Leaf Bacteriome in Sugar Beet Shows Differential Response against Beet curly top virus during Resistant and Susceptible Interactions
Abstract
:1. Introduction
2. Results
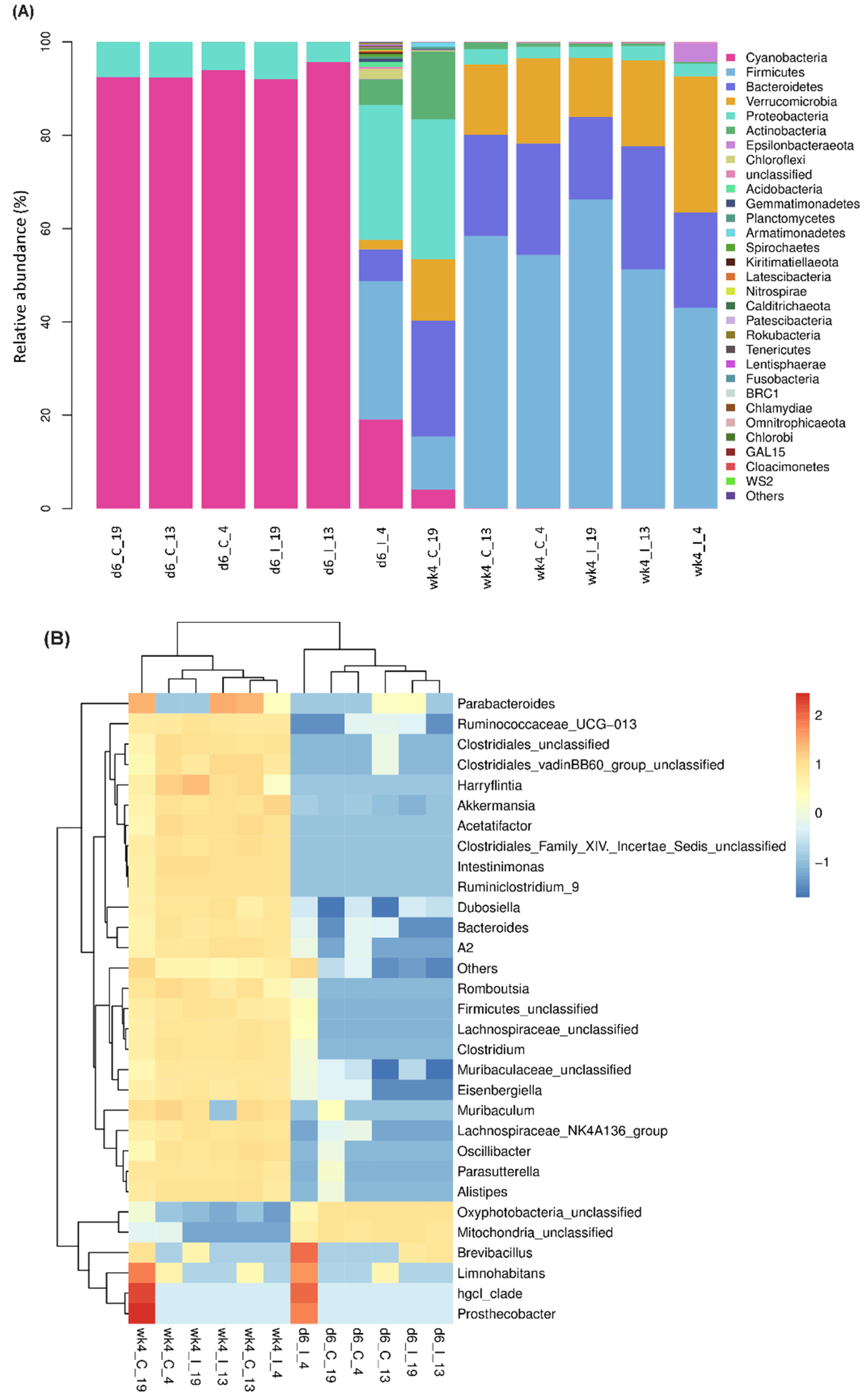
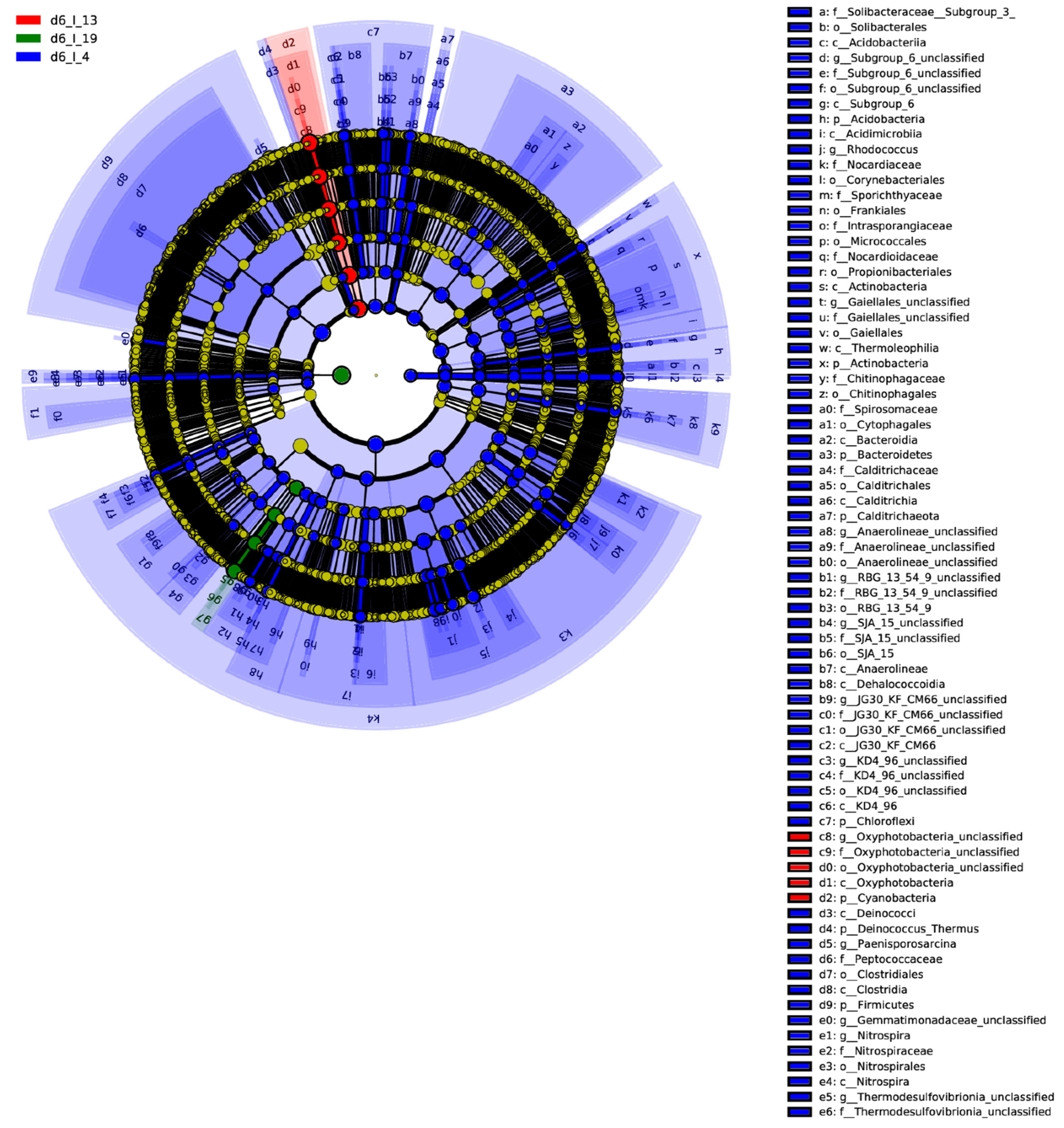
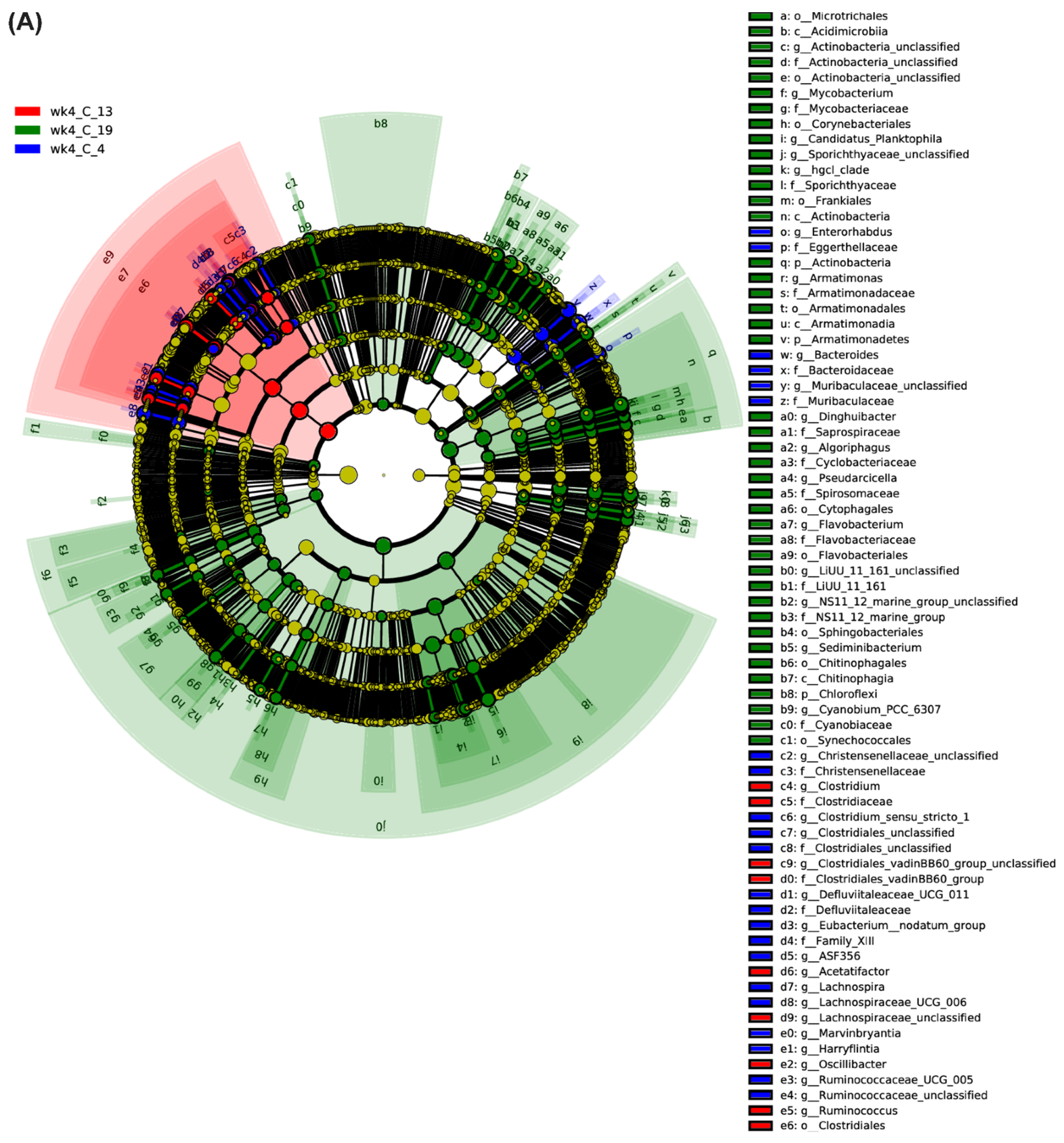
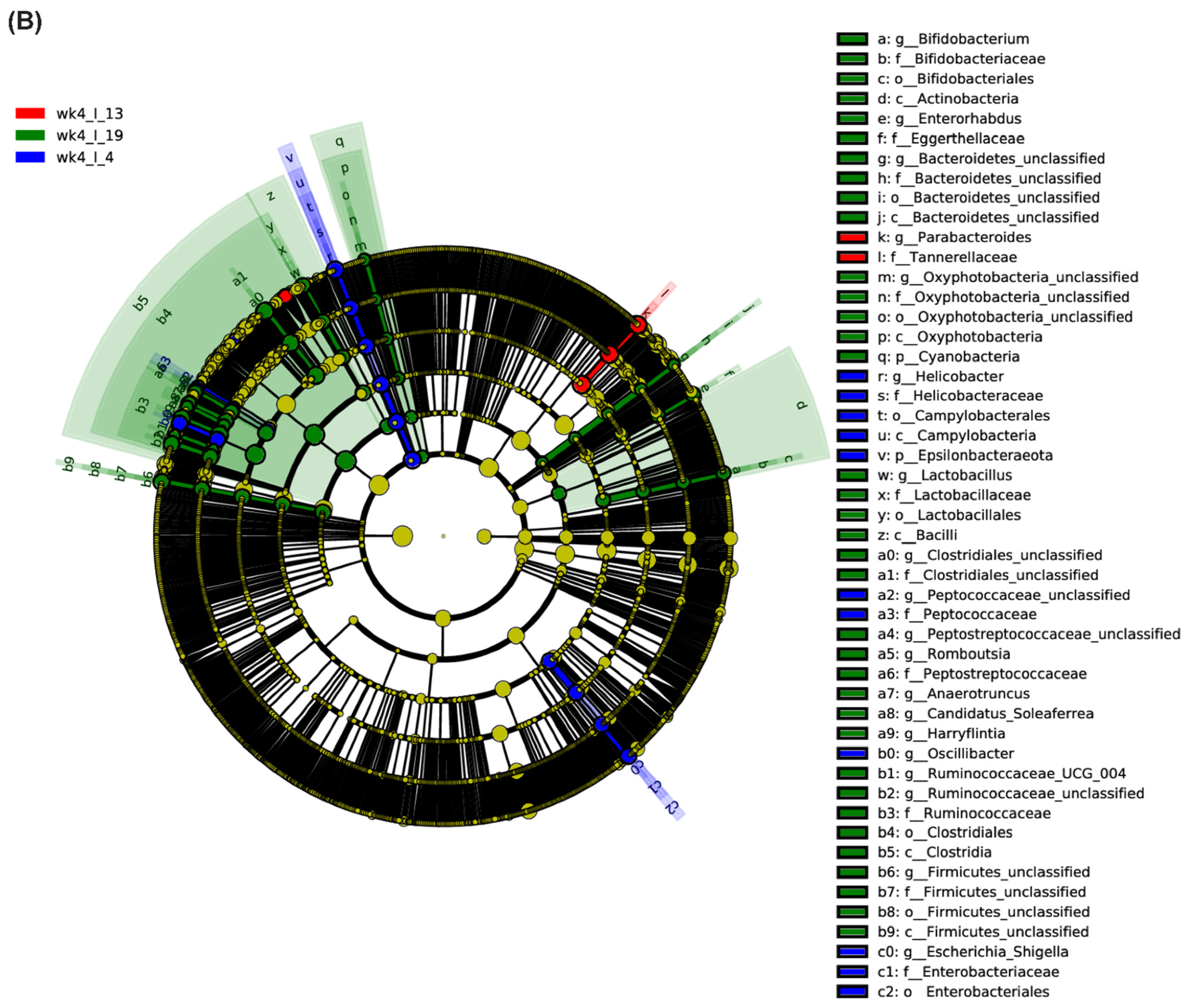
2.1. Leaf Bacterial Phyla and Genera Showed Significant Changes between the Resistant and Susceptible Lines with BCTV Infection
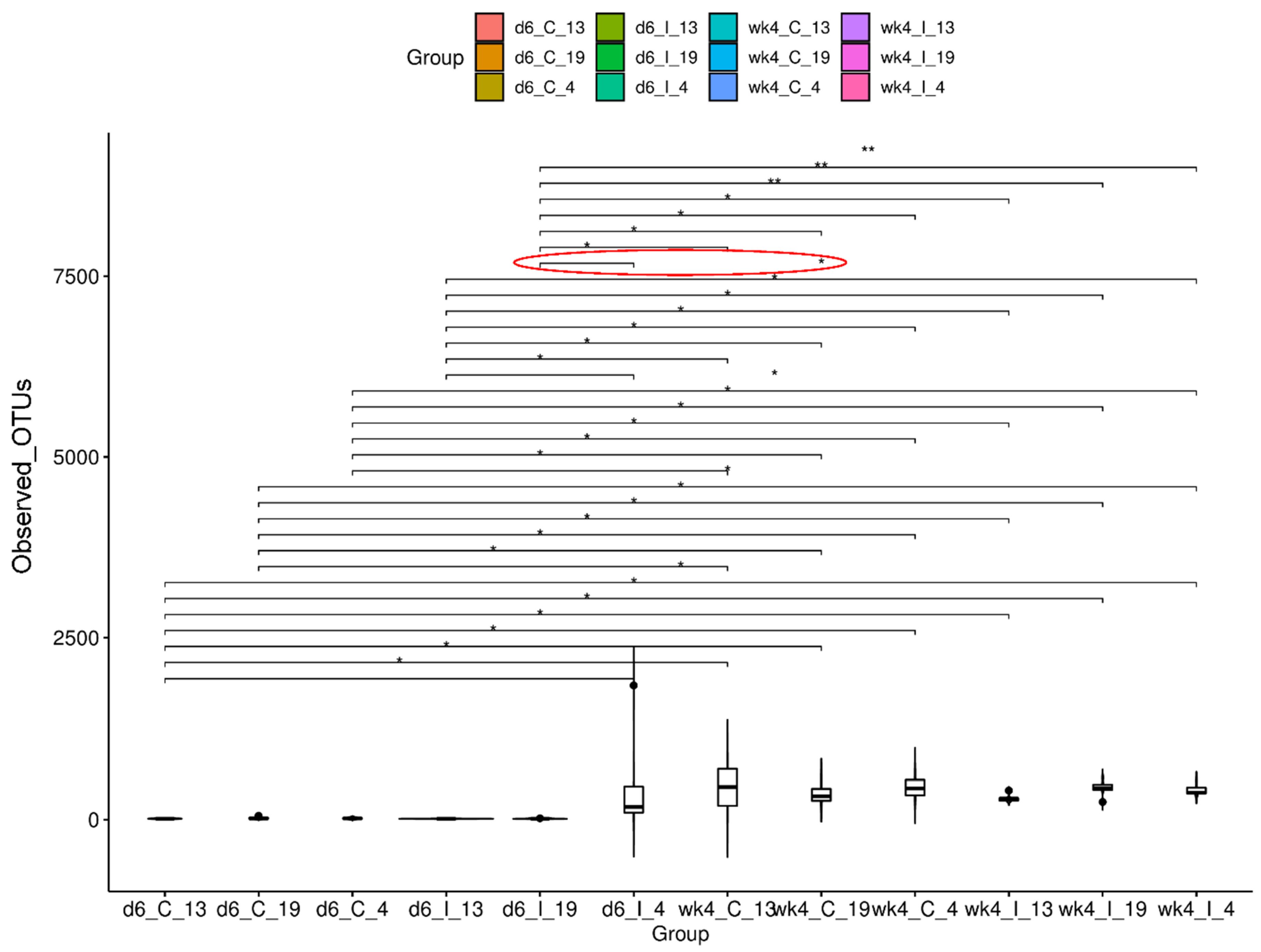
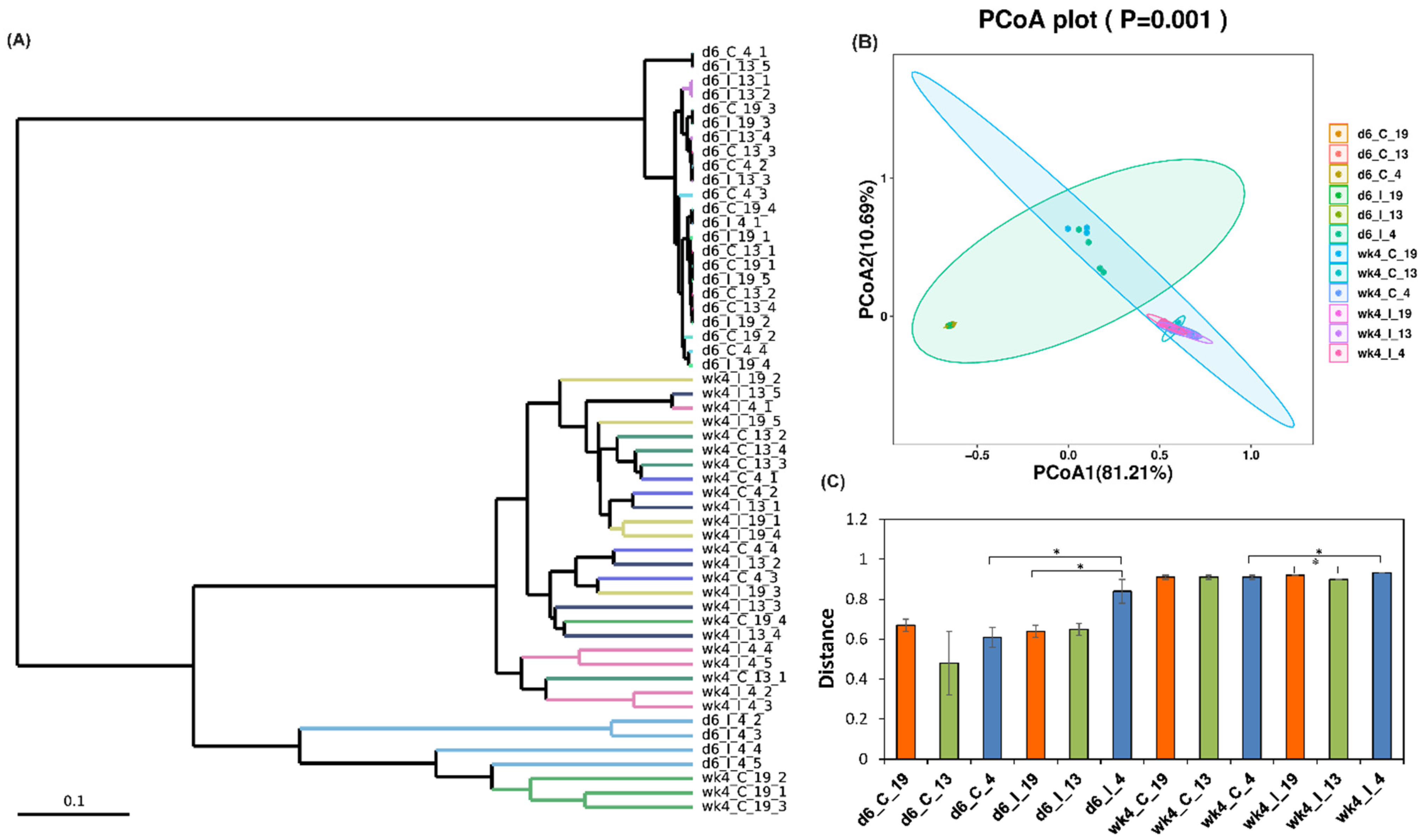
2.2. Leaf Bacterial Diversity Was Significantly Altered following BCTV Infection in the Resistant vs. Susceptible Lines
2.2.1. Alpha Diversity of Samples
2.2.2. Beta Diversity of Samples
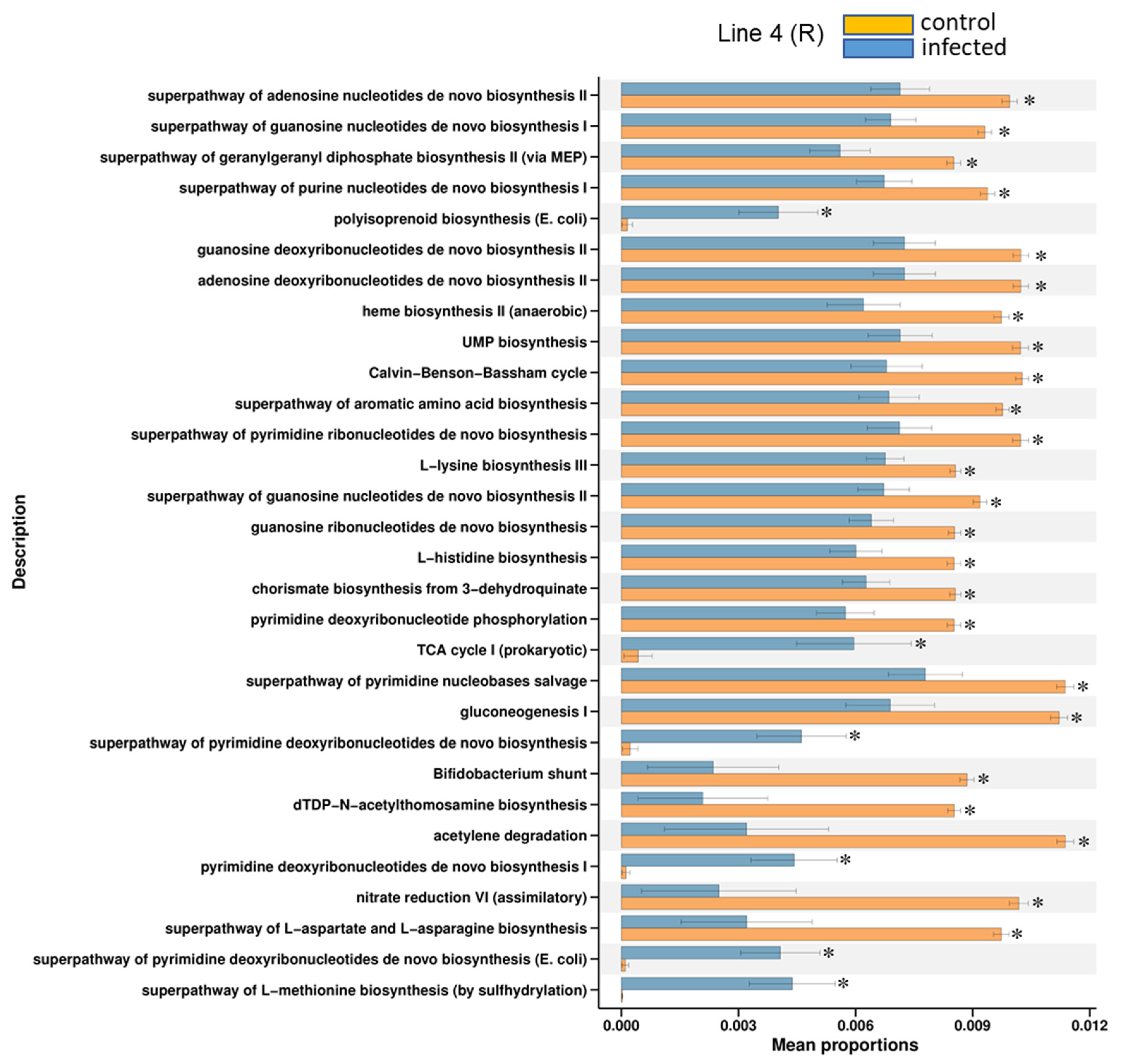
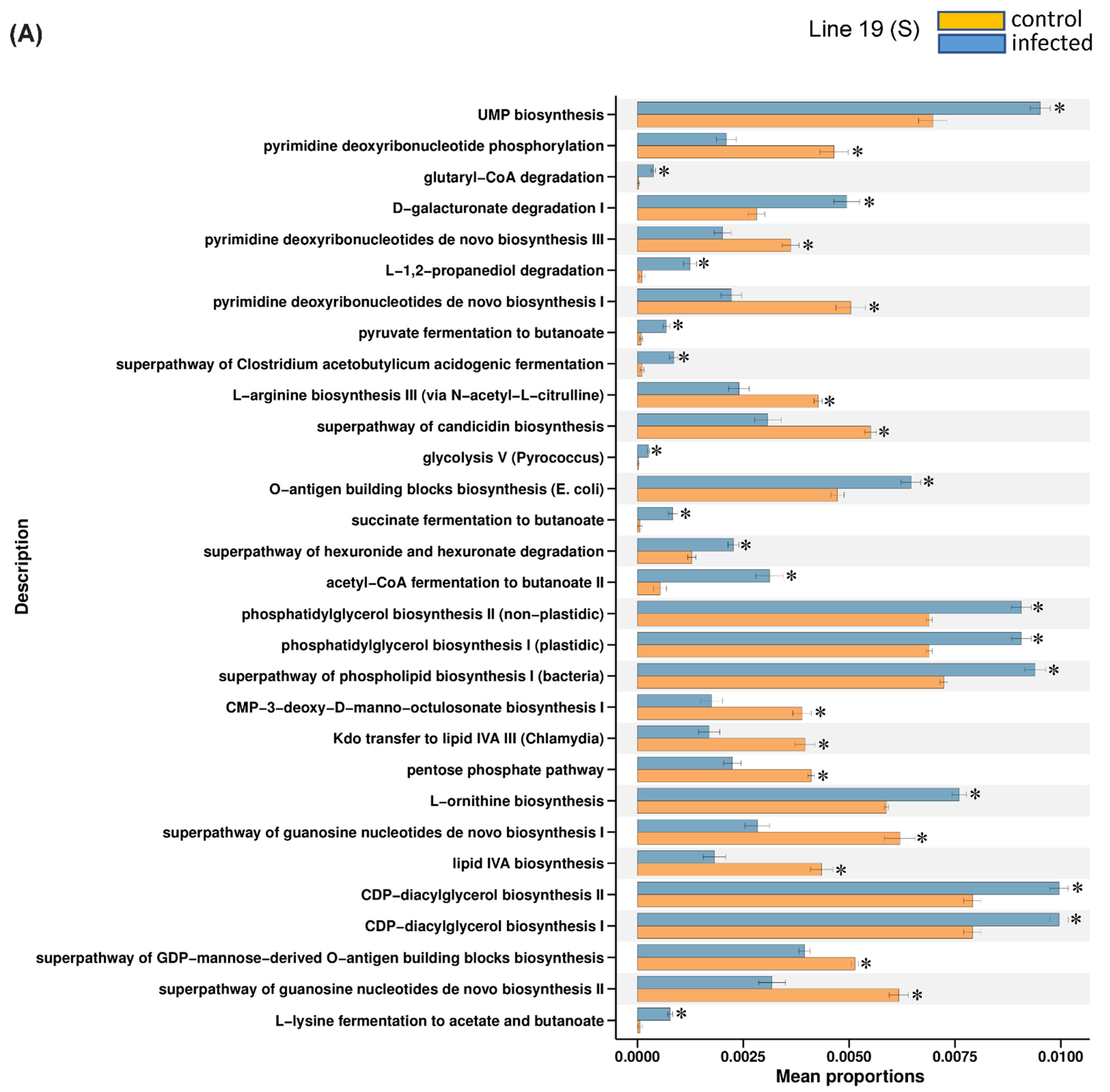
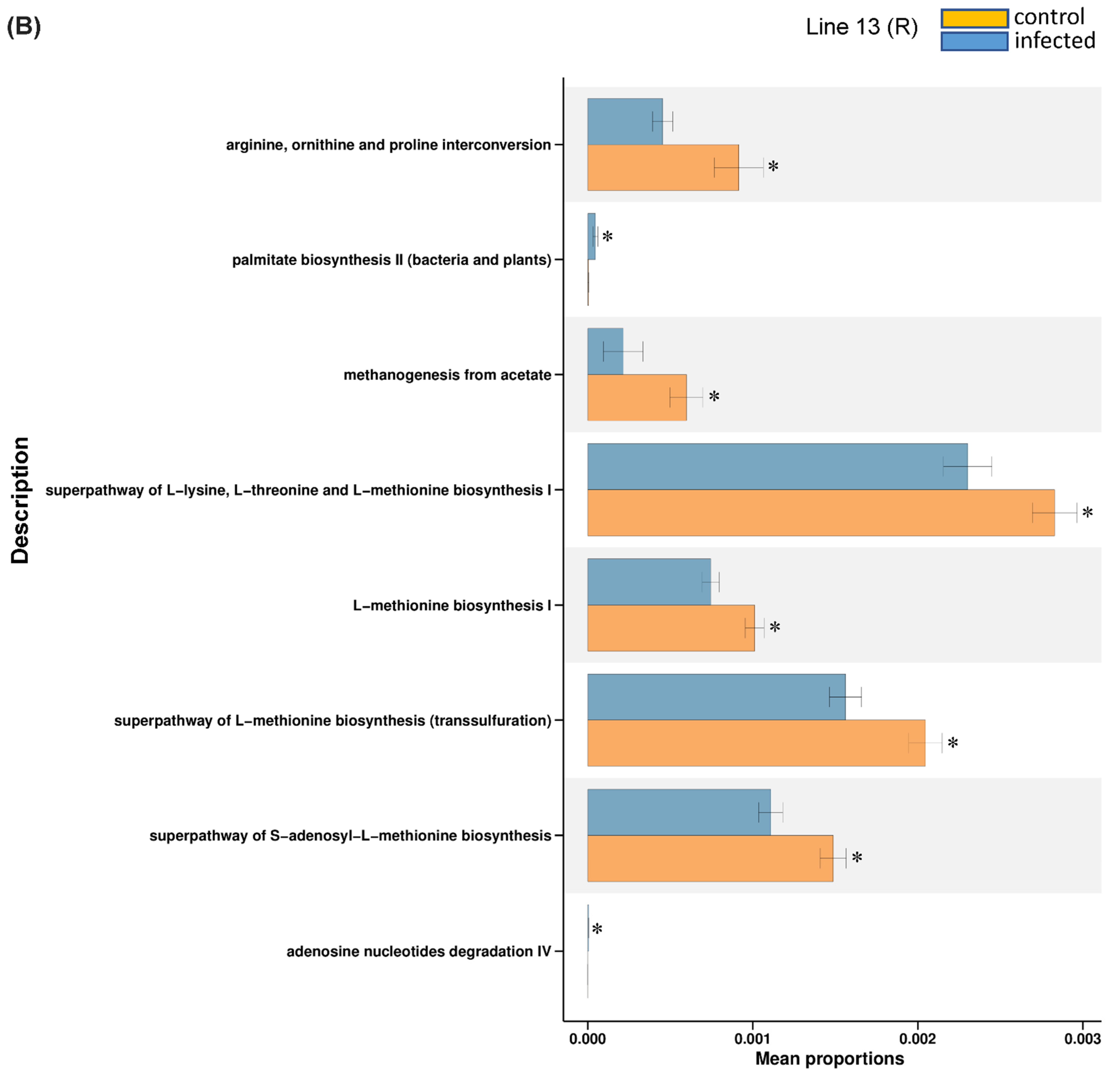
2.3. Predicted KEGG Pathways Were Differentially Regulated in the Resistant vs. Susceptible Lines
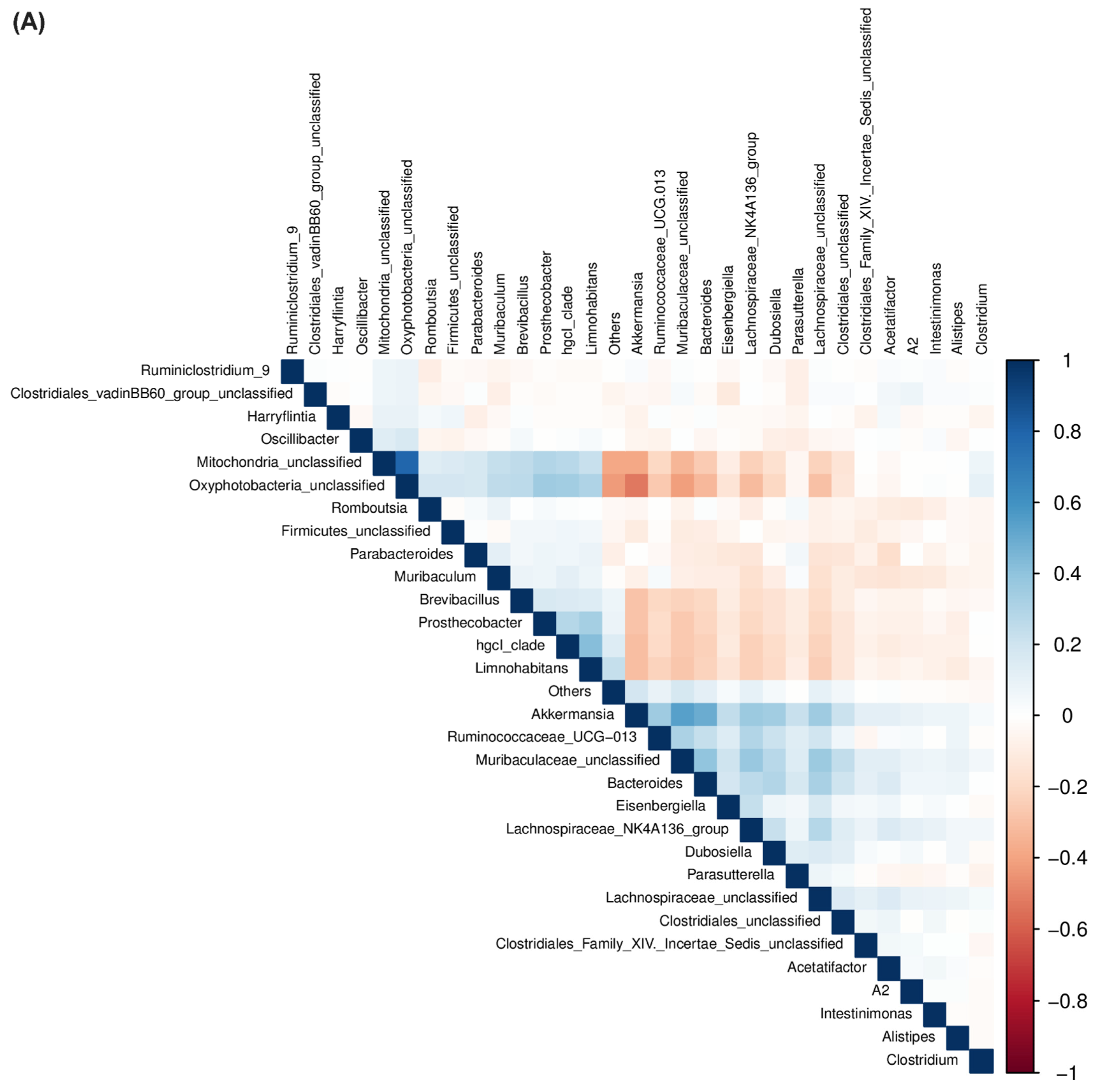
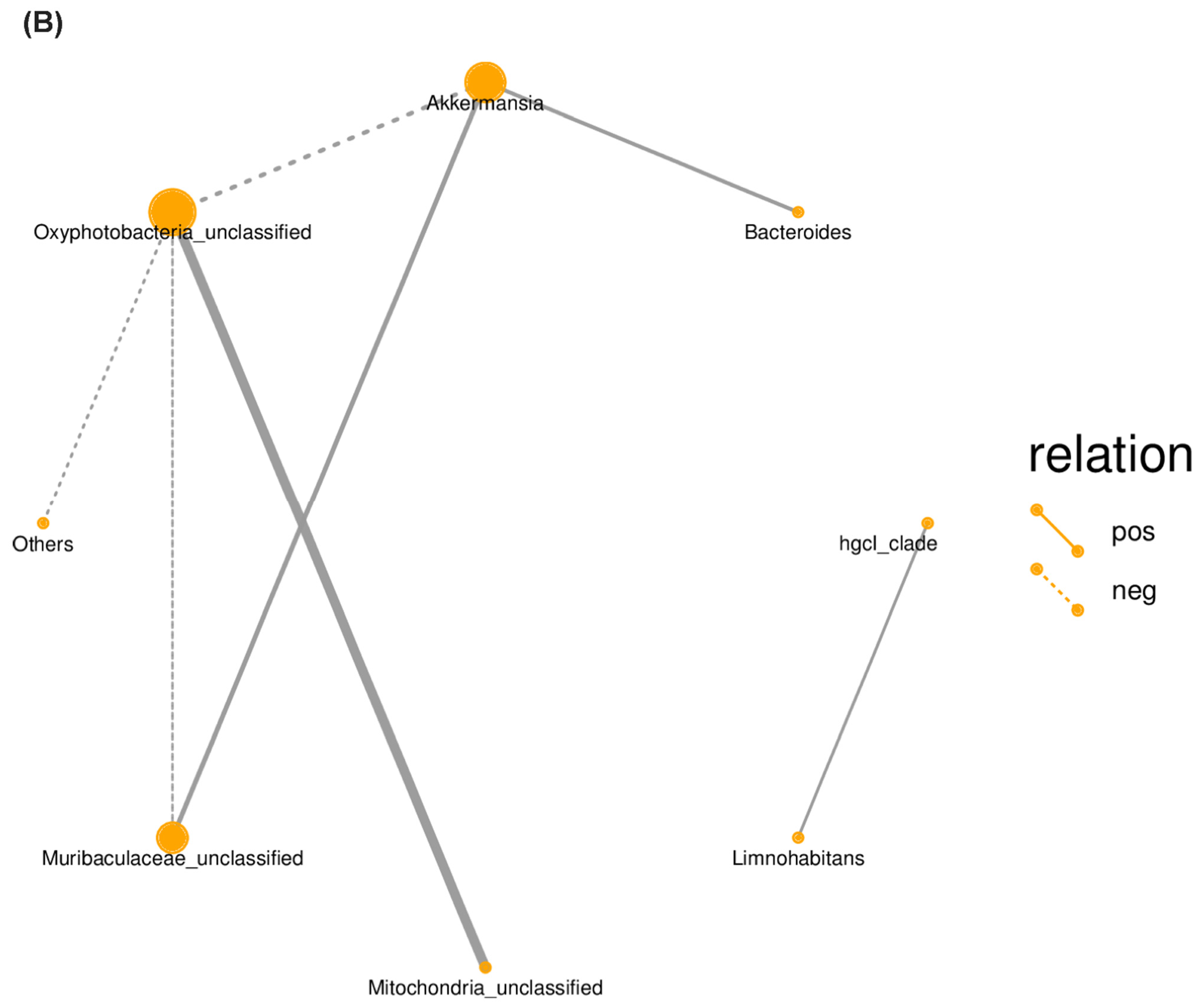
2.4. Correlation between Bacterial Communities across Treatments
2.5. Susceptible Lines Exhibited Higher BCTV-Related Symptoms vs. Resistant Lines
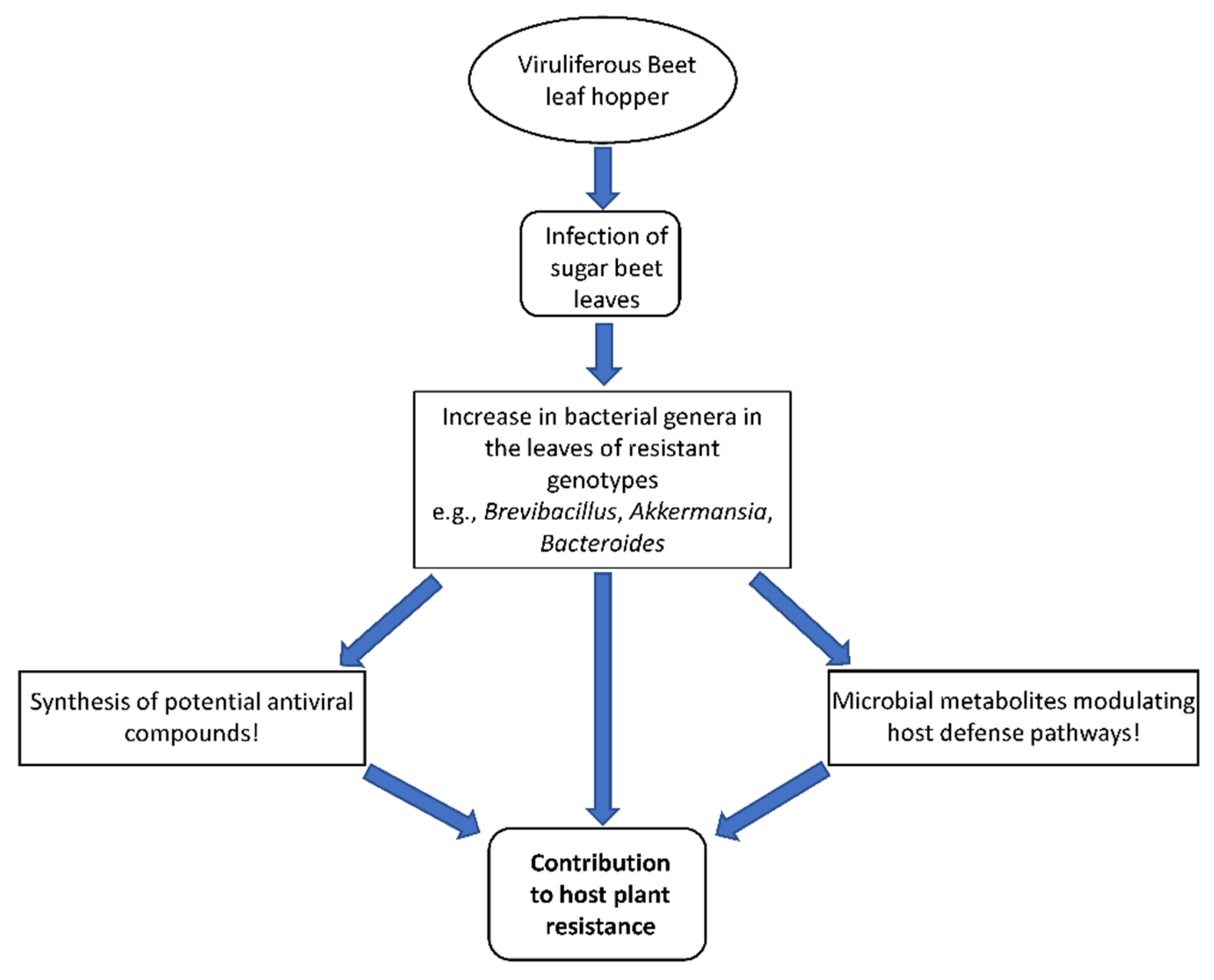
3. Discussion
4. Materials and Methods
4.1. Plant Growth Condition, Viral Infection of Sugar Beet Plants, and Sample Collection
