Understanding the Underlying Molecular Mechanisms of Meiotic Arrest during In Vitro Spermatogenesis in Rat Prepubertal Testicular Tissue
Abstract
1. Introduction
2. Results
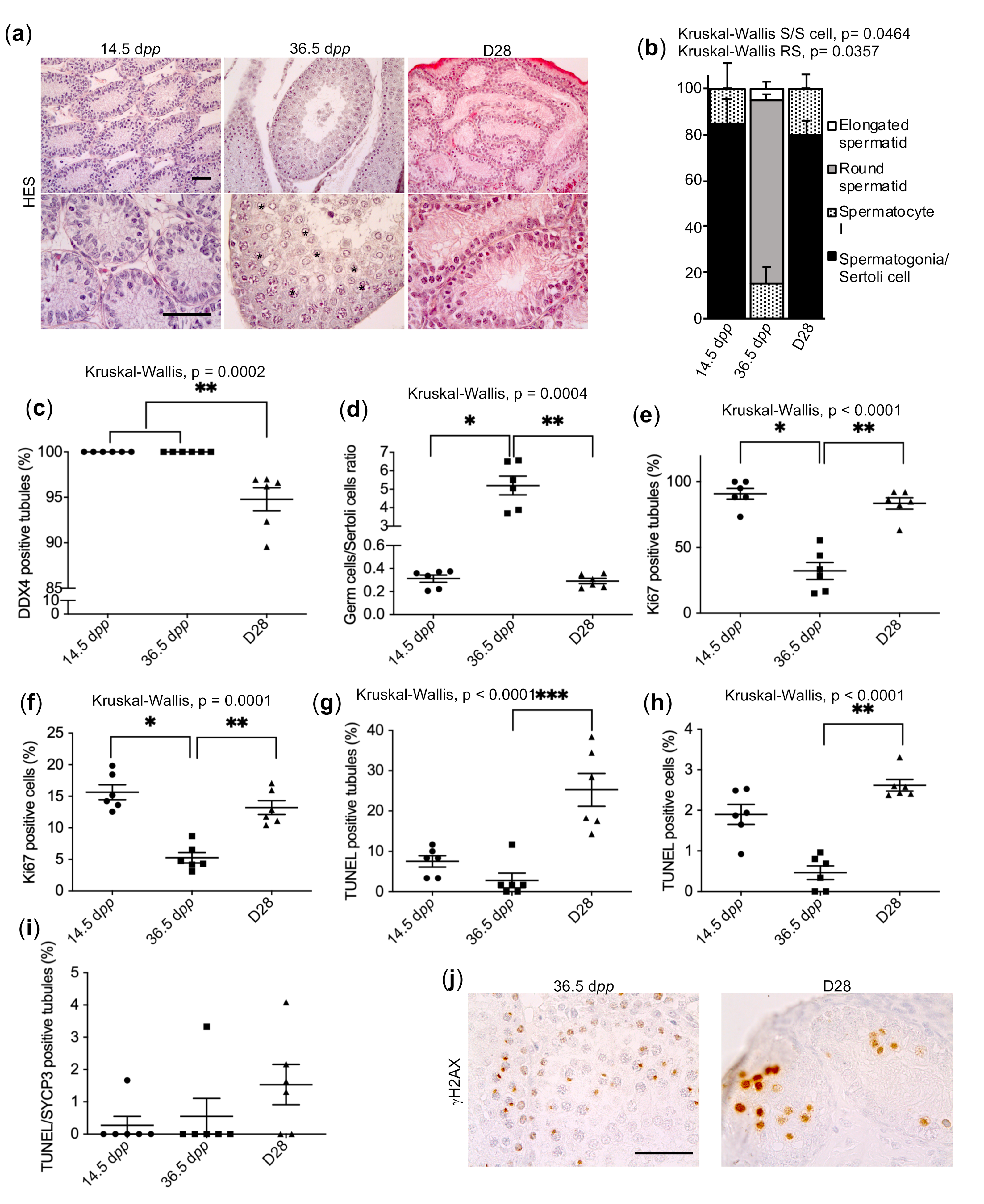
2.1. Pre-Meiotic and Meiotic Germ Cells Survive in Cultured Prepubertal Rat Testicular Tissue but Do Not Achieve Their Differentiation
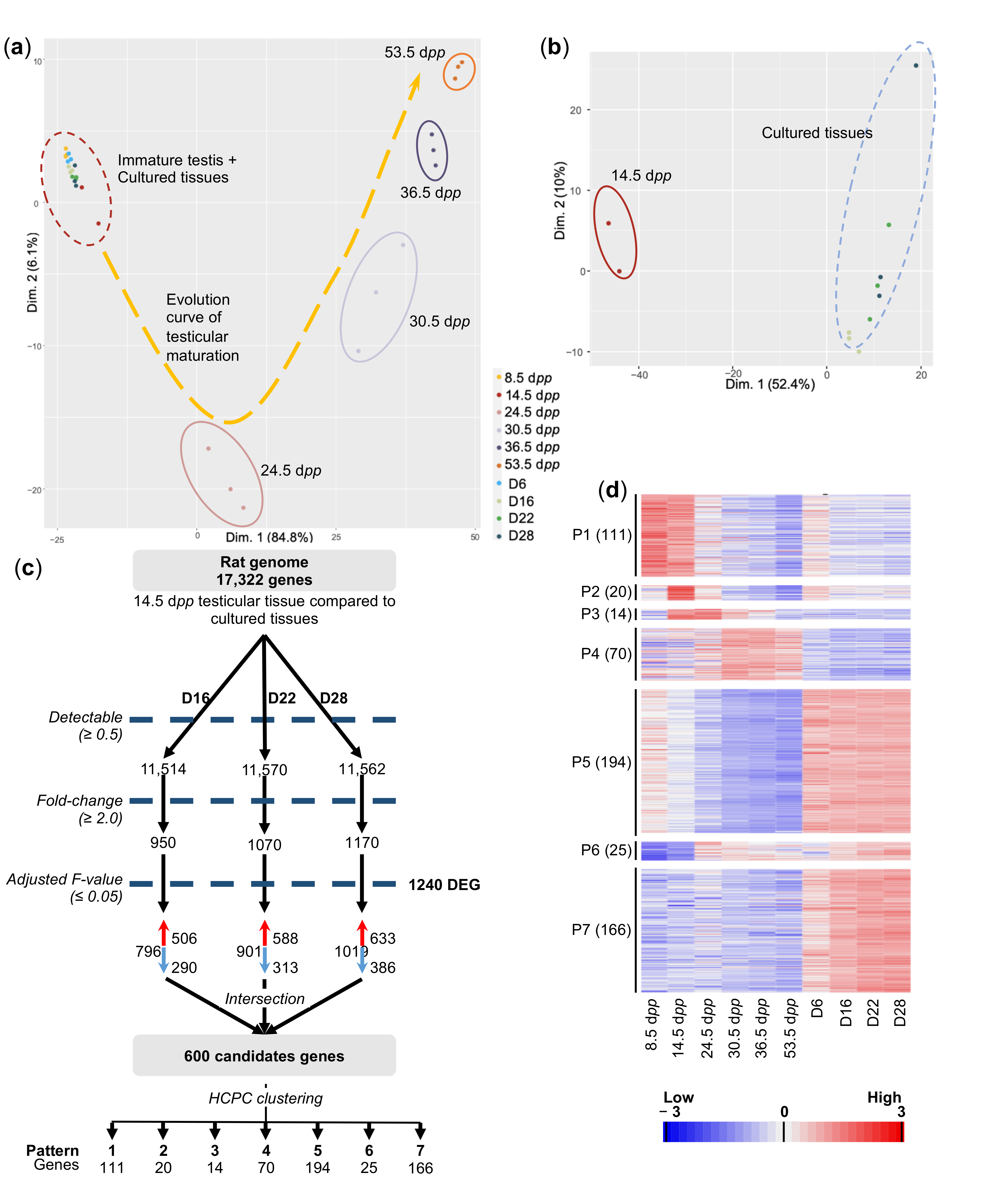
2.2. In Vitro- and In Vivo-Matured Prepubertal Testicular Tissues Have Distinct Transcriptomic Profiles
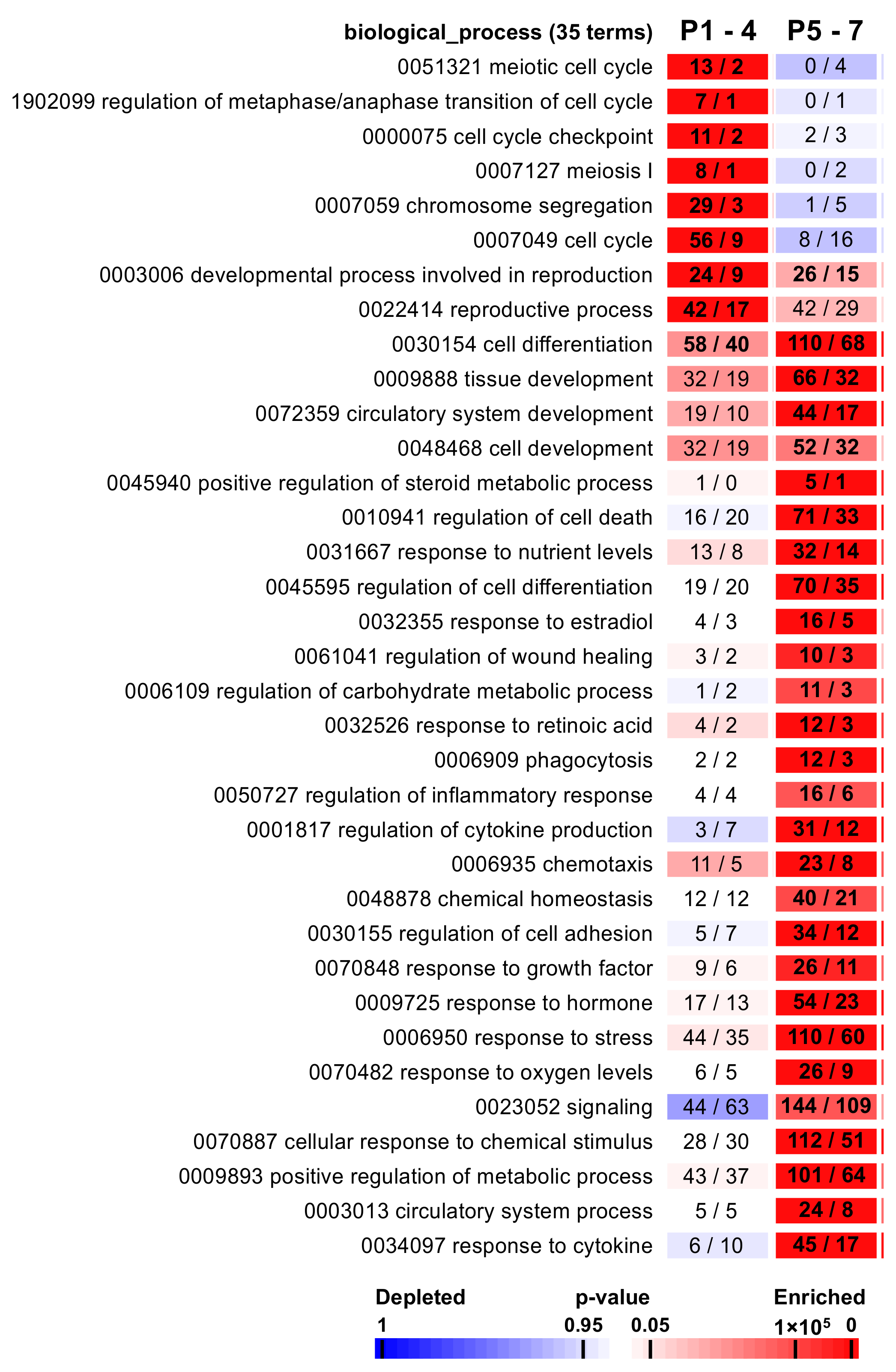
2.3. The Functional Analysis Revealed Associations with Specific Biological Processes
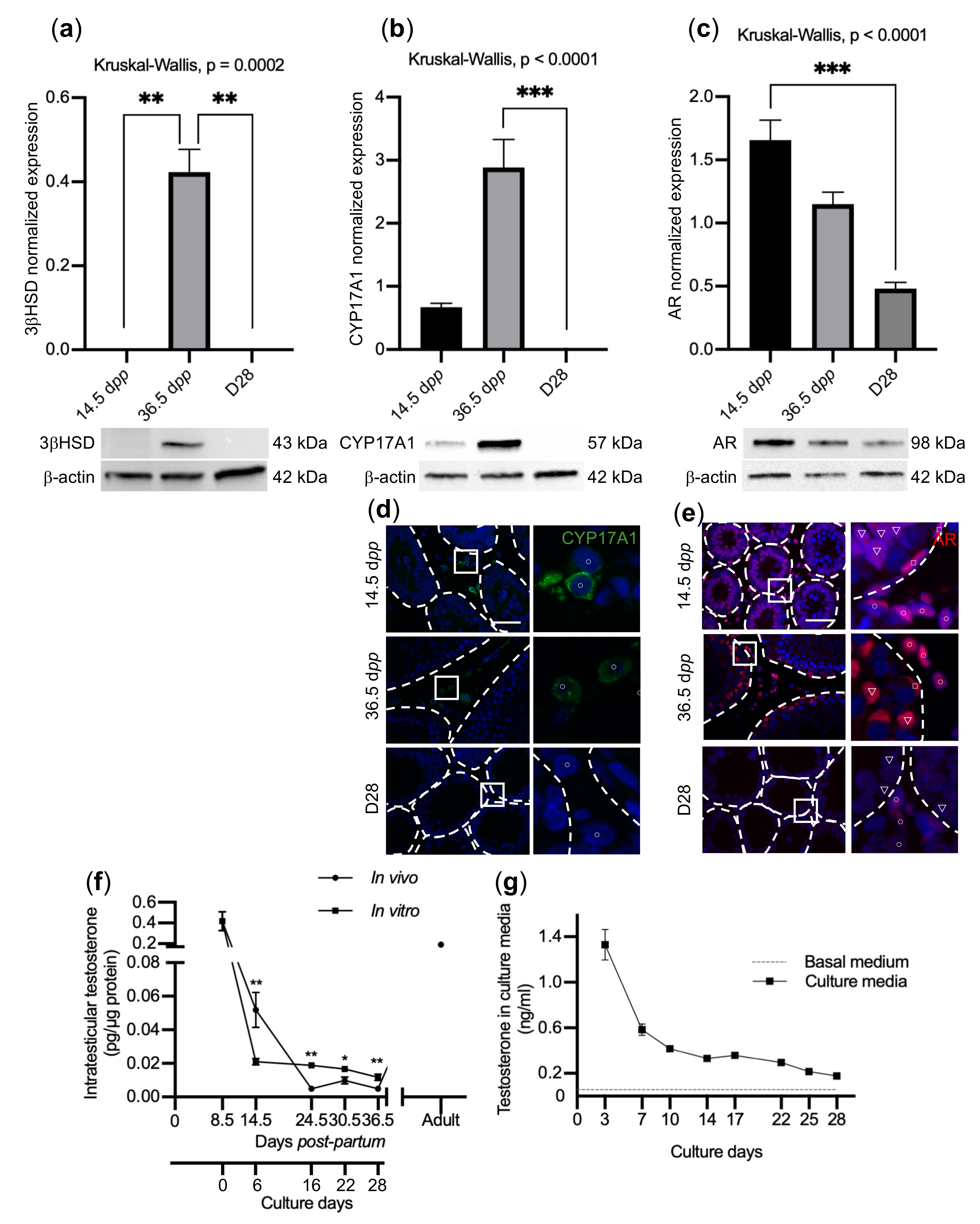
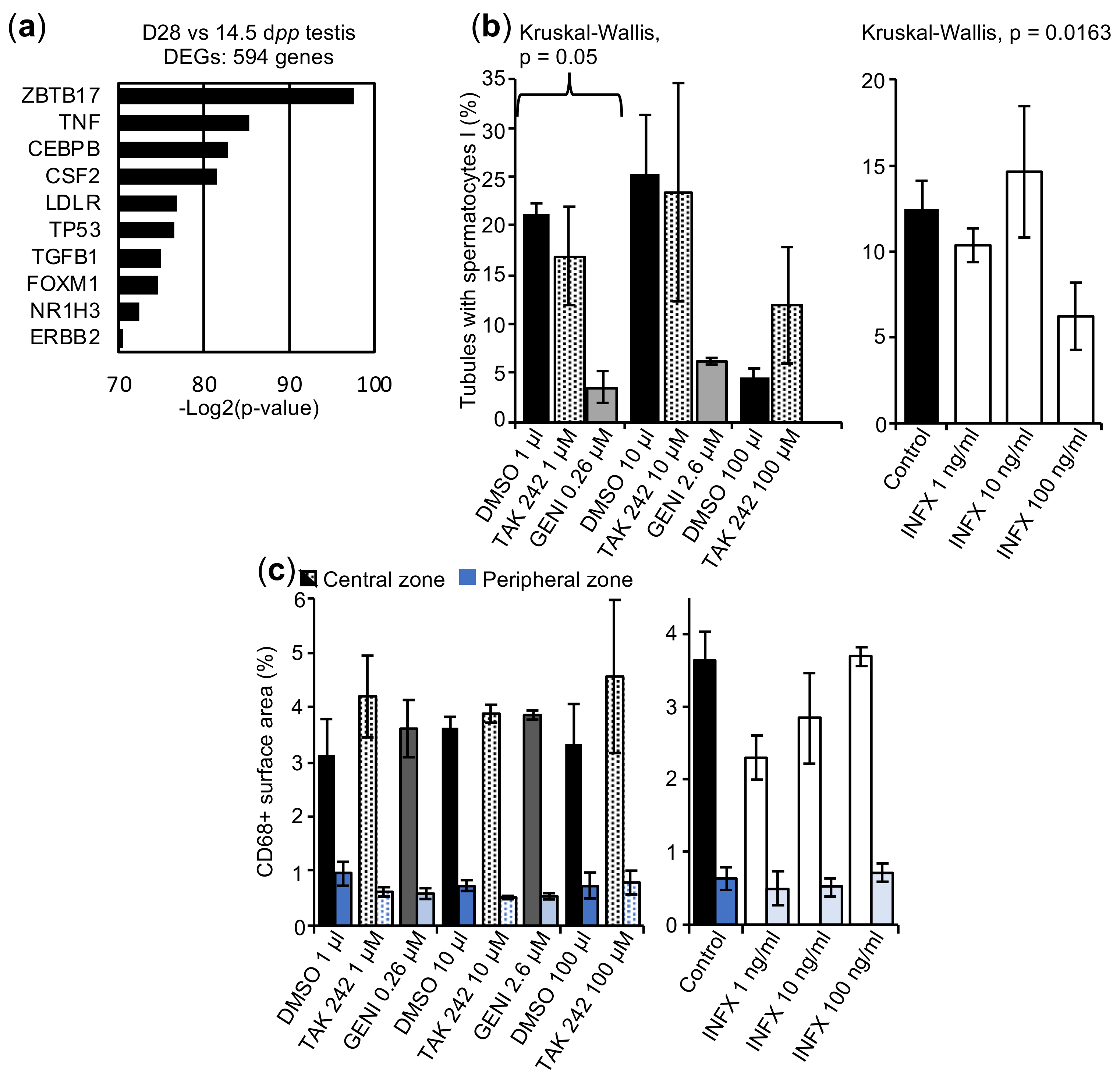
2.4. Impaired BTB Formation Could Impede Meiotic Progression
2.5. The In Vitro Spermatogenesis Process May Be Associated with an Inflammatory Process
3. Discussion
4. Materials and Methods
4.1. Animal Care and Handling
4.2. Animals and Tissue Samples
4.3. Organotypic Cultures
4.4. Histological and Immunohistochemical Analyses
4.4.1. HES
4.4.2. TUNEL-SYCP3
4.4.3. CD68
4.4.4. γH2AX
4.5. Chromosome Spreads
4.6. Analysis of Chromosome Synapsis
4.7. Classification of Prophase I Stages
4.8. Confocal Microscopy Analysis
4.9. RNA Extraction
4.10. RNA-Seq Library Preparation and Sequencing
4.11. RNA-Seq Analysis
4.11.1. Read Mapping
4.11.2. Transcriptome Quantification
4.11.3. Principal Component Analysis
4.11.4. Statistical Filtration and Clustering Analysis
4.12. Functional Analysis
4.13. Validation of the Transcriptome Data by RT-qPCR
4.13.1. Reverse Transcription
4.13.2. qPCR
4.14. BTB Integrity Assay
4.15. Hormone Measurement with LC-MS/MS
4.15.1. Testicular Tissue Preparation
4.15.2. Sample Preparation
4.15.3. 2D-UHPLC-MS/MS Conditions
4.16. Western Blot Analysis
4.17. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Delessard, M.; Saulnier, J.; Rives, A.; Dumont, L.; Rondanino, C.; Rives, N. Exposure to Chemotherapy During Childhood or Adulthood and Consequences on Spermatogenesis and Male Fertility. Int. J. Mol. Sci. 2020, 21, 1454. [Google Scholar] [CrossRef] [PubMed]
- Picton, H.M.; Wyns, C.; Anderson, R.A.; Goossens, E.; Jahnukainen, K.; Kliesch, S.; Mitchell, R.T.; Pennings, G.; Rives, N.; Tournaye, H.; et al. A European Perspective on Testicular Tissue Cryopreservation for Fertility Preservation in Prepubertal and Adolescent Boys. Hum. Reprod. 2015, 30, 2463–2475. [Google Scholar] [CrossRef] [PubMed]
- Onofre, J.; Baert, Y.; Faes, K.; Goossens, E. Cryopreservation of Testicular Tissue or Testicular Cell Suspensions: A Pivotal Step in Fertility Preservation. Hum. Reprod. Update 2016, 22, 744–761. [Google Scholar] [CrossRef] [PubMed]
- Huleihel, M.; Nourashrafeddin, S.; Plant, T.M. Application of Three-Dimensional Culture Systems to Study Mammalian Spermatogenesis, with an Emphasis on the Rhesus Monkey (Macaca Mulatta). Asian J. Androl. 2015, 17, 972–980. [Google Scholar] [CrossRef]
- Rezaei Topraggaleh, T.; Rezazadeh Valojerdi, M.; Montazeri, L.; Baharvand, H. A Testis-Derived Macroporous 3D Scaffold as a Platform for the Generation of Mouse Testicular Organoids. Biomater. Sci. 2019, 7, 1422–1436. [Google Scholar] [CrossRef]
- Sato, T.; Katagiri, K.; Gohbara, A.; Inoue, K.; Ogonuki, N.; Ogura, A.; Kubota, Y.; Ogawa, T. In Vitro Production of Functional Sperm in Cultured Neonatal Mouse Testes. Nature 2011, 471, 504–507. [Google Scholar] [CrossRef]
- Arkoun, B.; Dumont, L.; Milazzo, J.-P.; Way, A.; Bironneau, A.; Wils, J.; Macé, B.; Rives, N. Retinol Improves in Vitro Differentiation of Pre-Pubertal Mouse Spermatogonial Stem Cells into Sperm during the First Wave of Spermatogenesis. PLoS ONE 2015, 10, e0116660. [Google Scholar] [CrossRef]
- Oblette, A.; Rives-Feraille, A.; Dumont, L.; Delessard, M.; Saulnier, J.; Rives, N.; Rondanino, C. Dynamics of Epigenetic Modifications in ICSI Embryos from in Vitro-Produced Spermatozoa. Andrology 2020, 9, 640–656. [Google Scholar] [CrossRef]
- Pullium, J.K.; Milner, R.; Tuma, G.A.; Lin, P.H. Fertility after Homologous Prepubertal Testis Transplantation in the Dog. Transplant. Proc. 2008, 40, 2744–2749. [Google Scholar] [CrossRef]
- Fayomi, A.P.; Peters, K.; Sukhwani, M.; Valli-Pulaski, H.; Shetty, G.; Meistrich, M.L.; Houser, L.; Robertson, N.; Roberts, V.; Ramsey, C.; et al. Autologous Grafting of Cryopreserved Prepubertal Rhesus Testis Produces Sperm and Offspring. Science 2019, 363, 1314–1319. [Google Scholar] [CrossRef]
- Kanatsu-Shinohara, M.; Ogonuki, N.; Inoue, K.; Miki, H.; Ogura, A.; Toyokuni, S.; Shinohara, T. Long-Term Proliferation in Culture and Germline Transmission of Mouse Male Germline Stem Cells. Biol. Reprod. 2003, 69, 612–616. [Google Scholar] [CrossRef] [PubMed]
- Hermann, B.P.; Sukhwani, M.; Winkler, F.; Pascarella, J.N.; Peters, K.A.; Sheng, Y.; Valli, H.; Rodriguez, M.; Ezzelarab, M.; Dargo, G.; et al. Spermatogonial Stem Cell Transplantation into Rhesus Testes Regenerates Spermatogenesis Producing Functional Sperm. Cell Stem Cell 2012, 11, 715–726. [Google Scholar] [CrossRef] [PubMed]
- De Michele, F.; Poels, J.; Giudice, M.G.; De Smedt, F.; Ambroise, J.; Vermeulen, M.; Gruson, D.; Wyns, C. In-Vitro Formation of the Blood-Testis Barrier during Long-Term Organotypic Culture of Human Prepubertal Tissue: Comparison with a Large Cohort of Pre/Peripubertal Boys. Mol. Hum. Reprod. 2018, 24, 271–282. [Google Scholar] [CrossRef]
- Matsumura, T.; Sato, T.; Abe, T.; Sanjo, H.; Katagiri, K.; Kimura, H.; Fujii, T.; Tanaka, H.; Hirabayashi, M.; Ogawa, T. Rat in Vitro Spermatogenesis Promoted by Chemical Supplementations and Oxygen-Tension Control. Sci. Rep. 2021, 11, 3458. [Google Scholar] [CrossRef]
- Reda, A.; Hou, M.; Winton, T.R.; Chapin, R.E.; Söder, O.; Stukenborg, J.-B. In Vitro Differentiation of Rat Spermatogonia into Round Spermatids in Tissue Culture. Mol. Hum. Reprod. 2016, 22, 601–612. [Google Scholar] [CrossRef]
- Saulnier, J.; Oblette, A.; Delessard, M.; Dumont, L.; Rives, A.; Rives, N.; Rondanino, C. Improving Freezing Protocols and Organotypic Culture: A Histological Study on Rat Prepubertal Testicular Tissue. Ann. Biomed. Eng. 2021, 49, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Kurek, M.; Åkesson, E.; Yoshihara, M.; Oliver, E.; Cui, Y.; Becker, M.; Alves-Lopes, J.P.; Bjarnason, R.; Romerius, P.; Sundin, M.; et al. Spermatogonia Loss Correlates with LAMA 1 Expression in Human Prepubertal Testes Stored for Fertility Preservation. Cells 2021, 10, 241. [Google Scholar] [CrossRef] [PubMed]
