvesicular steatosis in nonalcoholic fatty liver disease. J. Hepatol. 2011, 55, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet, J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Koljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef] [PubMed]
- Ramette, A. Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 2007, 62, 142–160. [Google Scholar] [CrossRef] [Green Version]
- Zaura, E.; Keijser, B.J.; Huse, S.M.; Crielaard, W. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009, 9, 259. [Google Scholar] [CrossRef] [Green Version]


| Gene | Primer | Sequence |
|---|---|---|
| mouse β-actin NM_007393 | F | 5′-AGAGGGAAATCGTGCGTGAC-3′ |
| R | 5′-CAATAGTGATGACCTGGCCGT-3′ | |
| mouse IL-2 NM_008366 | F | 5′-CCTGAGCAGGATGGAGAATTACA-3′ |
| R | 5′-TCCAGAACATGCCGCAGAG-3′ | |
| mouse IL-6 NM_031168 | F | 5′-CTTCCATCCAGTTGCCTTCTTG-3′ |
| R | 5′-AATTAAGCCTCCGACTTGTGAAG-3′ | |
| mouse IL-10 NM_010548 | F | 5′-AAGGGTTACTTGGGTTGCCA-3′ |
| R | 5′-CCTGGGGCATCACTTCTACC-3′ | |
| mouse IL-21 NM_001291041 | F | 5′-GCATGGAGAGGACCCTTGTC-3′ |
| R | 5′-CTAATCAGGAGGCGATCTGGC-3′ | |
| mouse IL-23 NM_031252 | F | 5′-ATGCTGGATTGCAGAGCAGTA-3′ |
| R | 5′-ACGGGGCACATTATTTTTAGTCT-3′ | |
| mouse TGF-β NM_011577 | F | 5′-GTGTGGAGCAACATGTGGAACTCTA-3′ |
| R | 5′-CGCTGAATCGAAAGCCCTGTA-3′ | |
| mouse Claudin-1 NM_016674 | F | 5′-TGGTAATTGGCATCCTGCTG-3′ |
| R | 5′-CAGCCATCCACATCTTCTGC-3′ | |
| mouse Occludin NM_008756 | F | 5′-GTACCCACCAGTGACCAACA-3′ |
| R | 5′-GTTGCTGGAGCTTAGCCTGT-3′ | |
| mouse ZO-1 NM_001163574 | F | 5′-CGAGGCATCATCCCAAATAAGAAC-3′ |
| R | 5′-TCCAGAAGTCTGCCCGATCAC-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Lan, X.; Peng, X.; Shi, S.; Zhao, Y.; Liu, W.; Luo, Q.; Jia, L.; Feng, B.; Chen, Z.; et al. Polysaccharide from Salviae miltiorrhizae Radix et Rhizoma Attenuates the Progress of Obesity-Induced Non-Alcoholic Fatty Liver Disease through Modulating Intestinal Microbiota-Related Gut–Liver Axis. Int. J. Mol. Sci. 2022, 23, 10620. https://doi.org/10.3390/ijms231810620
Li L, Lan X, Peng X, Shi S, Zhao Y, Liu W, Luo Q, Jia L, Feng B, Chen Z, et al. Polysaccharide from Salviae miltiorrhizae Radix et Rhizoma Attenuates the Progress of Obesity-Induced Non-Alcoholic Fatty Liver Disease through Modulating Intestinal Microbiota-Related Gut–Liver Axis. International Journal of Molecular Sciences. 2022; 23(18):10620. https://doi.org/10.3390/ijms231810620
Chicago/Turabian StyleLi, Lixia, Xinting Lan, Xi Peng, Shuai Shi, Yanlin Zhao, Wentao Liu, Qihui Luo, Lanlan Jia, Bin Feng, Zhengli Chen, and et al. 2022. "Polysaccharide from Salviae miltiorrhizae Radix et Rhizoma Attenuates the Progress of Obesity-Induced Non-Alcoholic Fatty Liver Disease through Modulating Intestinal Microbiota-Related Gut–Liver Axis" International Journal of Molecular Sciences 23, no. 18: 10620. https://doi.org/10.3390/ijms231810620
APA StyleLi, L., Lan, X., Peng, X., Shi, S., Zhao, Y., Liu, W., Luo, Q., Jia, L., Feng, B., Chen, Z., Zou, Y., & Huang, C. (2022). Polysaccharide from Salviae miltiorrhizae Radix et Rhizoma Attenuates the Progress of Obesity-Induced Non-Alcoholic Fatty Liver Disease through Modulating Intestinal Microbiota-Related Gut–Liver Axis. International Journal of Molecular Sciences, 23(18), 10620. https://doi.org/10.3390/ijms231810620






