Proteoglycans: Systems-Level Insight into Their Expression in Healthy and Diseased Placentas
Abstract
1. Introduction
2. Results and Discussion
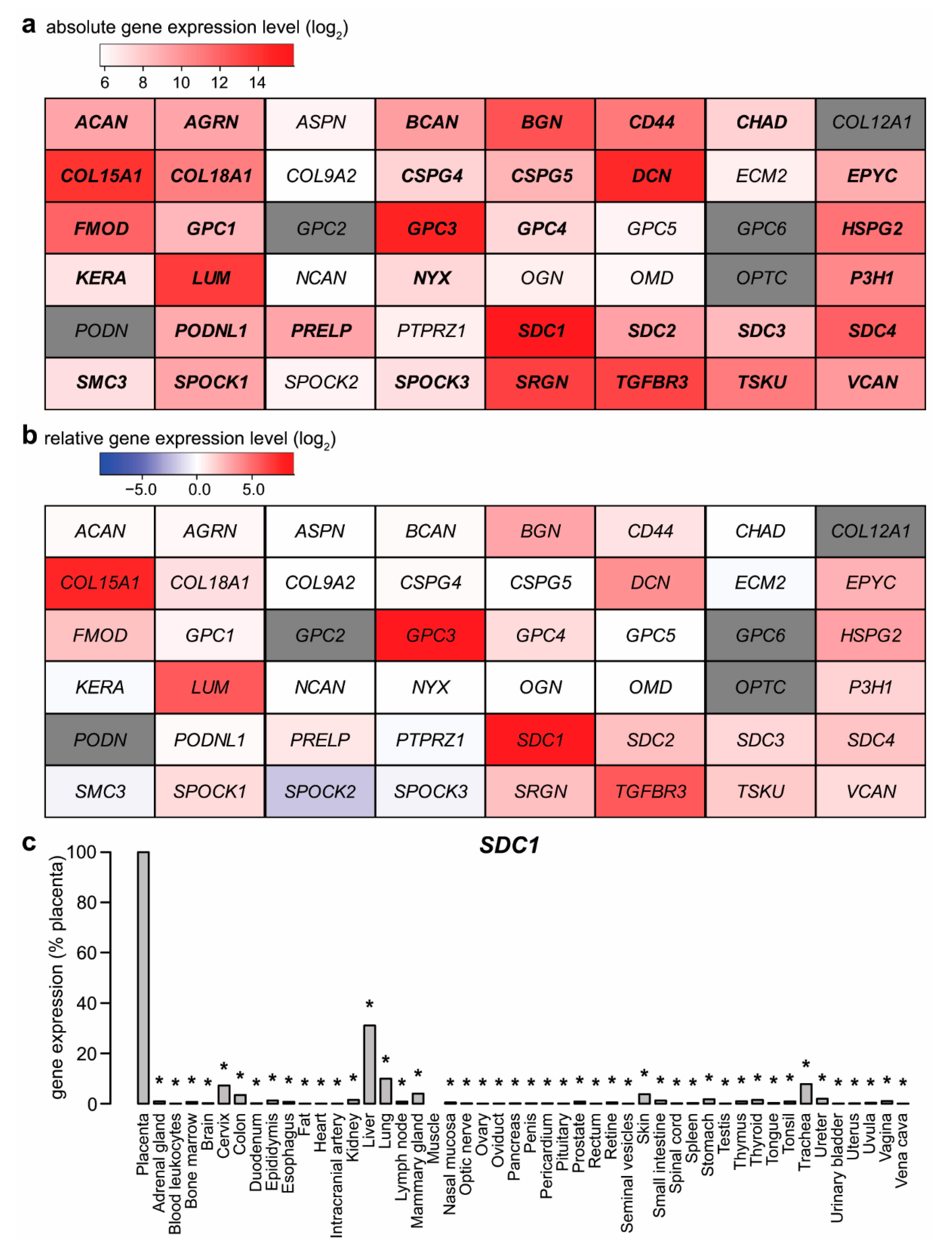
2.1. Expression of Proteoglycans by the Placenta
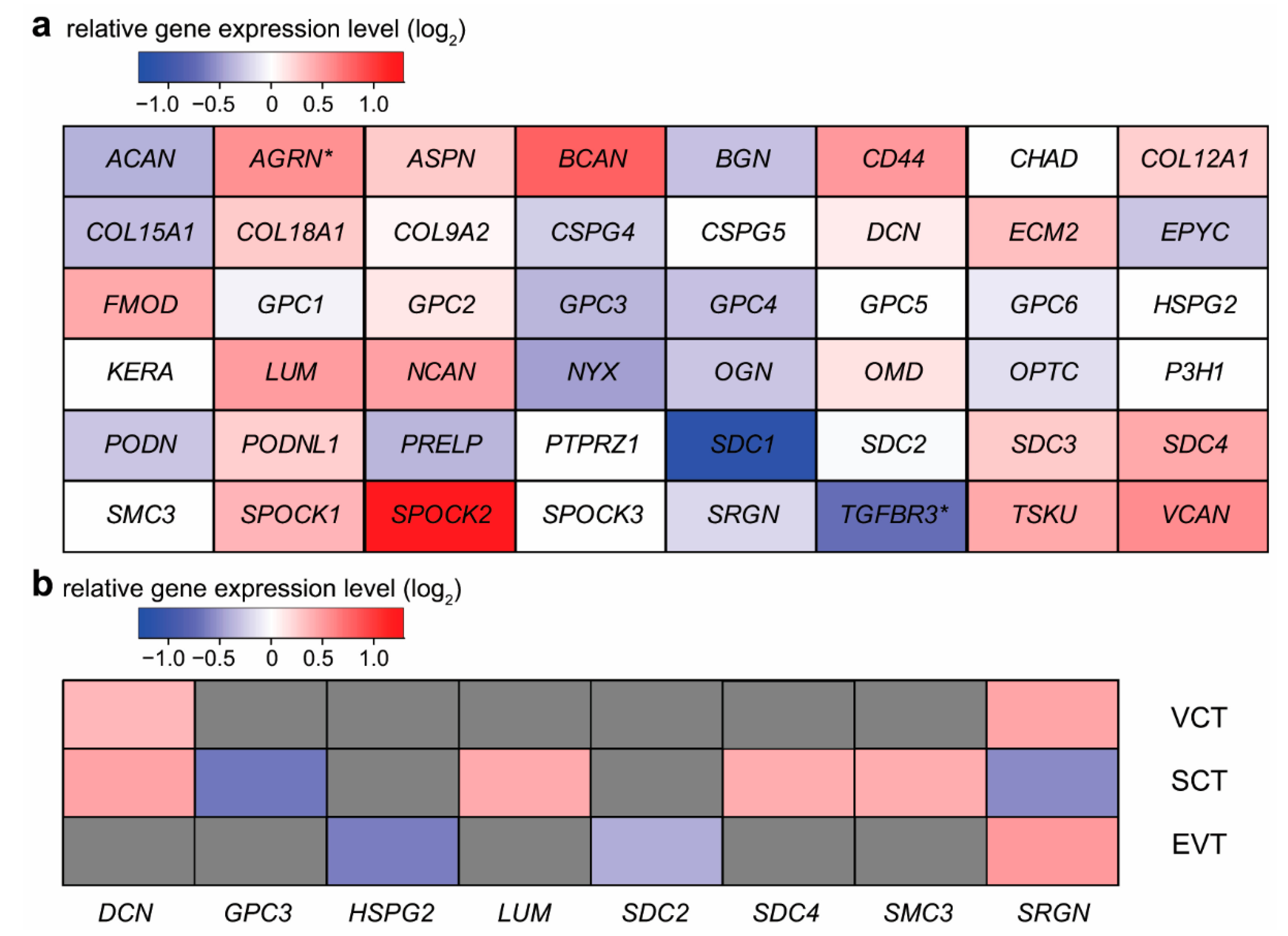
2.2. Expression Changes of Proteoglycans during Syncytiotrophoblast Differentiation
2.3. Expression Changes of Proteoglycans during Extravillous Trophoblast Differentiation
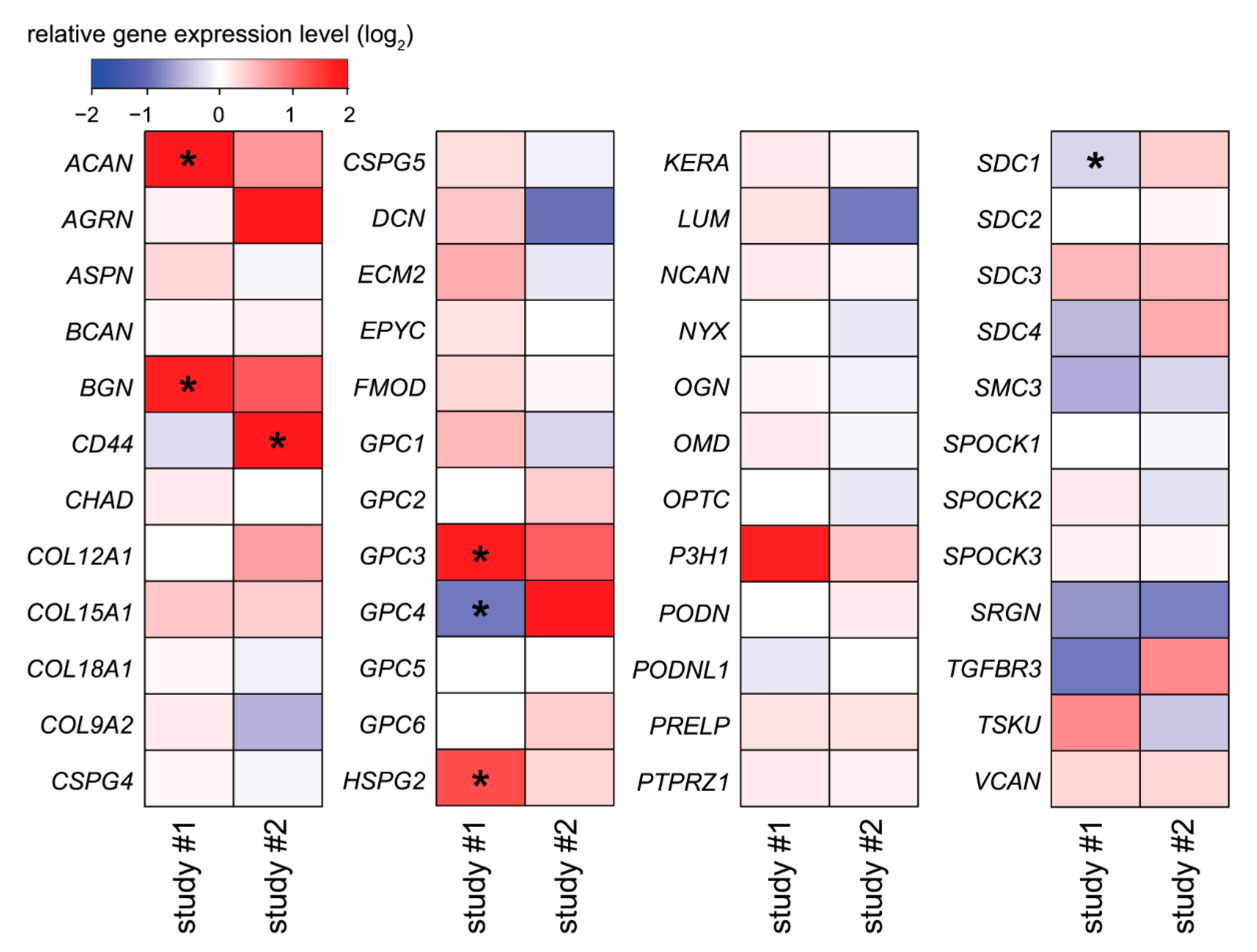
2.4. Placental Expression Changes of Proteoglycans in Pre-Eclampsia and Fetal Growth Restriction
2.5. Potential Functional Significance of Syndecan-1 in the Placenta
3. Summary and Conclusions
4. Materials and Methods
4.1. Placental Expression of Proteoglycan Coding Genes
4.2. Isolation of Primary Trophoblasts and Cell Culturing
4.3. Total RNA Isolation, cDNA Generation, and RT-qPCR
4.4. Human 48-Tissue cDNA Panel Expression Profiling
4.5. Syndecan-1 Immunoassay
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Iozzo, R.V.; Schaefer, L. Proteoglycan form and function: A comprehensive nomenclature of proteoglycans. Matrix Biol. 2015, 42, 11–55. [Google Scholar] [CrossRef] [PubMed]
- Niemann, C.U.; Cowland, J.B.; Klausen, P.; Askaa, J.; Calafat, J.; Borregaard, N. Localization of serglycin in human neutrophil granulocytes and their precursors. J. Leukoc. Biol. 2004, 76, 406–415. [Google Scholar] [CrossRef] [PubMed]
- Couchman, J.R.; Woods, A. Syndecans, signaling, and cell adhesion. J. Cell. Biochem. 1996, 61, 578–584. [Google Scholar] [CrossRef]
- Jokimaa, V.; Inki, P.; Kujari, H.; Hirvonen, O.; Ekholm, E.; Anttila, L. Expression of syndecan-1 in human placenta and decidua. Placenta 1998, 19, 157–163. [Google Scholar] [CrossRef]
- Crescimanno, C.; Marzioni, D.; Paradinas, F.J.; Schrurs, B.; Muhlhauser, J.; Todros, T.; Newlands, E.; David, G.; Castellucci, M. Expression pattern alterations of syndecans and glypican-1 in normal and pathological trophoblast. J. Pathol. 1999, 189, 600–608. [Google Scholar] [CrossRef]
- Marzioni, D.; Crescimanno, C.; Zaccheo, D.; Coppari, R.; Underhill, C.B.; Castellucci, M. Hyaluronate and CD44 expression patterns in the human placenta throughout pregnancy. Eur. J. Histochem. 2001, 45, 131–140. [Google Scholar] [CrossRef]
- Khan, S.; Blackburn, M.; Mao, D.L.; Huber, R.; Schlessinger, D.; Fant, M. Glypican-3 (GPC3) expression in human placenta: Localization to the differentiated syncytiotrophoblast. Histol. Histopathol. 2001, 16, 71–78. [Google Scholar] [CrossRef]
- Van Sinderen, M.; Cuman, C.; Winship, A.; Menkhorst, E.; Dimitriadis, E. The chrondroitin sulfate proteoglycan (CSPG4) regulates human trophoblast function. Placenta 2013, 34, 907–912. [Google Scholar] [CrossRef]
- Afratis, N.A.; Nikitovic, D.; Multhaupt, H.A.; Theocharis, A.D.; Couchman, J.R.; Karamanos, N.K. Syndecans-key regulators of cell signaling and biological functions. FEBS J. 2017, 284, 27–41. [Google Scholar] [CrossRef]
- Jeyarajah, M.J.; Jaju Bhattad, G.; Kops, B.F.; Renaud, S.J. Syndecan-4 regulates extravillous trophoblast migration by coordinating protein kinase C activation. Sci. Rep. 2019, 9, 10175. [Google Scholar] [CrossRef]
- Goryszewska-Szczurek, E.; Baryla, M.; Kaczynski, P.; Waclawik, A. Prokineticin 1-prokineticin receptor 1 signaling in trophoblast promotes embryo implantation and placenta development. Sci. Rep. 2021, 11, 13715. [Google Scholar] [CrossRef] [PubMed]
- Halari, C.D.; Zheng, M.; Lala, P.K. Roles of two small leucine-rich proteoglycans decorin and biglycan in pregnancy and pregnancy-associated diseases. Int. J. Mol. Sci. 2021, 22, 10584. [Google Scholar] [CrossRef] [PubMed]