- Medrano, J.V.; Vilanova-Pérez, T.; Fornés-Ferrer, V.; Navarro-Gomezlechon, A.; Martínez-Triguero, M.L.; García, S.; Gómez-Chacón, J.; Povo, I.; Pellicer, A.; Andrés, M.M.; et al. Influence of Temperature, Serum, and Gonadotropin Supplementation in Short- and Long-Term Organotypic Culture of Human Immature Testicular Tissue. Fertil. Steril. 2018, 110, 1045–1057.E3. [Google Scholar] [CrossRef] [PubMed]
- Zickler, D.; Kleckner, N. Recombination, Pairing, and Synapsis of Homologs during Meiosis. Cold Spring Harb. Perspect. Biol. 2015, 7, a016626. [Google Scholar] [CrossRef]
- Gao, J.; Colaiácovo, M.P. Zipping and Unzipping: Protein Modifications Regulating Synaptonemal Complex Dynamics. Trends Genet. 2018, 34, 232–245. [Google Scholar] [CrossRef]
- Keeney, S.; Giroux, C.N.; Kleckner, N. Meiosis-Specific DNA Double-Strand Breaks Are Catalyzed by Spo11, a Member of a Widely Conserved Protein Family. Cell 1997, 88, 375–384. [Google Scholar] [CrossRef]
- Baudat, F.; Manova, K.; Yuen, J.P.; Jasin, M.; Keeney, S. Chromosome Synapsis Defects and Sexually Dimorphic Meiotic Progression in Mice Lacking Spo11. Mol. Cell 2000, 6, 989–998. [Google Scholar] [CrossRef]
- Carofiglio, F.; Sleddens-Linkels, E.; Wassenaar, E.; Inagaki, A.; van Cappellen, W.A.; Grootegoed, J.A.; Toth, A.; Baarends, W.M. Repair of Exogenous DNA Double-Strand Breaks Promotes Chromosome Synapsis in SPO11-Mutant Mouse Meiocytes, and Is Altered in the Absence of HORMAD1. DNA Repair 2018, 63, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Roeder, G.S.; Bailis, J.M. The Pachytene Checkpoint. Trends Genet. 2000, 16, 395–403. [Google Scholar] [CrossRef]
- De Rooij, D.G.; de Boer, P. Specific Arrests of Spermatogenesis in Genetically Modified and Mutant Mice. Cytogenet. Genome Res. 2003, 103, 267–276. [Google Scholar] [CrossRef]
- Stahl, P.J.; Mielnik, A.N.; Barbieri, C.E.; Schlegel, P.N.; Paduch, D.A. Deletion or Underexpression of the Y-Chromosome Genes CDY2 and HSFY Is Associated with Maturation Arrest in American Men with Nonobstructive Azoospermia. Asian J. Androl. 2012, 14, 676–682. [Google Scholar] [CrossRef]
- Yatsenko, A.N.; Georgiadis, A.P.; Röpke, A.; Berman, A.J.; Jaffe, T.; Olszewska, M.; Westernströer, B.; Sanfilippo, J.; Kurpisz, M.; Rajkovic, A.; et al. X-Linked TEX11 Mutations, Meiotic Arrest, and Azoospermia in Infertile Men. N. Engl. J. Med. 2015, 372, 2097–2107. [Google Scholar] [CrossRef]
- Kui, F.; Ye, H.; Chen, X.-L.; Zhang, J. Microarray Meta-Analysis Identifies Candidate Genes for Human Spermatogenic Arrest. Andrologia 2019, 51, e13301. [Google Scholar] [CrossRef]
- Wang, Y.-H.; Yan, M.; Zhang, X.; Liu, X.-Y.; Ding, Y.-F.; Lai, C.-P.; Tong, M.-H.; Li, J.-S. Rescue of Male Infertility through Correcting a Genetic Mutation Causing Meiotic Arrest in Spermatogonial Stem Cells. Asian J. Androl. 2021, 23, 590–599. [Google Scholar] [CrossRef]
- Ghieh, F.; Mitchell, V.; Mandon-Pepin, B.; Vialard, F. Genetic Defects in Human Azoospermia. Basic Clin. Androl. 2019, 29, 4. [Google Scholar] [CrossRef]
- Bolcun-Filas, E.; Hall, E.; Speed, R.; Taggart, M.; Grey, C.; de Massy, B.; Benavente, R.; Cooke, H.J. Mutation of the Mouse Syce1 Gene Disrupts Synapsis and Suggests a Link between Synaptonemal Complex Structural Components and DNA Repair. PLoS Genet. 2009, 5, e1000393. [Google Scholar] [CrossRef]
- Barchi, M.; Mahadevaiah, S.; Di Giacomo, M.; Baudat, F.; de Rooij, D.G.; Burgoyne, P.S.; Jasin, M.; Keeney, S. Surveillance of Different Recombination Defects in Mouse Spermatocytes Yields Distinct Responses despite Elimination at an Identical Developmental Stage. Mol. Cell. Biol. 2005, 25, 7203–7215. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, N.A.; Romanienko, P.J.; Khil, P.P.; Camerini-Otero, R.D. Gene Expression Profiles of Spo11-/- Mouse Testes with Spermatocytes Arrested in Meiotic Prophase I. Reproduction 2006, 132, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Mahadevaiah, S.K.; Bourc’his, D.; de Rooij, D.G.; Bestor, T.H.; Turner, J.M.A.; Burgoyne, P.S. Extensive Meiotic Asynapsis in Mice Antagonises Meiotic Silencing of Unsynapsed Chromatin and Consequently Disrupts Meiotic Sex Chromosome Inactivation. J. Cell Biol. 2008, 182, 263–276. [Google Scholar] [CrossRef] [PubMed]
- ElInati, E.; Russell, H.R.; Ojarikre, O.A.; Sangrithi, M.; Hirota, T.; de Rooij, D.G.; McKinnon, P.J.; Turner, J.M.A. DNA Damage Response Protein TOPBP1 Regulates X Chromosome Silencing in the Mammalian Germ Line. Proc. Natl. Acad. Sci. USA 2017, 114, 12536–12541. [Google Scholar] [CrossRef]
- Xu, X.; Aprelikova, O.; Moens, P.; Deng, C.-X.; Furth, P.A. Impaired Meiotic DNA-Damage Repair and Lack of Crossing-over during Spermatogenesis in BRCA1 Full-Length Isoform Deficient Mice. Development 2003, 130, 2001–2012. [Google Scholar] [CrossRef]
- Manfo, F.P.T.; Nantia, E.A.; Mathur, P.P. Effect of Environmental Contaminants on Mammalian Testis. Curr. Mol. Pharm. 2014, 7, 119–135. [Google Scholar] [CrossRef]
- Durairajanayagam, D.; Agarwal, A.; Ong, C. Causes, Effects and Molecular Mechanisms of Testicular Heat Stress. Reprod. Biomed. Online 2015, 30, 14–27. [Google Scholar] [CrossRef]
- Chen, Q.; Deng, T.; Han, D. Testicular Immunoregulation and Spermatogenesis. Semin. Cell Dev. Biol. 2016, 59, 157–165. [Google Scholar] [CrossRef]
- Li, H.; Palczewski, K.; Baehr, W.; Clagett-Dame, M. Vitamin A Deficiency Results in Meiotic Failure and Accumulation of Undifferentiated Spermatogonia in Prepubertal Mouse Testis. Biol. Reprod. 2011, 84, 336–341. [Google Scholar] [CrossRef]
- Chihara, M.; Otsuka, S.; Ichii, O.; Kon, Y. Vitamin A Deprivation Affects the Progression of the Spermatogenic Wave and Initial Formation of the Blood-Testis Barrier, Resulting in Irreversible Testicular Degeneration in Mice. J. Reprod. Dev. 2013, 59, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Cobellis, G.; Noviello, C.; Nino, F.; Romano, M.; Mariscoli, F.; Martino, A.; Parmeggiani, P.; Papparella, A. Spermatogenesis and Cryptorchidism. Front. Endocrinol. 2014, 5, 63. [Google Scholar] [CrossRef] [PubMed]
- Hazra, R.; Jimenez, M.; Desai, R.; Handelsman, D.J.; Allan, C.M. Sertoli Cell Androgen Receptor Expression Regulates Temporal Fetal and Adult Leydig Cell Differentiation, Function, and Population Size. Endocrinology 2013, 154, 3410–3422. [Google Scholar] [CrossRef] [PubMed]