4.2. Genomic DNA Extraction, PCR Amplification, and 16S rRNA Sequencing
4.3. Data Analysis
4.4. Statistical Analysis
4.5. Data Availability
4.6. Evaluation of BCTV Symptoms and RT-PCR Analysis of BCTV-Infected Sugar Beet Plants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Srivastava, S. Management of Sugarbeet Diseases. In Fruit and Vegetable Diseases; Springer: Dordrecht, The Netherlands, 2004; Volume 1, pp. 307–355. [Google Scholar]
- Panella, L.; Kaffka, S.R.; Lewellen, R.T.; Mitchell McGrath, J.; Metzger, M.S.; Strausbaugh, C.A. Sugarbeet. In Yield Gains in Major; U.S. Field Crops: Washington, DC, USA, 2014; pp. 357–395. [Google Scholar]
- Beam, K.; Ascencio-Ibanez, J.T. Geminivirus Resistance: A Minireview. Front. Plant Sci. 2020, 11, 1131. [Google Scholar] [CrossRef]
- Bennett, C.W. The Curly Top Disease of Sugar Beet and Other Plants; The American Phytopathological Society (APS): St. Paul, MN, USA, 1971; Volume 7, pp. 1–4. [Google Scholar]
- Chiginsky, J.; Langemeier, K.; MacWilliams, J.; Albrecht, T.; Cranshaw, W.; Fulladolsa, A.C.; Kapuscinski, M.; Stenglein, M.; Nachappa, P. First Insights into the Virus and Viroid Communities in Hemp (Cannabis sativa). Front. Agron. 2021, 3, 778433. [Google Scholar] [CrossRef]