- Apps, R.; Sharkey, A.; Gardner, L.; Male, V.; Trotter, M.; Miller, N.; North, R.; Founds, S.; Moffett, A. Genome-wide expression profile of first trimester villous and extravillous human trophoblast cells. Placenta 2011, 32, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Guimond, M.J.; Chakraborty, C.; Lala, P.K. Control of proliferation, migration, and invasiveness of human extravillous trophoblast by decorin, a decidual product. Biol. Reprod. 2002, 67, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Ball, M.; Carmody, M.; Wynne, F.; Dockery, P.; Aigner, A.; Cameron, I.; Higgins, J.; Smith, S.D.; Aplin, J.D.; Moore, T. Expression of pleiotrophin and its receptors in human placenta suggests roles in trophoblast life cycle and angiogenesis. Placenta 2009, 30, 649–653. [Google Scholar] [CrossRef]
- Lala, N.; Girish, G.V.; Cloutier-Bosworth, A.; Lala, P.K. Mechanisms in decorin regulation of vascular endothelial growth factor-induced human trophoblast migration and acquisition of endothelial phenotype. Biol. Reprod. 2012, 87, 59. [Google Scholar] [CrossRef]
- Hayashida, K.; Parks, W.C.; Park, P.W. Syndecan-1 shedding facilitates the resolution of neutrophilic inflammation by removing sequestered CXC chemokines. Blood 2009, 114, 3033–3043. [Google Scholar] [CrossRef]
- Teng, Y.H.; Aquino, R.S.; Park, P.W. Molecular functions of syndecan-1 in disease. Matrix Biol. 2012, 31, 3–16. [Google Scholar] [CrossRef]
- Jokimaa, V.I.; Kujari, H.P.; Ekholm, E.M.; Inki, P.L.; Anttila, L. Placental expression of syndecan 1 is diminished in preeclampsia. Am. J. Obstet. Gynecol. 2000, 183, 1495–1498. [Google Scholar] [CrossRef]
- Chui, A.; Murthi, P.; Brennecke, S.P.; Ignjatovic, V.; Monagle, P.T.; Said, J.M. The expression of placental proteoglycans in pre-eclampsia. Gynecol. Obstet. Investig. 2012, 73, 277–284. [Google Scholar] [CrossRef]
- Szabo, S.; Xu, Y.; Romero, R.; Fule, T.; Karaszi, K.; Bhatti, G.; Varkonyi, T.; Varkonyi, I.; Krenacs, T.; Dong, Z.; et al. Changes of placental syndecan-1 expression in preeclampsia and HELLP syndrome. Virchows Arch. 2013, 463, 445–458. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Yanoma, S.; Nagashima, Y.; Okamoto, N.; Ishikawa, H.; Haruki, A.; Miyagi, E.; Takahashi, T.; Hirahara, F.; Miyagi, Y. Paradoxical discrepancy between the serum level and the placental intensity of PP5/TFPI-2 in preeclampsia and/or intrauterine growth restriction: Possible interaction and correlation with glypican-3 hold the key. Placenta 2007, 28, 224–232. [Google Scholar] [CrossRef] [PubMed]
- Chui, A.; Zainuddin, N.; Rajaraman, G.; Murthi, P.; Brennecke, S.P.; Ignjatovic, V.; Monagle, P.T.; Said, J.M. Placental syndecan expression is altered in human idiopathic fetal growth restriction. Am. J. Pathol. 2012, 180, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Gunatillake, T.; Chui, A.; Fitzpatrick, E.; Ignjatovic, V.; Monagle, P.; Whitelock, J.; Zanten, D.; Eijsink, J.; Borg, A.; Stevenson, J.; et al. Decreased placental glypican expression is associated with human fetal growth restriction. Placenta 2019, 76, 6–9. [Google Scholar] [CrossRef]
- Chen, C.P.; Chang, S.C.; Vivian Yang, W.C. High glucose alters proteoglycan expression and the glycosaminoglycan composition in placentas of women with gestational diabetes mellitus and in cultured trophoblasts. Placenta 2007, 28, 97–106. [Google Scholar] [CrossRef]
- Murthi, P.; van Zanten, D.E.; Eijsink, J.J.; Borg, A.J.; Stevenson, J.L.; Kalionis, B.; Chui, A.K.; Said, J.M.; Brennecke, S.P.; Erwich, J.J. Decorin expression is decreased in first trimester placental tissue from pregnancies with small for gestation age infants at birth. Placenta 2016, 45, 58–62. [Google Scholar] [CrossRef]
- Kramer, K.L. Specific sides to multifaceted glycosaminoglycans are observed in embryonic development. Semin. Cell Dev. Biol. 2010, 21, 631–637. [Google Scholar] [CrossRef]