- Jeyaraj, D.A.; Grossman, G.; Weaver, C.; Petrusz, P. Dynamics of Testicular Germ Cell Proliferation in Normal Mice and Transgenic Mice Overexpressing Rat Androgen-Binding Protein: A Flow Cytometric Evaluation. Biol. Reprod. 2002, 66, 877–885. [Google Scholar] [CrossRef][Green Version]
- Gangwar, P.K.; Sankhwar, S.N.; Pant, S.; Krishna, A.; Singh, B.P.; Mahdi, A.A.; Singh, R. Increased Gonadotropins and Prolactin Are Linked to Infertility in Males. Bioinformation 2020, 16, 176–182. [Google Scholar] [CrossRef]
- Welsh, M.; Saunders, P.T.K.; Atanassova, N.; Sharpe, R.M.; Smith, L.B. Androgen Action via Testicular Peritubular Myoid Cells Is Essential for Male Fertility. FASEB J. 2009, 23, 4218–4230. [Google Scholar] [CrossRef]
- Xu, Q.; Lin, H.-Y.; Yeh, S.-D.; Yu, I.-C.; Wang, R.-S.; Chen, Y.-T.; Zhang, C.; Altuwaijri, S.; Chen, L.-M.; Chuang, K.-H.; et al. Infertility with Defective Spermatogenesis and Steroidogenesis in Male Mice Lacking Androgen Receptor in Leydig Cells. Endocrine 2007, 32, 96–106. [Google Scholar] [CrossRef]
- Yao, J.; Zuo, H.; Gao, J.; Wang, M.; Wang, D.; Li, X. The Effects of IGF-1 on Mouse Spermatogenesis Using an Organ Culture Method. Biochem. Biophys. Res. Commun. 2017, 491, 840–847. [Google Scholar] [CrossRef]
- Abe, T.; Nishimura, H.; Sato, T.; Suzuki, H.; Ogawa, T.; Suzuki, T. Transcriptome Analysis Reveals Inadequate Spermatogenesis and Immediate Radical Immune Reactions during Organ Culture in Vitro Spermatogenesis. Biochem. Biophys. Res. Commun. 2020, 530, 732–738. [Google Scholar] [CrossRef]
- Morohaku, K.; Tanimoto, R.; Sasaki, K.; Kawahara-Miki, R.; Kono, T.; Hayashi, K.; Hirao, Y.; Obata, Y. Complete in Vitro Generation of Fertile Oocytes from Mouse Primordial Germ Cells. Proc. Natl. Acad. Sci. USA 2016, 113, 9021–9026. [Google Scholar] [CrossRef]
- Burgoyne, P.S.; Mahadevaiah, S.K.; Turner, J.M.A. The Consequences of Asynapsis for Mammalian Meiosis. Nat. Rev. Genet. 2009, 10, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Rodríguez, J. ΓH2AX Marks the Main Events of the Spermatogenic Process. Microsc. Res. Tech. 2009, 72, 823–832. [Google Scholar] [CrossRef] [PubMed]
- Fijak, M.; Bhushan, S.; Meinhardt, A. Immunoprivileged Sites: The Testis. Methods Mol. Biol. 2011, 677, 459–470. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.-H.; Bukhari, I.; Zheng, W.; Yin, S.; Wang, Z.; Cooke, H.J.; Shi, Q.-H. Blood-Testis Barrier and Spermatogenesis: Lessons from Genetically-Modified Mice. Asian J. Androl. 2014, 16, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Mruk, D.D.; Cheng, C.Y. The Mammalian Blood-Testis Barrier: Its Biology and Regulation. Endocr. Rev. 2015, 36, 564–591. [Google Scholar] [CrossRef]
- Aghazadeh, Y.; Zirkin, B.R.; Papadopoulos, V. Pharmacological Regulation of the Cholesterol Transport Machinery in Steroidogenic Cells of the Testis. Vitam. Horm. 2015, 98, 189–227. [Google Scholar] [CrossRef]
- Sansone, A.; Kliesch, S.; Isidori, A.M.; Schlatt, S. AMH and INSL3 in Testicular and Extragonadal Pathophysiology: What Do We Know? Andrology 2019, 7, 131–138. [Google Scholar] [CrossRef]
- Toshima, J.; Ohashi, K.; Okano, I.; Nunoue, K.; Kishioka, M.; Kuma, K.; Miyata, T.; Hirai, M.; Baba, T.; Mizuno, K. Identification and Characterization of a Novel Protein Kinase, TESK1, Specifically Expressed in Testicular Germ Cells. J. Biol. Chem. 1995, 270, 31331–31337. [Google Scholar] [CrossRef]
- Toshima, J.; Koji, T.; Mizuno, K. Stage-Specific Expression of Testis-Specific Protein Kinase 1 (TESK1) in Rat Spermatogenic Cells. Biochem. Biophys. Res. Commun. 1998, 249, 107–112. [Google Scholar] [CrossRef]
- Toshima, J.; Toshima, J.Y.; Suzuki, M.; Noda, T.; Mizuno, K. Cell-Type-Specific Expression of a TESK1 Promoter-Linked LacZ Gene in Transgenic Mice. Biochem. Biophys. Res. Commun. 2001, 286, 566–573. [Google Scholar] [CrossRef]
- Pati, D.; Meistrich, M.L.; Plon, S.E. Human Cdc34 and Rad6B Ubiquitin-Conjugating Enzymes Target Repressors of Cyclic AMP-Induced Transcription for Proteolysis. Mol. Cell. Biol. 1999, 19, 5001–5013. [Google Scholar] [CrossRef] [PubMed]
- Butz, N.; Ruetz, S.; Natt, F.; Hall, J.; Weiler, J.; Mestan, J.; Ducarre, M.; Grossenbacher, R.; Hauser, P.; Kempf, D.; et al. The Human Ubiquitin-Conjugating Enzyme Cdc34 Controls Cellular Proliferation through Regulation of P27Kip1 Protein Levels. Exp. Cell Res. 2005, 303, 482–493. [Google Scholar] [CrossRef] [PubMed]
- Legesse-Miller, A.; Elemento, O.; Pfau, S.J.; Forman, J.J.; Tavazoie, S.; Coller, H.A. Let-7 Overexpression Leads to an Increased Fraction of Cells in G2/M, Direct down-Regulation of Cdc34, and Stabilization of Wee1 Kinase in Primary Fibroblasts. J. Biol. Chem. 2009, 284, 6605–6609. [Google Scholar] [CrossRef] [PubMed]
- Hara-Yokoyama, M.; Kurihara, H.; Ichinose, S.; Matsuda, H.; Ichinose, S.; Kurosawa, M.; Tada, N.; Iwahara, C.; Terasawa, K.; Podyma-Inoue, K.A.; et al. KIF11 as a Potential Marker of Spermatogenesis Within Mouse Seminiferous Tubule Cross-Sections. J. Histochem. Cytochem. 2019, 67, 813–824. [Google Scholar] [CrossRef]
- Blangy, A.; Lane, H.A.; d’Hérin, P.; Harper, M.; Kress, M.; Nigg, E.A. Phosphorylation by P34cdc2 Regulates Spindle Association of Human Eg5, a Kinesin-Related Motor Essential for Bipolar Spindle Formation in Vivo. Cell 1995, 83, 1159–1169. [Google Scholar] [CrossRef]
- Kapitein, L.C.; Peterman, E.J.G.; Kwok, B.H.; Kim, J.H.; Kapoor, T.M.; Schmidt, C.F. The Bipolar Mitotic Kinesin Eg5 Moves on Both Microtubules That It Crosslinks. Nature 2005, 435, 114–118. [Google Scholar] [CrossRef]
- Ben-Eliezer, I.; Pomerantz, Y.; Galiani, D.; Nevo, N.; Dekel, N. Appropriate Expression of Ube2C and Ube2S Controls the Progression of the First Meiotic Division. FASEB J. 2015, 29, 4670–4681. [Google Scholar] [CrossRef]
- Peters, J.-M. The Anaphase-Promoting Complex: Proteolysis in Mitosis and Beyond. Mol. Cell 2002, 9, 931–943. [Google Scholar] [CrossRef]
- Hao, Z.; Zhang, H.; Cowell, J. Ubiquitin-Conjugating Enzyme UBE2C: Molecular Biology, Role in Tumorigenesis, and Potential as a Biomarker. Tumour Biol. 2012, 33, 723–730. [Google Scholar] [CrossRef]
- Adams, R.R.; Eckley, D.M.; Vagnarelli, P.; Wheatley, S.P.; Gerloff, D.L.; Mackay, A.M.; Svingen, P.A.; Kaufmann, S.H.; Earnshaw, W.C. Human INCENP Colocalizes with the Aurora-B/AIRK2 Kinase on Chromosomes and Is Overexpressed in Tumour Cells. Chromosoma 2001, 110, 65–74. [Google Scholar] [CrossRef]