- Melgarejo, T.; Chen, L.; Rojas, M.; Schilder, A.; Gilbertson, R. Curly top disease of hemp (Cannabis sativa) in California is caused by mild-type strains of Beet curly top virus often in mixed infection. Plant Dis. 2022; Online ahead of print. [Google Scholar] [CrossRef]
- Strausbaugh, C.A.; Eujayl, I.A.; Wintermantel, W.M. Beet curly top virus Strains Associated with Sugar Beet in Idaho, Oregon, and a Western U.S. Collection. Plant Dis. 2017, 101, 1373–1382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swisher Grimm, K.D.; Crosslin, J.; Cooper, R.; Frost, K.; du Toit, L.J.; Wohleb, C.H. First Report of Curly Top of Coriandrum sativum Caused by Beet curly top virus in the Columbia Basin of Washington State. Plant Dis. 2021, 105, 3313. [Google Scholar] [CrossRef] [PubMed]
- Strausbaugh, C.A.; Wenninger, E.J.; Eujayl, I.A. Management of Severe Curly Top in Sugar Beet with Insecticides. Plant Dis. 2012, 96, 1159–1164. [Google Scholar] [CrossRef] [Green Version]
- Strausbaugh, C.A.; Gillen, A.M.; Gallian, J.J.; Camp, S.; Stander, J.R. Influence of Host Resistance and Insecticide Seed Treatments on Curly Top in Sugar Beets. Plant Dis. 2006, 90, 1539–1544. [Google Scholar] [CrossRef] [Green Version]
- Viric Gasparic, H.; Grubelic, M.; Dragovic Uzelac, V.; Bazok, R.; Cacija, M.; Drmic, Z.; Lemic, D. Neonicotinoid Residues in Sugar Beet Plants and Soil under Different Agro-Climatic Conditions. Agriculture 2020, 10, 484. [Google Scholar] [CrossRef]