- Costanzo, V.; Bardelli, A.; Siena, S.; Abrignani, S. Exploring the links between cancer and placenta development. Open Biol. 2018, 8, 180081. [Google Scholar] [CrossRef]
- Sanderson, R.D. Heparan sulfate proteoglycans in invasion and metastasis. Semin. Cell Dev. Biol. 2001, 12, 89–98. [Google Scholar] [CrossRef]
- Timar, J.; Lapis, K.; Dudas, J.; Sebestyen, A.; Kopper, L.; Kovalszky, I. Proteoglycans and tumor progression: Janus-faced molecules with contradictory functions in cancer. Semin. Cancer Biol. 2002, 12, 173–186. [Google Scholar] [CrossRef]
- Edwards, I.J. Proteoglycans in prostate cancer. Nat. Rev. Urol. 2012, 9, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Theocharis, A.D.; Skandalis, S.S.; Neill, T.; Multhaupt, H.A.; Hubo, M.; Frey, H.; Gopal, S.; Gomes, A.; Afratis, N.; Lim, H.C.; et al. Insights into the key roles of proteoglycans in breast cancer biology and translational medicine. Biochim. Biophys. Acta 2015, 1855, 276–300. [Google Scholar] [CrossRef] [PubMed]
- Szatmari, T.; Otvos, R.; Hjerpe, A.; Dobra, K. Syndecan-1 in cancer: Implications for cell signaling, differentiation, and prognostication. Dis. Mark. 2015, 2015, 796052. [Google Scholar] [CrossRef] [PubMed]
- Baghy, K.; Tatrai, P.; Regos, E.; Kovalszky, I. Proteoglycans in liver cancer. World J. Gastroenterol. 2016, 22, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, L.; Tredup, C.; Gubbiotti, M.A.; Iozzo, R.V. Proteoglycan neofunctions: Regulation of inflammation and autophagy in cancer biology. FEBS J. 2017, 284, 10–26. [Google Scholar] [CrossRef]
- Tanaka, Y.; Tateishi, R.; Koike, K. Proteoglycans are attractive biomarkers and therapeutictargets in hepatocellular carcinoma. Int. J. Mol. Sci. 2018, 19, 3070. [Google Scholar] [CrossRef]
- Xu, L.; Tang, L.; Zhang, L. Proteoglycans as miscommunication biomarkers for cancer diagnosis. Prog. Mol. Biol. Transl. Sci. 2019, 162, 59–92. [Google Scholar] [CrossRef]
- Rigoglio, N.N.; Rabelo, A.C.S.; Borghesi, J.; de Sa Schiavo Matias, G.; Fratini, P.; Prazeres, P.; Pimentel, C.; Birbrair, A.; Miglino, M.A. The tumor microenvironment: Focus on extracellular matrix. Adv. Exp. Med. Biol. 2020, 1245, 1–38. [Google Scholar] [CrossRef]
- Teixeira, F.; Gotte, M. Involvement of syndecan-1 and heparanase in cancer and inflammation. Adv. Exp. Med. Biol. 2020, 1221, 97–135. [Google Scholar] [CrossRef]
- Reszegi, A.; Horvath, Z.; Feher, H.; Wichmann, B.; Tatrai, P.; Kovalszky, I.; Baghy, K. Protective role of decorin in primary hepatocellular carcinoma. Front. Oncol. 2020, 10, 645. [Google Scholar] [CrossRef]
- Gubbiotti, M.A.; Buraschi, S.; Kapoor, A.; Iozzo, R.V. Proteoglycan signaling in tumor angiogenesis and endothelial cell autophagy. Semin. Cancer Biol. 2020, 62, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Espinoza-Sanchez, N.A.; Gotte, M. Role of cell surface proteoglycans in cancer immunotherapy. Semin. Cancer Biol. 2020, 62, 48–67. [Google Scholar] [CrossRef] [PubMed]
- Reszegi, A.; Horvath, Z.; Karaszi, K.; Regos, E.; Postnikova, V.; Tatrai, P.; Kiss, A.; Schaff, Z.; Kovalszky, I.; Baghy, K. The protective role of decorin in hepatic metastasis of colorectal carcinoma. Biomolecules 2020, 10, 1199. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Hu, M.; Huang, K.; Lin, S.; Du, H. Roles of proteoglycans and glycosaminoglycans in cancer development and progression. Int. J. Mol. Sci. 2020, 21, 5983. [Google Scholar] [CrossRef]
- De Pasquale, V.; Pavone, L.M. Heparan sulfate proteoglycan signaling in tumor microenvironment. Int. J. Mol. Sci. 2020, 21, 6588. [Google Scholar] [CrossRef]
- Hollosi, P.; Vancza, L.; Karaszi, K.; Dobos, K.; Peterfia, B.; Tatrai, E.; Tatrai, P.; Szarvas, T.; Paku, S.; Szilak, L.; et al. Syndecan-1 promotes hepatocyte-like differentiation of hepatoma cells targeting Ets-1 and AP-1. Biomolecules 2020, 10, 1356. [Google Scholar] [CrossRef]