- Parra, M.T.; Viera, A.; Gómez, R.; Page, J.; Carmena, M.; Earnshaw, W.C.; Rufas, J.S.; Suja, J.A. Dynamic Relocalization of the Chromosomal Passenger Complex Proteins Inner Centromere Protein (INCENP) and Aurora-B Kinase during Male Mouse Meiosis. J. Cell Sci. 2003, 116, 961. [Google Scholar] [CrossRef] [PubMed]
- Goto, H.; Kiyono, T.; Tomono, Y.; Kawajiri, A.; Urano, T.; Furukawa, K.; Nigg, E.A.; Inagaki, M. Complex Formation of Plk1 and INCENP Required for Metaphase-Anaphase Transition. Nat. Cell Biol. 2006, 8, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, N.; Yanagisawa, M.; Nishimune, Y.; Obinata, M.; Matsui, Y. Murine Polo like Kinase 1 Gene Is Expressed in Meiotic Testicular Germ Cells and Oocytes. Mol. Reprod. Dev. 1995, 41, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Jordan, P.W.; Karppinen, J.; Handel, M.A. Polo-like Kinase Is Required for Synaptonemal Complex Disassembly and Phosphorylation in Mouse Spermatocytes. J. Cell Sci. 2012, 125, 5061. [Google Scholar] [CrossRef]
- Liu, D.; Davydenko, O.; Lampson, M.A. Polo-like Kinase-1 Regulates Kinetochore-Microtubule Dynamics and Spindle Checkpoint Silencing. J. Cell Biol. 2012, 198, 491–499. [Google Scholar] [CrossRef]
- Mondal, G.; Ohashi, A.; Yang, L.; Rowley, M.; Couch, F.J. Tex14, a Plk1-Regulated Protein, Is Required for Kinetochore-Microtubule Attachment and Regulation of the Spindle Assembly Checkpoint. Mol. Cell 2012, 45, 680–695. [Google Scholar] [CrossRef]
- Giménez-Abián, J.F.; Sumara, I.; Hirota, T.; Hauf, S.; Gerlich, D.; de la Torre, C.; Ellenberg, J.; Peters, J.-M. Regulation of Sister Chromatid Cohesion between Chromosome Arms. Curr. Biol. 2004, 14, 1187–1193. [Google Scholar] [CrossRef]
- Matsumura, S.; Toyoshima, F.; Nishida, E. Polo-like Kinase 1 Facilitates Chromosome Alignment during Prometaphase through BubR1. J. Biol. Chem. 2007, 282, 15217–15227. [Google Scholar] [CrossRef]
- Kong, M.; Barnes, E.A.; Ollendorff, V.; Donoghue, D.J. Cyclin F Regulates the Nuclear Localization of Cyclin B1 through a Cyclin-Cyclin Interaction. EMBO J. 2000, 19, 1378–1388. [Google Scholar] [CrossRef]
- Sun, Y.; Fu, L.; Xue, F.; Li, Y.; Xu, H.; Chen, J. Digital Gene Expression Profiling and Validation Study Highlight Cyclin F as an Important Regulator for Sperm Motility of Chickens. Poult. Sci. 2019, 98, 5118–5126. [Google Scholar] [CrossRef]
- Rother, K.; Dengl, M.; Lorenz, J.; Tschöp, K.; Kirschner, R.; Mössner, J.; Engeland, K. Gene Expression of Cyclin-Dependent Kinase Subunit Cks2 Is Repressed by the Tumor Suppressor P53 but Not by the Related Proteins P63 or P73. FEBS Lett. 2007, 581, 1166–1172. [Google Scholar] [CrossRef] [PubMed]
- Spruck, C.H.; de Miguel, M.P.; Smith, A.P.L.; Ryan, A.; Stein, P.; Schultz, R.M.; Lincoln, A.J.; Donovan, P.J.; Reed, S.I. Requirement of Cks2 for the First Metaphase/Anaphase Transition of Mammalian Meiosis. Science 2003, 300, 647–650. [Google Scholar] [CrossRef] [PubMed]
- Fischer, K.A.; Van Leyen, K.; Lovercamp, K.W.; Manandhar, G.; Sutovsky, M.; Feng, D.; Safranski, T.; Sutovsky, P. 15-Lipoxygenase Is a Component of the Mammalian Sperm Cytoplasmic Droplet. Reproduction 2005, 130, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Bromfield, E.G.; Mihalas, B.P.; Dun, M.D.; Aitken, R.J.; McLaughlin, E.A.; Walters, J.L.H.; Nixon, B. Inhibition of Arachidonate 15-Lipoxygenase Prevents 4-Hydroxynonenal-Induced Protein Damage in Male Germ Cells. Biol. Reprod. 2017, 96, 598–609. [Google Scholar] [CrossRef] [PubMed]
- Bromfield, E.G.; Walters, J.L.H.; Cafe, S.L.; Bernstein, I.R.; Stanger, S.J.; Anderson, A.L.; Aitken, R.J.; McLaughlin, E.A.; Dun, M.D.; Gadella, B.M.; et al. Differential Cell Death Decisions in the Testis: Evidence for an Exclusive Window of Ferroptosis in Round Spermatids. Mol. Hum. Reprod. 2019, 25, 241–256. [Google Scholar] [CrossRef]
- Walters, J.L.H.; De Iuliis, G.N.; Dun, M.D.; Aitken, R.J.; McLaughlin, E.A.; Nixon, B.; Bromfield, E.G. Pharmacological Inhibition of Arachidonate 15-Lipoxygenase Protects Human Spermatozoa against Oxidative Stress. Biol. Reprod. 2018, 98, 784–794. [Google Scholar] [CrossRef]
- Izumi, H.; Matsumoto, Y.; Ikeuchi, T.; Saya, H.; Kajii, T.; Matsuura, S. BubR1 Localizes to Centrosomes and Suppresses Centrosome Amplification via Regulating Plk1 Activity in Interphase Cells. Oncogene 2009, 28, 2806–2820. [Google Scholar] [CrossRef]
- Jeganathan, K.B.; van Deursen, J.M. Differential Mitotic Checkpoint Protein Requirements in Somatic and Germ Cells. Biochem. Soc. Trans. 2006, 34, 583–586. [Google Scholar] [CrossRef]
- Choi, E.; Lee, H. Chromosome Damage in Mitosis Induces BubR1 Activation and Prometaphase Arrest. FEBS Lett. 2008, 582, 1700–1706. [Google Scholar] [CrossRef]
- Parra, M.T.; Gómez, R.; Viera, A.; Llano, E.; Pendás, A.M.; Rufas, J.S.; Suja, J.A. Sequential Assembly of Centromeric Proteins in Male Mouse Meiosis. PLoS Genet. 2009, 5, e1000417. [Google Scholar] [CrossRef]
- Ravnik, S.E.; Wolgemuth, D.J. The Developmentally Restricted Pattern of Expression in the Male Germ Line of a Murine Cyclin A, Cyclin A2, Suggests Roles in Both Mitotic and Meiotic Cell Cycles. Dev. Biol. 1996, 173, 69–78. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ravnik, S.E.; Wolgemuth, D.J. Regulation of Meiosis during Mammalian Spermatogenesis: The A-Type Cyclins and Their Associated Cyclin-Dependent Kinases Are Differentially Expressed in the Germ-Cell Lineage. Dev. Biol. 1999, 207, 408–418. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, C.; Murphy, M.; Kubelka, M.; Ravnik, S.E.; Hawkins, C.F.; Wolgemuth, D.J.; Carrington, M. A Distinct Cyclin A Is Expressed in Germ Cells in the Mouse. Development 1996, 122, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Wang, X.; Chung, S.S.W.; Sicinski, P.; Shang, E.; Wolgemuth, D.J. Cyclin A2 Is Essential for Mouse Gonocyte Maturation. Cell Cycle 2020, 19, 1654–1664. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-H.; Yuen, W.S.; Adhikari, D.; Flegg, J.A.; FitzHarris, G.; Conti, M.; Sicinski, P.; Nabti, I.; Marangos, P.; Carroll, J. Cyclin A2 Modulates Kinetochore-Microtubule Attachment in Meiosis II. J. Cell Biol. 2017, 216, 3133–3143. [Google Scholar] [CrossRef] [PubMed]
- Baumann, C.; De La Fuente, R. Role of Polycomb Group Protein Cbx2/M33 in Meiosis Onset and Maintenance of Chromosome Stability in the Mammalian Germline. Genes 2011, 2, 59–80. [Google Scholar] [CrossRef]