- Thompson, H.; Vaughan, S.; Mahlein, A.K.; Ladewig, E.; Kenter, C. Is there a risk to honeybees from use of thiamethoxam as a sugar beet seed treatment? Integr. Environ. Assess. Manag. 2022, 18, 709–721. [Google Scholar] [CrossRef]
- Liu, H.; Brettell, L.E.; Qiu, Z.; Singh, B.K. Microbiome-Mediated Stress Resistance in Plants. Trends Plant Sci. 2020, 25, 733–743. [Google Scholar] [CrossRef]
- Dastogeer, K.; Tumpa, F.; Sultana, A.; Akter, M.A.; Chakraborty, A. Plant microbiome–an account of the factors that shape community composition and diversity. Curr. Plant Biol. 2020, 23, 100161. [Google Scholar] [CrossRef]
- Vannier, N.; Agler, M.; Hacquard, S. Microbiota-mediated disease resistance in plants. PLoS Pathog. 2019, 15, e1007740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Cook, J.; Nearing, J.T.; Zhang, J.; Raudonis, R.; Glick, B.R.; Langille, M.G.I.; Cheng, Z. Harnessing the plant microbiome to promote the growth of agricultural crops. Microbiol. Res. 2021, 245, 126690. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, R.; Elena, S.F. The Interplay between the Host Microbiome and Pathogenic Viral Infections. mBio 2021, 12, e0249621. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Liu, M.; Guo, L.; Yang, X.; Qiu, D. A Novel Protein Elicitor (PeBA1) from Bacillus amyloliquefaciens NC6 Induces Systemic Resistance in Tobacco. Int. J. Biol. Sci. 2016, 12, 757–767. [Google Scholar] [CrossRef] [Green Version]