- Li, N.; Spetz, M.R.; Ho, M. The role of glypicans in cancer progression and therapy. J. Histochem. Cytochem. 2020, 68, 841–862. [Google Scholar] [CrossRef]
- Vancza, L.; Karaszi, K.; Peterfia, B.; Turiak, L.; Dezso, K.; Sebestyen, A.; Reszegi, A.; Petovari, G.; Kiss, A.; Schaff, Z.; et al. SPOCK1 promotes the development of hepatocellular carcinoma. Front. Oncol. 2022, 12, 819883. [Google Scholar] [CrossRef]
- Su, A.I.; Wiltshire, T.; Batalov, S.; Lapp, H.; Ching, K.A.; Block, D.; Zhang, J.; Soden, R.; Hayakawa, M.; Kreiman, G.; et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA 2004, 101, 6062–6067. [Google Scholar] [CrossRef]
- Wu, C.; Orozco, C.; Boyer, J.; Leglise, M.; Goodale, J.; Batalov, S.; Hodge, C.L.; Haase, J.; Janes, J.; Huss, J.W., 3rd; et al. BioGPS: An extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009, 10, R130. [Google Scholar] [CrossRef]
- Than, N.G.; Romero, R.; Tarca, A.L.; Kekesi, K.A.; Xu, Y.; Xu, Z.; Juhasz, K.; Bhatti, G.; Leavitt, R.J.; Gelencser, Z.; et al. Integrated systems biology approach identifies novel maternal and placental pathways of preeclampsia. Front. Immunol. 2018, 9, 1661. [Google Scholar] [CrossRef] [PubMed]
- Szilagyi, A.; Gelencser, Z.; Romero, R.; Xu, Y.; Kiraly, P.; Demeter, A.; Palhalmi, J.; Gyorffy, B.A.; Juhasz, K.; Hupuczi, P.; et al. Placenta-specific genes, their regulation during villous trophoblast differentiation and dysregulation in preterm preeclampsia. Int. J. Mol. Sci. 2020, 21, 628. [Google Scholar] [CrossRef] [PubMed]
- Romero, R.; Erez, O.; Maymon, E.; Chaemsaithong, P.; Xu, Z.; Pacora, P.; Chaiworapongsa, T.; Done, B.; Hassan, S.S.; Tarca, A.L. The maternal plasma proteome changes as a function of gestational age in normal pregnancy: A longitudinal study. Am. J. Obstet. Gynecol. 2017, 217, 67.e1–67.e21. [Google Scholar] [CrossRef] [PubMed]
- Azar, C.; Valentine, M.; Trausch-Azar, J.; Druley, T.; Nelson, D.M.; Schwartz, A.L. RNA-Seq identifies genes whose proteins are transformative in the differentiation of cytotrophoblast to syncytiotrophoblast, in human primary villous and BeWo trophoblasts. Sci. Rep. 2018, 8, 5142. [Google Scholar] [CrossRef]
- Vento-Tormo, R.; Efremova, M.; Botting, R.A.; Turco, M.Y.; Vento-Tormo, M.; Meyer, K.B.; Park, J.E.; Stephenson, E.; Polanski, K.; Goncalves, A.; et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 2018, 563, 347–353. [Google Scholar] [CrossRef]
- Karlsson, M.; Zhang, C.; Mear, L.; Zhong, W.; Digre, A.; Katona, B.; Sjostedt, E.; Butler, L.; Odeberg, J.; Dusart, P.; et al. A single-cell type transcriptomics map of human tissues. Sci. Adv. 2021, 7, eabh2169. [Google Scholar] [CrossRef]
- Pique-Regi, R.; Romero, R.; Tarca, A.L.; Sendler, E.D.; Xu, Y.; Garcia-Flores, V.; Leng, Y.; Luca, F.; Hassan, S.S.; Gomez-Lopez, N. Single cell transcriptional signatures of the human placenta in term and preterm parturition. Elife 2019, 8, e52004. [Google Scholar] [CrossRef]
- Meinhardt, G.; Haider, S.; Kunihs, V.; Saleh, L.; Pollheimer, J.; Fiala, C.; Hetey, S.; Feher, Z.; Szilagyi, A.; Than, N.G.; et al. Pivotal role of the transcriptional co-activator YAP in trophoblast stemness of the developing human placenta. Proc. Natl. Acad. Sci. USA 2020, 117, 13562–13570. [Google Scholar] [CrossRef]
- Hofmann-Kiefer, K.F.; Knabl, J.; Martinoff, N.; Schiessl, B.; Conzen, P.; Rehm, M.; Becker, B.F.; Chappell, D. Increased serum concentrations of circulating glycocalyx components in HELLP syndrome compared to healthy pregnancy: An observational study. Reprod. Sci 2013, 20, 318–325. [Google Scholar] [CrossRef]
- Gandley, R.E.; Althouse, A.; Jeyabalan, A.; Bregand-White, J.M.; McGonigal, S.; Myerski, A.C.; Gallaher, M.; Powers, R.W.; Hubel, C.A. Low soluble syndecan-1 precedes preeclampsia. PLoS ONE 2016, 11, e0157608. [Google Scholar] [CrossRef]