- Lee, W.-Y.; Do, J.T.; Park, C.; Kim, J.H.; Chung, H.-J.; Kim, K.-W.; Gil, C.-H.; Kim, N.-H.; Song, H. Identification of Putative Biomarkers for the Early Stage of Porcine Spermatogonial Stem Cells Using Next-Generation Sequencing. PLoS ONE 2016, 11, e0147298. [Google Scholar] [CrossRef]
- Robinson, L.L.; Sznajder, N.A.; Riley, S.C.; Anderson, R.A. Matrix Metalloproteinases and Tissue Inhibitors of Metalloproteinases in Human Fetal Testis and Ovary. Mol. Hum. Reprod. 2001, 7, 641–648. [Google Scholar] [CrossRef]
- Kumar, S.; Tinson, A.; Mulligan, B.P.; Ojha, S. Gelatin Binding Proteins in Reproductive Physiology. Indian J. Microbiol. 2016, 56, 383–393. [Google Scholar] [CrossRef][Green Version]
- Kurzawski, M.; Kaczmarek, M.; Kłysz, M.; Malinowski, D.; Kazienko, A.; Kurzawa, R.; Droździk, M. MMP2, MMP9 and TIMP2 Polymorphisms Affect Sperm Parameters but Not Fertility in Polish Males. Andrologia 2017, 49, e12654. [Google Scholar] [CrossRef]
- Lydka, M.; Bilinska, B.; Cheng, C.Y.; Mruk, D.D. Tumor Necrosis Factor α-Mediated Restructuring of the Sertoli Cell Barrier in Vitro Involves Matrix Metalloprotease 9 (MMP9), Membrane-Bound Intercellular Adhesion Molecule-1 (ICAM-1) and the Actin Cytoskeleton. Spermatogenesis 2012, 2, 294–303. [Google Scholar] [CrossRef] [PubMed]
- Siu, M.K.Y.; Lee, W.M.; Cheng, C.Y. The Interplay of Collagen IV, Tumor Necrosis Factor-α, Gelatinase B (Matrix Metalloprotease-9), and Tissue Inhibitor of Metalloproteases-1 in the Basal Lamina Regulates Sertoli Cell-Tight Junction Dynamics in the Rat Testis. Endocrinology 2003, 144, 371–387. [Google Scholar] [CrossRef] [PubMed]
- Siu, M.K.Y.; Cheng, C.Y. Interactions of Proteases, Protease Inhibitors, and the Beta1 Integrin/Laminin Gamma3 Protein Complex in the Regulation of Ectoplasmic Specialization Dynamics in the Rat Testis. Biol. Reprod. 2004, 70, 945–964. [Google Scholar] [CrossRef]
- Siu, M.K.Y.; Cheng, C.Y. Extracellular Matrix and Its Role in Spermatogenesis. Adv. Exp. Med. Biol. 2008, 636, 74–91. [Google Scholar] [CrossRef] [PubMed]
- Mohagheghi, S.; Khodadadi, I.; Karami, M.; Amiri, I.; Tavilani, H. Gene Polymorphism of Matrix Metalloproteinase 9 in Asthenozoospermic Male Subjects. Int. J. Fertil. Steril. 2018, 11, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Riaz, M.A.; Stammler, A.; Borgers, M.; Konrad, L. Clusterin Signals via ApoER2/VLDLR and Induces Meiosis of Male Germ Cells. Am. J. Transl. Res. 2017, 9, 1266–1276. [Google Scholar] [PubMed]
- Tacken, P.J.; van der Zee, A.; Beumer, T.L.; Florijn, R.J.; Gijpels, M.J.; Havekes, L.M.; Frants, R.R.; van Dijk, K.W.; Hofker, M.H. Effective Generation of Very Low Density Lipoprotein Receptor Transgenic Mice by Overlapping Genomic DNA Fragments: High Testis Expression and Disturbed Spermatogenesis. Transgenic Res. 2001, 10, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, L.; Ma, T.; Gao, L.; Yang, L.; Wu, M.; Pang, Z.; Wang, X.; Yao, Q.; Xiao, Y.; et al. Bisphenol A Attenuates Testosterone Production in Leydig Cells via the Inhibition of NR1D1 Signaling. Chemosphere 2021, 263, 128020. [Google Scholar] [CrossRef]
- Ferder, I.C.; Fung, L.; Ohguchi, Y.; Zhang, X.; Lassen, K.G.; Capen, D.; Brown, D.; Xavier, R.J.; Wang, N. Meiotic Gatekeeper STRA8 Suppresses Autophagy by Repressing Nr1d1 Expression during Spermatogenesis in Mice. PLoS Genet. 2019, 15, e1008084. [Google Scholar] [CrossRef]
- Rappaport, M.S.; Smith, E.P. Insulin-like Growth Factor (IGF) Binding Protein 3 in the Rat Testis: Follicle-Stimulating Hormone Dependence of MRNA Expression and Inhibition of IGF-I Action on Cultured Sertoli Cells. Biol. Reprod. 1995, 52, 419–425. [Google Scholar] [CrossRef][Green Version]
- Lin, T.; Wang, D.; Nagpal, M.L.; Shimasaki, S.; Ling, N. Expression and Regulation of Insulin-like Growth Factor-Binding Protein-1, -2, -3, and -4 Messenger Ribonucleic Acids in Purified Rat Leydig Cells and Their Biological Effects. Endocrinology 1993, 132, 1898–1904. [Google Scholar] [CrossRef] [PubMed]
- Lue, Y.; Swerdloff, R.; Liu, Q.; Mehta, H.; Hikim, A.S.; Lee, K.-W.; Jia, Y.; Hwang, D.; Cobb, L.J.; Cohen, P.; et al. Opposing Roles of Insulin-like Growth Factor Binding Protein 3 and Humanin in the Regulation of Testicular Germ Cell Apoptosis. Endocrinology 2010, 151, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; You, S.; Li, W.; Wang, D.; Nagpal, M.L.; Mi, Y.; Liang, P.; Lin, T. Expression and Regulation of Interferon-Gamma-Inducible Protein 10 Gene in Rat Leydig Cells. Endocrinology 1998, 139, 3637–3645. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Le Goffic, R.; Mouchel, T.; Aubry, F.; Patard, J.-J.; Ruffault, A.; Jégou, B.; Samson, M. Production of the Chemokines Monocyte Chemotactic Protein-1, Regulated on Activation Normal T Cell Expressed and Secreted Protein, Growth-Related Oncogene, and Interferon-Gamma-Inducible Protein-10 Is Induced by the Sendai Virus in Human and Rat Testicular Cells. Endocrinology 2002, 143, 1434–1440. [Google Scholar] [CrossRef][Green Version]
- Aubry, F.; Habasque, C.; Satie, A.P.; Jégou, B.; Samson, M. Expression and Regulation of the CXC-Chemokines, GRO/KC and IP-10/Mob-1 in Rat Seminiferous Tubules. Eur. Cytokine Netw. 2000, 11, 690–698. [Google Scholar]
- Jiang, Q.; Wang, F.; Shi, L.; Zhao, X.; Gong, M.; Liu, W.; Song, C.; Li, Q.; Chen, Y.; Wu, H.; et al. C-X-C Motif Chemokine Ligand 10 Produced by Mouse Sertoli Cells in Response to Mumps Virus Infection Induces Male Germ Cell Apoptosis. Cell Death Dis. 2017, 8, e3146. [Google Scholar] [CrossRef]
- Dong, Y.-S.; Hou, W.-G.; Li, Y.; Liu, D.-B.; Hao, G.-Z.; Zhang, H.-F.; Li, J.-C.; Zhao, J.; Zhang, S.; Liang, G.-B.; et al. Unexpected Requirement for a Binding Partner of the Syntaxin Family in Phagocytosis by Murine Testicular Sertoli Cells. Cell Death Differ. 2016, 23, 787–800. [Google Scholar] [CrossRef]
- Merkl, M.; Ertl, R.; Handschuh, S.; Aurich, C.; Schäfer-Somi, S. The Cholesterol Transporter ABCA1 Is Expressed in Stallion Spermatozoa and Reproductive Tract Tissues. Theriogenology 2016, 85, 1080–1089. [Google Scholar] [CrossRef]
- Stocchi, L.; Giardina, E.; Varriale, L.; Sechi, A.; Vagnini, A.; Parri, G.; Valentini, M.; Capalbo, M. Can Tangier Disease Cause Male Infertility? A Case Report and an Overview on Genetic Causes of Male Infertility and Hormonal Axis Involved. Mol. Genet. Metab. 2018, 123, 43–49. [Google Scholar] [CrossRef]