- Bonanomi, G.; Alioto, D.; Minutolo, M.; Marra, R.; Cesarano, G.; Vinale, F. Organic Amendments Modulate Soil Microbiota and Reduce Virus Disease Incidence in the TSWV-Tomato Pathosystem. Pathogens 2020, 9, 379. [Google Scholar] [CrossRef]
- Chaudhry, V.; Runge, P.; Sengupta, P.; Doehlemann, G.; Parker, J.E.; Kemen, E. Shaping the leaf microbiota: Plant-microbe-microbe interactions. J. Exp. Bot. 2021, 72, 36–56. [Google Scholar] [CrossRef]
- Zachow, C.; Muller, H.; Tilcher, R.; Berg, G. Differences between the rhizosphere microbiome of Beta vulgaris ssp. maritima-ancestor of all beet crops-and modern sugar beets. Front. Microbiol. 2014, 5, 415. [Google Scholar] [CrossRef] [Green Version]
- Kusstatscher, P.; Zachow, C.; Harms, K.; Maier, J.; Eigner, H.; Berg, G.; Cernava, T. Microbiome-driven identification of microbial indicators for postharvest diseases of sugar beets. Microbiome 2019, 7, 112. [Google Scholar] [CrossRef]
- Kusstatscher, P.; Cernava, T.; Harms, K.; Maier, J.; Eigner, H.; Berg, G.; Zachow, C. Disease Incidence in Sugar Beet Fields Is Correlated with Microbial Diversity and Distinct Biological Markers. Phytobiomes J. 2019, 3, 22–30. [Google Scholar] [CrossRef] [Green Version]