- Kuessel, L.; Husslein, H.; Montanari, E.; Kundi, M.; Himmler, G.; Binder, J.; Schiefer, J.; Zeisler, H. Dynamics of soluble syndecan-1 in maternal serum during and after pregnancies complicated by preeclampsia: A nested case control study. Clin. Chem. Lab. Med. 2019, 58, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Pollheimer, J.; Vondra, S.; Baltayeva, J.; Beristain, A.G.; Knofler, M. Regulation of placental extravillous trophoblasts by the maternal uterine environment. Front. Immunol. 2018, 9, 2597. [Google Scholar] [CrossRef] [PubMed]
- Bilban, M.; Haslinger, P.; Prast, J.; Klinglmuller, F.; Woelfel, T.; Haider, S.; Sachs, A.; Otterbein, L.E.; Desoye, G.; Hiden, U.; et al. Identification of novel trophoblast invasion-related genes: Heme oxygenase-1 controls motility via peroxisome proliferator-activated receptor gamma. Endocrinology 2009, 150, 1000–1013. [Google Scholar] [CrossRef] [PubMed]
- Espinoza, J.; Romero, R.; Mee Kim, Y.; Kusanovic, J.P.; Hassan, S.; Erez, O.; Gotsch, F.; Than, N.G.; Papp, Z.; Jai Kim, C. Normal and abnormal transformation of the spiral arteries during pregnancy. J. Perinat. Med. 2006, 34, 447–458. [Google Scholar] [CrossRef]
- Brosens, I.; Pijnenborg, R.; Vercruysse, L.; Romero, R. The “Great Obstetrical Syndromes” are associated with disorders of deep placentation. Am. J. Obstet. Gynecol. 2011, 204, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, H.; Ota, S.; Suzuki, M.; Kato, T.; Sekiya, T.; Kurahashi, H.; Udagawa, Y. Comparative gene expression profiling of placentas from patients with severe pre-eclampsia and unexplained fetal growth restriction. Reprod. Biol. Endocrinol. 2011, 9, 107. [Google Scholar] [CrossRef] [PubMed]
- Chaiworapongsa, T.; Chaemsaithong, P.; Yeo, L.; Romero, R. Pre-eclampsia part 1: Current understanding of its pathophysiology. Nat. Rev. Nephrol. 2014, 10, 466–480. [Google Scholar] [CrossRef]
- Szalai, G.; Xu, Y.; Romero, R.; Chaiworapongsa, T.; Xu, Z.; Chiang, P.J.; Ahn, H.; Sundell, B.; Plazyo, O.; Jiang, Y.; et al. In vivo experiments reveal the good, the bad and the ugly faces of sFlt-1 in pregnancy. PLoS ONE 2014, 9, e110867. [Google Scholar] [CrossRef]
- Szalai, G.; Romero, R.; Chaiworapongsa, T.; Xu, Y.; Wang, B.; Ahn, H.; Xu, Z.; Chiang, P.J.; Sundell, B.; Wang, R.; et al. Full-length human placental sFlt-1-e15a isoform induces distinct maternal phenotypes of preeclampsia in mice. PLoS ONE 2015, 10, e0119547. [Google Scholar] [CrossRef]
- Zhang, T.; Bian, Q.; Chen, Y.; Wang, X.; Yu, S.; Liu, S.; Ji, P.; Li, L.; Shrestha, M.; Dong, S.; et al. Dissecting human trophoblast cell transcriptional heterogeneity in preeclampsia using single-cell RNA sequencing. Mol. Genet. Genom. Med. 2021, 9, e1730. [Google Scholar] [CrossRef]
- Gong, S.; Gaccioli, F.; Dopierala, J.; Sovio, U.; Cook, E.; Volders, P.J.; Martens, L.; Kirk, P.D.W.; Richardson, S.; Smith, G.C.S.; et al. The RNA landscape of the human placenta in health and disease. Nat. Commun. 2021, 12, 2639. [Google Scholar] [CrossRef] [PubMed]
- Than, N.G.; Posta, M.; Gyorffy, D.; Orosz, L.; Orosz, G.; Rossi, S.W.; Ambrus-Aikelin, G.; Szilagyi, A.; Nagy, S.; Hupuczi, P.; et al. Early pathways, biomarkers, and four distinct molecular subclasses of preeclampsia: The intersection of clinical, pathological, and high-dimensional biology studies. Placenta 2022, in press. [CrossRef] [PubMed]