- Xia, Y.; Sidis, Y.; Schneyer, A. Overexpression of Follistatin-like 3 in Gonads Causes Defects in Gonadal Development and Function in Transgenic Mice. Mol. Endocrinol. 2004, 18, 979–994. [Google Scholar] [CrossRef]
- Tsuchida, K.; Arai, K.Y.; Kuramoto, Y.; Yamakawa, N.; Hasegawa, Y.; Sugino, H. Identification and Characterization of a Novel Follistatin-like Protein as a Binding Protein for the TGF-Beta Family. J. Biol. Chem. 2000, 275, 40788–40796. [Google Scholar] [CrossRef] [PubMed]
- Oldknow, K.J.; Seebacher, J.; Goswami, T.; Villen, J.; Pitsillides, A.A.; O’Shaughnessy, P.J.; Gygi, S.P.; Schneyer, A.L.; Mukherjee, A. Follistatin-like 3 (FSTL3) Mediated Silencing of Transforming Growth Factor β (TGFβ) Signaling Is Essential for Testicular Aging and Regulating Testis Size. Endocrinology 2013, 154, 1310–1320. [Google Scholar] [CrossRef] [PubMed]
- Boyer, A.; Yeh, J.R.; Zhang, X.; Paquet, M.; Gaudin, A.; Nagano, M.C.; Boerboom, D. CTNNB1 Signaling in Sertoli Cells Downregulates Spermatogonial Stem Cell Activity via WNT4. PLoS ONE 2012, 7, e29764. [Google Scholar] [CrossRef]
- Jordan, B.K.; Shen, J.H.-C.; Olaso, R.; Ingraham, H.A.; Vilain, E. Wnt4 Overexpression Disrupts Normal Testicular Vasculature and Inhibits Testosterone Synthesis by Repressing Steroidogenic Factor 1/Beta-Catenin Synergy. Proc. Natl. Acad. Sci. USA 2003, 100, 10866–10871. [Google Scholar] [CrossRef] [PubMed]
- Orwig, K.E.; Ryu, B.-Y.; Master, S.R.; Phillips, B.T.; Mack, M.; Avarbock, M.R.; Chodosh, L.; Brinster, R.L. Genes Involved in Post-Transcriptional Regulation Are Overrepresented in Stem/Progenitor Spermatogonia of Cryptorchid Mouse Testes. Stem Cells 2008, 26, 927–938. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-J.; Lee, W.-Y.; Park, C.; Hong, K.; Song, H. CD14 Is a Unique Membrane Marker of Porcine Spermatogonial Stem Cells, Regulating Their Differentiation. Sci. Rep. 2019, 9, 9980. [Google Scholar] [CrossRef]
- Hu, J.; Zhang, Y.-Q.; Liu, X.-P.; Wang, R.-A.; Jin, Y.; Xu, R.-J. Expression and Localization of Smad1, Smad2 and Smad4 Proteins in Rat Testis during Postnatal Development. Asian J. Androl. 2003, 5, 51–55. [Google Scholar]
- Palladino, M.A.; Fasano, G.A.; Patel, D.; Dugan, C.; London, M. Effects of Lipopolysaccharide-Induced Inflammation on Hypoxia and Inflammatory Gene Expression Pathways of the Rat Testis. Basic Clin. Androl. 2018, 28, 14. [Google Scholar] [CrossRef]
- Winnall, W.R.; Hedger, M.P. Phenotypic and Functional Heterogeneity of the Testicular Macrophage Population: A New Regulatory Model. J. Reprod. Immunol. 2013, 97, 147–158. [Google Scholar] [CrossRef]
- Murphy, K.; Carvajal, L.; Medico, L.; Pepling, M. Expression of Stat3 in Germ Cells of Developing and Adult Mouse Ovaries and Testes. Gene Expr. Patterns 2005, 5, 475–482. [Google Scholar] [CrossRef]
- Nagasawa, K.; Imura-Kishi, K.; Uchida, A.; Hiramatsu, R.; Kurohmaru, M.; Kanai, Y. Regionally Distinct Patterns of STAT3 Phosphorylation in the Seminiferous Epithelia of Mouse Testes. Mol. Reprod. Dev. 2018, 85, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Saxena, N.K.; Vertino, P.M.; Anania, F.A.; Sharma, D. Leptin-Induced Growth Stimulation of Breast Cancer Cells Involves Recruitment of Histone Acetyltransferases and Mediator Complex to CYCLIN D1 Promoter via Activation of Stat3. J. Biol. Chem. 2007, 282, 13316–13325. [Google Scholar] [CrossRef] [PubMed]
- Oatley, J.M.; Brinster, R.L. The Germline Stem Cell Niche Unit in Mammalian Testes. Physiol. Rev. 2012, 92, 577–595. [Google Scholar] [CrossRef] [PubMed]
- Sisakhtnezhad, S.; Heshmati, P. Comparative Analysis of Single-Cell RNA Sequencing Data from Mouse Spermatogonial and Mesenchymal Stem Cells to Identify Differentially Expressed Genes and Transcriptional Regulators of Germline Cells. J. Cell Physiol. 2018, 233, 5231–5242. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-X.; Xu, L.-L.; Wang, X.-C.; Qin, H.-Y.; Wang, J.-L. Involvement of C-Src/STAT3 Signal in EGF-Induced Proliferation of Rat Spermatogonial Stem Cells. Mol. Cell. Biochem. 2011, 358, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Haraguchi, S.; Ikeda, M.; Akagi, S.; Hirao, Y. Dynamic Changes in PStat3 Are Involved in Meiotic Spindle Assembly in Mouse Oocytes. Int. J. Mol. Sci. 2020, 21, 1220. [Google Scholar] [CrossRef]
- Kominami, E.; Tsukahara, T.; Bando, Y.; Katunuma, N. Distribution of Cathepsins B and H in Rat Tissues and Peripheral Blood Cells. J. Biochem. 1985, 98, 87–93. [Google Scholar] [CrossRef]
- Haraguchi, C.M.; Ishido, K.; Kominami, E.; Yokota, S. Expression of Cathepsin H in Differentiating Rat Spermatids: Immunoelectron Microscopic Study. Histochem. Cell Biol. 2003, 120, 63–71. [Google Scholar] [CrossRef]
- Habasque, C.; Satie, A.-P.; Aubry, F.; Jégou, B.; Samson, M. Expression of Fractalkine in the Rat Testis: Molecular Cloning of a Novel Alternative Transcript of Its Gene That Is Differentially Regulated by pro-Inflammatory Cytokines. Mol. Hum. Reprod. 2003, 9, 449–455. [Google Scholar] [CrossRef][Green Version]
- Truman, L.A.; Ford, C.A.; Pasikowska, M.; Pound, J.D.; Wilkinson, S.J.; Dumitriu, I.E.; Melville, L.; Melrose, L.A.; Ogden, C.A.; Nibbs, R.; et al. CX3CL1/Fractalkine Is Released from Apoptotic Lymphocytes to Stimulate Macrophage Chemotaxis. Blood 2008, 112, 5026–5036. [Google Scholar] [CrossRef]
- Ergün, S.; Kiliç, N.; Fiedler, W.; Mukhopadhyay, A.K. Vascular Endothelial Growth Factor and Its Receptors in Normal Human Testicular Tissue. Mol. Cell. Endocrinol. 1997, 131, 9–20. [Google Scholar] [CrossRef]
- Bott, R.C.; Clopton, D.T.; Fuller, A.M.; McFee, R.M.; Lu, N.; McFee, R.M.; Cupp, A.S. KDR-LacZ-Expressing Cells Are Involved in Ovarian and Testis-Specific Vascular Development, Suggesting a Role for VEGFA in the Regulation of This Vasculature. Cell Tissue Res. 2010, 342, 117–130. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Caires, K.C.; de Avila, J.; McLean, D.J. Vascular Endothelial Growth Factor Regulates Germ Cell Survival during Establishment of Spermatogenesis in the Bovine Testis. Reproduction 2009, 138, 667–677. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Yang, S.; Zhu, Y.; Zou, S.; Li, P.; Wang, J.; Zhu, Z.; Huang, Y.; He, Z.; Li, Z. VEGF/VEGFR2 Signaling Regulates Germ Cell Proliferation in Vitro and Promotes Mouse Testicular Regeneration in Vivo. Cells Tissues Organs 2016, 201, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Fon Tacer, K.; Kalanj-Bognar, S.; Waterman, M.R.; Rozman, D. Lanosterol Metabolism and Sterol Regulatory Element Binding Protein (SREBP) Expression in Male Germ Cell Maturation. J. Steroid Biochem. Mol. Biol. 2003, 85, 429–438. [Google Scholar] [CrossRef]