- Majumdar, R.; Galewski, P.J.; Eujayl, I.; Minocha, R.; Vincill, E.; Strausbaugh, C.A. Regulatory Roles of Small Non-coding RNAs in Sugar Beet Resistance Against Beet curly top virus. Front. Plant Sci. 2021, 12, 780877. [Google Scholar] [CrossRef]
- Lee, S.M.; Kong, H.G.; Song, G.C.; Ryu, C.M. Disruption of Firmicutes and Actinobacteria abundance in tomato rhizosphere causes the incidence of bacterial wilt disease. ISME J. 2021, 15, 330–347. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Zhao, Y.; Yang, Y.; Gong, W.; Sun, X.; Yang, L.; Zhang, Q.; Jin, M. Akkermansia muciniphila Improves Host Defense Against Influenza Virus Infection. Front. Microbiol. 2021, 11, 586476. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jiao, Y.; Shi, J.; Xie, J.; Yin, J.; Zhao, X.; Chen, H. BLB8, an antiviral protein from Brevibacillus laterosporus strain B8, inhibits Tobacco mosaic virus infection by triggering immune response in tobacco. Pest. Manag. Sci. 2021, 77, 4383–4392. [Google Scholar] [CrossRef]
- Javed, K.; Qiu, D. Protein Elicitor PeBL1 of Brevibacillus laterosporus Enhances Resistance Against Myzus persicae in Tomato. Pathogens 2020, 9, 57. [Google Scholar] [CrossRef] [Green Version]
- Bajda, A.; Konopka-Postupolska, D.; Krzymowska, M.; Hennig, J.; Skorupinska-Tudek, K.; Surmacz, L.; Wojcik, J.; Matysiak, Z.; Chojnacki, T.; Skorzynska-Polit, E.; et al. Role of polyisoprenoids in tobacco resistance against biotic stresses. Physiol. Plant 2009, 135, 351–364. [Google Scholar] [CrossRef] [PubMed]