- Sitras, V.; Paulssen, R.; Leirvik, J.; Vartun, A.; Acharya, G. Placental gene expression profile in intrauterine growth restriction due to placental insufficiency. Reprod. Sci. 2009, 16, 701–711. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Tsai, S.Q.; Hardison, N.E.; James, A.H.; Motsinger-Reif, A.A.; Thames, B.; Stone, E.A.; Deng, C.; Piedrahita, J.A. Differentially expressed microRNAs and affected biological pathways revealed by modulated modularity clustering (MMC) analysis of human preeclamptic and IUGR placentas. Placenta 2013, 34, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Medina-Bastidas, D.; Guzman-Huerta, M.; Borboa-Olivares, H.; Ruiz-Cruz, C.; Parra-Hernandez, S.; Flores-Pliego, A.; Salido-Guadarrama, I.; Camargo-Marin, L.; Arambula-Meraz, E.; Estrada-Gutierrez, G. Placental microarray profiling reveals common mRNA and lncRNA expression patterns in preeclampsia and intrauterine growth restriction. Int. J. Mol. Sci. 2020, 21, 3597. [Google Scholar] [CrossRef]
- Majewska, M.; Lipka, A.; Paukszto, L.; Jastrzebski, J.P.; Szeszko, K.; Gowkielewicz, M.; Lepiarczyk, E.; Jozwik, M.; Majewski, M.K. Placenta transcriptome profiling in intrauterine growth restriction (IUGR). Int. J. Mol. Sci. 2019, 20, 1510. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, C.L.; McNamara, H.; Walker, S.P.; Alexiadis, M.; Fuller, P.J.; Vickers, D.K.; Hannan, N.J.; Hastie, R.; Tuohey, L.; Kaitu’u-Lino, T.J.; et al. Identifying late-onset fetal growth restriction by measuring circulating placental RNA in the maternal blood at 28 weeks’ gestation. Am. J. Obstet. Gynecol. 2016, 214, 521.e521–521.e528. [Google Scholar] [CrossRef]
- Szenasi, N.L.; Toth, E.; Balogh, A.; Juhasz, K.; Karaszi, K.; Ozohanics, O.; Gelencser, Z.; Kiraly, P.; Hargitai, B.; Drahos, L.; et al. Proteomic identification of membrane-associated placental protein 4 (MP4) as perlecan and characterization of its placental expression in normal and pathologic pregnancies. PeerJ 2019, 7, e6982. [Google Scholar] [CrossRef]
- Heyer-Chauhan, N.; Ovbude, I.J.; Hills, A.A.; Sullivan, M.H.; Hills, F.A. Placental syndecan-1 and sulphated glycosaminoglycans are decreased in preeclampsia. J. Perinat. Med. 2014, 42, 329–338. [Google Scholar] [CrossRef]
- Alici Davutoglu, E.; Akkaya Firat, A.; Ozel, A.; Yilmaz, N.; Uzun, I.; Temel Yuksel, I.; Madazli, R. Evaluation of maternal serum hypoxia inducible factor-1alpha, progranulin and syndecan-1 levels in pregnancies with early- and late-onset preeclampsia. J. Matern. Fetal. Neonatal. Med. 2018, 31, 1976–1982. [Google Scholar] [CrossRef]
- Kornacki, J.; Wirstlein, P.; Wender-Ozegowska, E. Levels of syndecan-1 and hyaluronan in early- and late-onset preeclampsia. Pregnancy Hypertens 2019, 18, 108–111. [Google Scholar] [CrossRef] [PubMed]
- Hassani Lahsinoui, H.; Amraoui, F.; Spijkers, L.J.A.; Veenboer, G.J.M.; Peters, S.L.M.; van Vlies, N.; Vogt, L.; Ris-Stalpers, C.; van den Born, B.J.H.; Afink, G.B. Soluble syndecan-1 and glycosaminoglycans in preeclamptic and normotensive pregnancies. Sci. Rep. 2021, 11, 4387. [Google Scholar] [CrossRef] [PubMed]
- Than, N.G.; Romero, R.; Goodman, M.; Weckle, A.; Xing, J.; Dong, Z.; Xu, Y.; Tarquini, F.; Szilagyi, A.; Gal, P.; et al. A primate subfamily of galectins expressed at the maternal-fetal interface that promote immune cell death. Proc. Natl. Acad. Sci. USA 2009, 106, 9731–9736. [Google Scholar] [CrossRef] [PubMed]
- Than, N.G.; Romero, R.; Meiri, H.; Erez, O.; Xu, Y.; Tarquini, F.; Barna, L.; Szilagyi, A.; Ackerman, R.; Sammar, M.; et al. PP13, maternal ABO blood groups and the risk assessment of pregnancy complications. PLoS ONE 2011, 6, e21564. [Google Scholar] [CrossRef] [PubMed]
- Than, N.G.; Balogh, A.; Romero, R.; Karpati, E.; Erez, O.; Szilagyi, A.; Kovalszky, I.; Sammar, M.; Gizurarson, S.; Matko, J.; et al. Placental Protein 13 (PP13)—A placental immunoregulatory galectin protecting pregnancy. Front. Immunol. 2014, 5, 348. [Google Scholar] [CrossRef] [PubMed]
- Than, N.G.; Romero, R.; Balogh, A.; Karpati, E.; Mastrolia, S.A.; Staretz-Chacham, O.; Hahn, S.; Erez, O.; Papp, Z.; Kim, C.J. Galectins: Double-edged swords in the cross-roads of pregnancy complications and female reproductive tract inflammation and neoplasia. J. Pathol. Transl. Med. 2015, 49, 181–208. [Google Scholar] [CrossRef]