- Brown, M.S.; Goldstein, J.L. A Proteolytic Pathway That Controls the Cholesterol Content of Membranes, Cells, and Blood. Proc. Natl. Acad. Sci. USA 1999, 96, 11041–11048. [Google Scholar] [CrossRef]
- Osborne, T.F. Sterol Regulatory Element-Binding Proteins (SREBPs): Key Regulators of Nutritional Homeostasis and Insulin Action. J. Biol. Chem. 2000, 275, 32379–32382. [Google Scholar] [CrossRef]
- Vannelli, B.G.; Orlando, C.; Barni, T.; Natali, A.; Serio, M.; Balboni, G.C. Immunostaining of Transferrin and Transferrin Receptor in Human Seminiferous Tubules. Fertil. Steril. 1986, 45, 536–541. [Google Scholar] [CrossRef]
- Forti, G.; Barni, T.; Vannelli, B.G.; Balboni, G.C.; Orlando, C.; Serio, M. Sertoli Cell Proteins in the Human Seminiferous Tubule. J. Steroid Biochem. 1989, 32, 135–144. [Google Scholar] [CrossRef]
- Petrie, R.G.; Morales, C.R. Receptor-Mediated Endocytosis of Testicular Transferrin by Germinal Cells of the Rat Testis. Cell Tissue Res. 1992, 267, 45–55. [Google Scholar] [CrossRef]
- Bhattacharya, I.; Basu, S.; Pradhan, B.S.; Sarkar, H.; Nagarajan, P.; Majumdar, S.S. Testosterone Augments FSH Signaling by Upregulating the Expression and Activity of FSH-Receptor in Pubertal Primate Sertoli Cells. Mol. Cell. Endocrinol. 2019, 482, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Lécureuil, C.; Staub, C.; Fouchécourt, S.; Maurel, M.C.; Fontaine, I.; Martinat, N.; Gauthier, C.; Daudignon, A.; Delaleu, B.; Sow, A.; et al. Transferrin Overexpression Alters Testicular Function in Aged Mice. Mol. Reprod. Dev. 2007, 74, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Rondanino, C.; Maouche, A.; Dumont, L.; Oblette, A.; Rives, N. Establishment, Maintenance and Functional Integrity of the Blood-Testis Barrier in Organotypic Cultures of Fresh and Frozen/Thawed Prepubertal Mouse Testes. Mol. Hum. Reprod. 2017, 23, 304–320. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.B.; Walker, W.H. The Regulation of Spermatogenesis by Androgens. Semin. Cell Dev. Biol. 2014, 30, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Jan, S.Z.; Jongejan, A.; Korver, C.M.; van Daalen, S.K.M.; van Pelt, A.M.M.; Repping, S.; Hamer, G. Distinct Prophase Arrest Mechanisms in Human Male Meiosis. Development 2018, 145, dev160614. [Google Scholar] [CrossRef]
- Hung, A.J.; King, P.; Schlegel, P.N. Uniform Testicular Maturation Arrest: A Unique Subset of Men with Nonobstructive Azoospermia. J. Urol. 2007, 178, 608–612. [Google Scholar] [CrossRef]
- Jiao, S.-Y.; Yang, Y.-H.; Chen, S.-R. Molecular Genetics of Infertility: Loss-of-Function Mutations in Humans and Corresponding Knockout/Mutated Mice. Hum. Reprod. Update 2021, 27, 154–189. [Google Scholar] [CrossRef]
- Gat, Y.; Gornish, M.; Chakraborty, J.; Perlow, A.; Levinger, U.; Pasqualotto, F. Azoospermia and Maturation Arrest: Malfunction of Valves in Erect Poster of Humans Leads to Hypoxia in Sperm Production Site. Andrologia 2010, 42, 389–394. [Google Scholar] [CrossRef]
- Weedin, J.W.; Bennett, R.C.; Fenig, D.M.; Lamb, D.J.; Lipshultz, L.I. Early versus Late Maturation Arrest: Reproductive Outcomes of Testicular Failure. J. Urol. 2011, 186, 621–626. [Google Scholar] [CrossRef]
- França, L.R.; Hess, R.A.; Dufour, J.M.; Hofmann, M.C.; Griswold, M.D. The Sertoli Cell: One Hundred Fifty Years of Beauty and Plasticity. Andrology 2016, 4, 189–212. [Google Scholar] [CrossRef]
- Meng, J.; Holdcraft, R.W.; Shima, J.E.; Griswold, M.D.; Braun, R.E. Androgens Regulate the Permeability of the Blood-Testis Barrier. Proc. Natl. Acad. Sci. USA 2005, 102, 16696–16700. [Google Scholar] [CrossRef] [PubMed]
- Stanton, P.G. Regulation of the Blood-Testis Barrier. Semin. Cell Dev. Biol. 2016, 59, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Cooke, P.S.; Walker, W.H. Male Fertility in Mice Requires Classical and Nonclassical Androgen Signaling. Cell Rep. 2021, 36, 109557. [Google Scholar] [CrossRef] [PubMed]
- De Gendt, K.; Swinnen, J.V.; Saunders, P.T.K.; Schoonjans, L.; Dewerchin, M.; Devos, A.; Tan, K.; Atanassova, N.; Claessens, F.; Lécureuil, C.; et al. A Sertoli Cell-Selective Knockout of the Androgen Receptor Causes Spermatogenic Arrest in Meiosis. Proc. Natl. Acad. Sci. USA 2004, 101, 1327–1332. [Google Scholar] [CrossRef] [PubMed]
- Syriou, V.; Papanikolaou, D.; Kozyraki, A.; Goulis, D.G. Cytokines and Male Infertility. Eur. Cytokine Netw. 2018, 29, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zhu, W.; Xue, S.; Han, D. Testicular Defense Systems: Immune Privilege and Innate Immunity. Cell Mol. Immunol. 2014, 11, 428–437. [Google Scholar] [CrossRef]
- Arkoun, B.; Galas, L.; Dumont, L.; Rives, A.; Saulnier, J.; Delessard, M.; Rondanino, C.; Rives, N. Vitamin E but Not GSH Decreases Reactive Oxygen Species Accumulation and Enhances Sperm Production during In Vitro Maturation of Frozen-Thawed Prepubertal Mouse Testicular Tissue. IJMS 2019, 20, 5380. [Google Scholar] [CrossRef]
- Yan, H.H.N.; Mruk, D.D.; Lee, W.M.; Cheng, C.Y. Blood-Testis Barrier Dynamics Are Regulated by Testosterone and Cytokines via Their Differential Effects on the Kinetics of Protein Endocytosis and Recycling in Sertoli Cells. FASEB J. 2008, 22, 1945–1959. [Google Scholar] [CrossRef]
- Lie, P.P.Y.; Cheng, C.Y.; Mruk, D.D. Interleukin-1alpha Is a Regulator of the Blood-Testis Barrier. FASEB J. 2011, 25, 1244–1253. [Google Scholar] [CrossRef]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological Properties of Extracellular Vesicles and Their Physiological Functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef]
- Hedger, M.P.; Meinhardt, A. Cytokines and the Immune-Testicular Axis. J. Reprod. Immunol. 2003, 58, 1–26. [Google Scholar] [CrossRef]
- Pérez, C.V.; Theas, M.S.; Jacobo, P.V.; Jarazo-Dietrich, S.; Guazzone, V.A.; Lustig, L. Dual Role of Immune Cells in the Testis: Protective or Pathogenic for Germ Cells? Spermatogenesis 2013, 3, e23870. [Google Scholar] [CrossRef] [PubMed]