- Makinen, K.; De, S. The significance of methionine cycle enzymes in plant virus infections. Curr. Opin. Plant Biol. 2019, 50, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Spechenkova, N.; Fesenko, I.A.; Mamaeva, A.; Suprunova, T.P.; Kalinina, N.O.; Love, A.J.; Taliansky, M. The Resistance Responses of Potato Plants to Potato Virus Y Are Associated with an Increased Cellular Methionine Content and an Altered SAM:SAH Methylation Index. Viruses 2021, 13, 955. [Google Scholar] [CrossRef]
- Zeier, J. New insights into the regulation of plant immunity by amino acid metabolic pathways. Plant Cell Environ. 2013, 36, 2085–2103. [Google Scholar] [CrossRef]
- Xu, W.; Tian, H.; Song, H.; Wang, Q. Route Development, Antiviral Studies, Field Evaluation and Toxicity of An Antiviral Plant Protectant NK0238. Front. Agr. Sci. Eng. 2022, 9, 110–119. [Google Scholar] [CrossRef]
- Thijs, S.; Op De Beeck, M.; Beckers, B.; Truyens, S.; Stevens, V.; Van Hamme, J.D.; Weyens, N.; Vangronsveld, J. Comparative evaluation of four bacteria-specific primer pairs for 16S rRNA gene surveys. Front. Microbiol. 2017, 8, 494. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [Green Version]