- Than, N.G.; Abdul Rahman, O.; Magenheim, R.; Nagy, B.; Fule, T.; Hargitai, B.; Sammar, M.; Hupuczi, P.; Tarca, A.L.; Szabo, G.; et al. Placental protein 13 (galectin-13) has decreased placental expression but increased shedding and maternal serum concentrations in patients presenting with preterm pre-eclampsia and HELLP syndrome. Virchows Arch. 2008, 453, 387–400. [Google Scholar] [CrossRef]
- Balogh, A.; Pozsgay, J.; Matko, J.; Dong, Z.; Kim, C.J.; Varkonyi, T.; Sammar, M.; Rigo, J., Jr.; Meiri, H.; Romero, R.; et al. Placental protein 13 (PP13/galectin-13) undergoes lipid raft-associated subcellular redistribution in the syncytiotrophoblast in preterm preeclampsia and HELLP syndrome. Am. J. Obstet. Gynecol. 2011, 205, 156.e1–156.e14. [Google Scholar] [CrossRef]
- Than, N.G.; Romero, R.; Xu, Y.; Erez, O.; Xu, Z.; Bhatti, G.; Leavitt, R.; Chung, T.H.; El-Azzamy, H.; LaJeunesse, C.; et al. Evolutionary origins of the placental expression of chromosome 19 cluster galectins and their complex dysregulation in preeclampsia. Placenta 2014, 35, 855–865. [Google Scholar] [CrossRef]
- Burton, G.J.; Jauniaux, E. Placental oxidative stress: From miscarriage to preeclampsia. J. Soc. Gynecol. Investig. 2004, 11, 342–352. [Google Scholar] [CrossRef]
- Szabo, S.; Mody, M.; Romero, R.; Xu, Y.; Karaszi, K.; Mihalik, N.; Xu, Z.; Bhatti, G.; Fule, T.; Hupuczi, P.; et al. Activation of villous trophoblastic p38 and ERK1/2 signaling pathways in preterm preeclampsia and HELLP syndrome. Pathol. Oncol. Res. 2015, 21, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Garcha, D.; Walker, S.P.; MacDonald, T.M.; Hyett, J.; Jellins, J.; Myers, J.; Illanes, S.E.; Nien, J.K.; Schepeler, M.; Keenan, E.; et al. Circulating syndecan-1 is reduced in pregnancies with poor fetal growth and its secretion regulated by matrix metalloproteinases and the mitochondria. Sci. Rep. 2021, 11, 16595. [Google Scholar] [CrossRef] [PubMed]
- Bernfield, M.; Gotte, M.; Park, P.W.; Reizes, O.; Fitzgerald, M.L.; Lincecum, J.; Zako, M. Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 1999, 68, 729–777. [Google Scholar] [CrossRef] [PubMed]
- Prakash, G.J.; Suman, P.; Gupta, S.K. Relevance of syndecan-1 in the trophoblastic BeWo cell syncytialization. Am. J. Reprod. Immunol. 2011, 66, 385–393. [Google Scholar] [CrossRef]
- Iba, K.; Albrechtsen, R.; Gilpin, B.; Frohlich, C.; Loechel, F.; Zolkiewska, A.; Ishiguro, K.; Kojima, T.; Liu, W.; Langford, J.K.; et al. The cysteine-rich domain of human ADAM 12 supports cell adhesion through syndecans and triggers signaling events that lead to beta1 integrin-dependent cell spreading. J. Cell Biol. 2000, 149, 1143–1156. [Google Scholar] [CrossRef]
- Aghababaei, M.; Hogg, K.; Perdu, S.; Robinson, W.P.; Beristain, A.G. ADAM12-directed ectodomain shedding of E-cadherin potentiates trophoblast fusion. Cell Death Differ. 2015, 22, 1970–1984. [Google Scholar] [CrossRef]
- Wang, Z.; Li, R.; Tan, J.; Peng, L.; Wang, P.; Liu, J.; Xiong, H.; Jiang, B.; Chen, Y. Syndecan-1 acts in synergy with tight junction through Stat3 signaling to maintain intestinal mucosal barrier and prevent bacterial translocation. Inflamm. Bowel Dis. 2015, 21, 1894–1907. [Google Scholar] [CrossRef]
- Moore, K.H.; Murphy, H.A.; Chapman, H.; George, E.M. Syncytialization alters the extracellular matrix and barrier function of placental trophoblasts. Am. J. Physiol. Cell Physiol. 2021, 321, C694–C703. [Google Scholar] [CrossRef]
- Biadasiewicz, K.; Fock, V.; Dekan, S.; Proestling, K.; Velicky, P.; Haider, S.; Knofler, M.; Frohlich, C.; Pollheimer, J. Extravillous trophoblast-associated ADAM12 exerts pro-invasive properties, including induction of integrin beta 1-mediated cellular spreading. Biol. Reprod. 2014, 90, 101. [Google Scholar] [CrossRef]
- Boeddeker, S.J.; Baston-Buest, D.M.; Altergot-Ahmad, O.; Kruessel, J.S.; Hess, A.P. Syndecan-1 knockdown in endometrial epithelial cells alters their apoptotic protein profile and enhances the inducibility of apoptosis. Mol. Hum. Reprod. 2014, 20, 567–578. [Google Scholar] [CrossRef][Green Version]
- Boeddeker, S.J.; Baston-Buest, D.M.; Fehm, T.; Kruessel, J.; Hess, A. Decidualization and syndecan-1 knock down sensitize endometrial stromal cells to apoptosis induced by embryonic stimuli. PLoS ONE 2015, 10, e0121103. [Google Scholar] [CrossRef] [PubMed]
- Baston-Bust, D.M.; Gotte, M.; Janni, W.; Krussel, J.S.; Hess, A.P. Syndecan-1 knock-down in decidualized human endometrial stromal cells leads to significant changes in cytokine and angiogenic factor expression patterns. Reprod. Biol. Endocrinol. 2010, 8, 133. [Google Scholar] [CrossRef] [PubMed]
- Knofler, M.; Pollheimer, J. Human placental trophoblast invasion and differentiation: A particular focus on Wnt signaling. Front. Genet. 2013, 4, 190. [Google Scholar] [CrossRef] [PubMed]
- Manon-Jensen, T.; Itoh, Y.; Couchman, J.R. Proteoglycans in health and disease: The multiple roles of syndecan shedding. FEBS J. 2010, 277, 3876–3889. [Google Scholar] [CrossRef]
- Daglar, K.; Kirbas, A.; Timur, H.; Ozturk Inal, Z.; Danisman, N. Placental levels of total oxidative and anti-oxidative status, ADAMTS-12 and decorin in early- and late-onset severe preeclampsia. J. Matern. Fetal. Neonatal. Med. 2016, 29, 4059–4064. [Google Scholar] [CrossRef]
- Varkonyi, T.; Nagy, B.; Fule, T.; Tarca, A.L.; Karaszi, K.; Schonleber, J.; Hupuczi, P.; Mihalik, N.; Kovalszky, I.; Rigo, J., Jr.; et al. Microarray profiling reveals that placental transcriptomes of early-onset HELLP syndrome and preeclampsia are similar. Placenta 2011, 32, S21–S29. [Google Scholar] [CrossRef]
- Sjostedt, E.; Zhong, W.; Fagerberg, L.; Karlsson, M.; Mitsios, N.; Adori, C.; Oksvold, P.; Edfors, F.; Limiszewska, A.; Hikmet, F.; et al. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science 2020, 367, eaay5947. [Google Scholar] [CrossRef]
- Kliman, H.J.; Nestler, J.E.; Sermasi, E.; Sanger, J.M.; Strauss, J.F., 3rd. Purification, characterization, and in vitro differentiation of cytotrophoblasts from human term placentae. Endocrinology 1986, 118, 1567–1582. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oravecz, O.; Balogh, A.; Romero, R.; Xu, Y.; Juhasz, K.; Gelencser, Z.; Xu, Z.; Bhatti, G.; Pique-Regi, R.; Peterfia, B.; et al. Proteoglycans: Systems-Level Insight into Their Expression in Healthy and Diseased Placentas. Int. J. Mol. Sci. 2022, 23, 5798. https://doi.org/10.3390/ijms23105798
Oravecz O, Balogh A, Romero R, Xu Y, Juhasz K, Gelencser Z, Xu Z, Bhatti G, Pique-Regi R, Peterfia B, et al. Proteoglycans: Systems-Level Insight into Their Expression in Healthy and Diseased Placentas. International Journal of Molecular Sciences. 2022; 23(10):5798. https://doi.org/10.3390/ijms23105798
Chicago/Turabian StyleOravecz, Orsolya, Andrea Balogh, Roberto Romero, Yi Xu, Kata Juhasz, Zsolt Gelencser, Zhonghui Xu, Gaurav Bhatti, Roger Pique-Regi, Balint Peterfia, and et al. 2022. "Proteoglycans: Systems-Level Insight into Their Expression in Healthy and Diseased Placentas" International Journal of Molecular Sciences 23, no. 10: 5798. https://doi.org/10.3390/ijms23105798
APA StyleOravecz, O., Balogh, A., Romero, R., Xu, Y., Juhasz, K., Gelencser, Z., Xu, Z., Bhatti, G., Pique-Regi, R., Peterfia, B., Hupuczi, P., Kovalszky, I., Murthi, P., Tarca, A. L., Papp, Z., Matko, J., & Than, N. G. (2022). Proteoglycans: Systems-Level Insight into Their Expression in Healthy and Diseased Placentas. International Journal of Molecular Sciences, 23(10), 5798. https://doi.org/10.3390/ijms23105798