- Friedman, J.; Alm, E.J. Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 2012, 8, e1002687. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Majumdar, R.; Strausbaugh, C.A.; Vincill, E.D.; Eujayl, I.; Galewski, P.J. Leaf Bacteriome in Sugar Beet Shows Differential Response against Beet curly top virus during Resistant and Susceptible Interactions. Int. J. Mol. Sci. 2022, 23, 8073. https://doi.org/10.3390/ijms23158073
Majumdar R, Strausbaugh CA, Vincill ED, Eujayl I, Galewski PJ. Leaf Bacteriome in Sugar Beet Shows Differential Response against Beet curly top virus during Resistant and Susceptible Interactions. International Journal of Molecular Sciences. 2022; 23(15):8073. https://doi.org/10.3390/ijms23158073
Chicago/Turabian StyleMajumdar, Rajtilak, Carl A. Strausbaugh, Eric D. Vincill, Imad Eujayl, and Paul J. Galewski. 2022. "Leaf Bacteriome in Sugar Beet Shows Differential Response against Beet curly top virus during Resistant and Susceptible Interactions" International Journal of Molecular Sciences 23, no. 15: 8073. https://doi.org/10.3390/ijms23158073
APA StyleMajumdar, R., Strausbaugh, C. A., Vincill, E. D., Eujayl, I., & Galewski, P. J. (2022). Leaf Bacteriome in Sugar Beet Shows Differential Response against Beet curly top virus during Resistant and Susceptible Interactions. International Journal of Molecular Sciences, 23(15), 8073. https://doi.org/10.3390/ijms23158073