- Winter, A.G.; Zhao, F.; Lee, R.K. Androgen Deficiency and Metabolic Syndrome in Men. Transl. Androl. Urol. 2014, 3, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Ham, S.W.; Jeon, H.-Y.; Jin, X.; Kim, E.-J.; Kim, J.-K.; Shin, Y.J.; Lee, Y.; Kim, S.H.; Lee, S.Y.; Seo, S.; et al. TP53 Gain-of-Function Mutation Promotes Inflammation in Glioblastoma. Cell Death Differ. 2019, 26, 409–425. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, S.; Williamson, C.; Bevan, C.L. Liver X Receptors and Male (In)Fertility. Int. J. Mol. Sci. 2019, 20, 5379. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yan, J.; Jiang, S.; Wen, J.; Chen, L.; Zhao, Y.; Lin, A. Site-Specific Ubiquitination Is Required for Relieving the Transcription Factor Miz1-Mediated Suppression on TNF-α-Induced JNK Activation and Inflammation. Proc. Natl. Acad. Sci. USA 2012, 109, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Zahid, M.D.K.; Rogowski, M.; Ponce, C.; Choudhury, M.; Moustaid-Moussa, N.; Rahman, S.M. CCAAT/Enhancer-Binding Protein Beta (C/EBPβ) Knockdown Reduces Inflammation, ER Stress, and Apoptosis, and Promotes Autophagy in OxLDL-Treated RAW264.7 Macrophage Cells. Mol. Cell Biochem. 2020, 463, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Zeng, R.-M.; Lu, X.-H.; Lin, J.; Hu, J.; Rong, Z.-J.; Xu, W.-C.; Liu, Z.-W.; Zeng, W.-T. Knockdown of FOXM1 Attenuates Inflammatory Response in Human Osteoarthritis Chondrocytes. Int. Immunopharmacol. 2019, 68, 74–80. [Google Scholar] [CrossRef]
- Zhou, M.; Shi, J.; Lan, S.; Gong, X. FOXM1 Regulates the Proliferation, Apoptosis and Inflammatory Response of Keratinocytes through the NF-ΚB Signaling Pathway. Hum. Exp. Toxicol. 2021, 40, 1130–1140. [Google Scholar] [CrossRef]
- Clermont, Y. Kinetics of Spermatogenesis in Mammals: Seminiferous Epithelium Cycle and Spermatogonial Renewal. Physiol. Rev. 1972, 52, 198–236. [Google Scholar] [CrossRef]
- Peters, A.; Plug, A.; van Vugt, M.; de Boer, P. A Drying-down Technique for the Spreading of Mammalian Meiocytes from the Male and Female Germline. Chromosome Res. Int. J. Mol. Supramol. Evol. Asp. Chromosome Biol. 1997, 5, 66–68. [Google Scholar] [CrossRef] [PubMed]
- Milazzo, J.P.; Rives, N.; Mousset-Siméon, N.; Macé, B. Chromosome Constitution and Apoptosis of Immature Germ Cells Present in Sperm of Two 47,XYY Infertile Males. Hum. Reprod. 2006, 21, 1749–1758. [Google Scholar] [CrossRef] [PubMed]
- Ishishita, S.; Matsuda, Y.; Kitada, K. Genetic Evidence Suggests That Spata22 Is Required for the Maintenance of Rad51 Foci in Mammalian Meiosis. Sci. Rep. 2014, 4, 6148. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Pauli, A.; Valen, E.; Lin, M.F.; Garber, M.; Vastenhouw, N.L.; Levin, J.Z.; Fan, L.; Sandelin, A.; Rinn, J.L.; Regev, A.; et al. Systematic Identification of Long Noncoding RNAs Expressed during Zebrafish Embryogenesis. Genome Res. 2012, 22, 577–591. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential Gene and Transcript Expression Analysis of RNA-Seq Experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Chalmel, F.; Lardenois, A.; Evrard, B.; Rolland, A.D.; Sallou, O.; Dumargne, M.-C.; Coiffec, I.; Collin, O.; Primig, M.; Jégou, B. High-Resolution Profiling of Novel Transcribed Regions during Rat Spermatogenesis. Biol. Reprod. 2014, 91, 5. [Google Scholar] [CrossRef]
- Zimmermann, C.; Stévant, I.; Borel, C.; Conne, B.; Pitetti, J.-L.; Calvel, P.; Kaessmann, H.; Jégou, B.; Chalmel, F.; Nef, S. Research Resource: The Dynamic Transcriptional Profile of Sertoli Cells during the Progression of Spermatogenesis. Mol. Endocrinol. 2015, 29, 627–642. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-Level Expression Analysis of RNA-Seq Experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Soft. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Chalmel, F.; Primig, M. The Annotation, Mapping, Expression and Network (AMEN) Suite of Tools for Molecular Systems Biology. BMC Bioinform. 2008, 9, 86. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.-K.; Wang, S.-J.; Huang, Y.-Q.; Zhao, R.-Q.; Zhu, Q.-F.; Lin, W.-Z. Identification and Validation of Suitable Reference Genes for RT-QPCR Analysis in Mouse Testis Development. Mol. Genet. Genom. 2014, 289, 1157–1169. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Darde, T.A.; Sallou, O.; Becker, E.; Evrard, B.; Monjeaud, C.; Le Bras, Y.; Jégou, B.; Collin, O.; Rolland, A.D.; Chalmel, F. The ReproGenomics Viewer: An Integrative Cross-Species Toolbox for the Reproductive Sci.ence Community. Nucleic Acids Res. 2015, 43, W109–W116. [Google Scholar] [CrossRef] [PubMed]
- Darde, T.A.; Lecluze, E.; Lardenois, A.; Stévant, I.; Alary, N.; Tüttelmann, F.; Collin, O.; Nef, S.; Jégou, B.; Rolland, A.D.; et al. The ReproGenomics Viewer: A Multi-Omics and Cross-Species Resource Compatible with Single-Cell Studies for the Reproductive Sci.ence Community. Bioinformatics 2019, 35, 3133–3139. [Google Scholar] [CrossRef] [PubMed]


| Patterns 1–4 | ||||||||
|---|---|---|---|---|---|---|---|---|
| ID | Gene | Adjusted F-Value | Pattern | Fold Change (Log(2)) | Cell Type (Testis) | Function in Spermatogenesis | ||
| D16 | D22 | D28 | ||||||
| 25146 | Cyp17a1 | 4.10 × 105 | P4 | Down (−7.47) | Down (−7.13) | Down (−7.46) | Leydig cells [56] | Conversion of progesterone into dehydroepiandrosterone [56] |
| 114215 | Insl3 | 4.10 × 105 | P4 | Down (−5.65) | Down (−4.63) | Down (−4.24) | Leydig cells and post-meiotic cells [57] | Testicular descent [57] |
| 29460 | Tesk1 | 5.20 × 105 | P4 | Up (1.43) | Up (1.60) | Up (1.78) | Spermatocytes (late pachytene) to round spermatids [58,59,60] | Cytoskeleton reorganization [60] |
| 362187 | Ccdc34 | 9.00 × 105 | P4 | Down (−1.39) | Down (−1.51) | Down (−1.38) | Meiotic and post-meiotic cells [61] | Regulation of cell cycle G2/M [61,62,63] |
| 171304 | Kif11 | 1.10 × 104 | P1 | Down (−1.47) | Down (−1.53) | Down (−1.81) | Spermatogonia, spermatocytes, spermatids, and Sertoli cells [64] | Chromosomes separation [65,66] |
| 296368 | Ube2c | 1.10 × 104 | P4 | Down (−2.14) | Down (−2.05) | Down (−2.24) | Meiotic cells [67] | Regulation of metaphase/anaphase transition [68,69] |
| 293733 | Incenp | 1.20 × 104 | P1 | Down (−1.40) | Down (−1.25) | Down (−1.44) | Dividing cells [70,71,72] | Metaphase/anaphase progression [72] Chromatids cohesion [71] |
| 25515 | Plk1 | 1.20 × 104 | P4 | Down (−1.82) | Down (−1.86) | Down (−1.77) | Spermatocytes (diplotene), spermatocytes II, and round spermatids [73,74] | Metaphase/anaphase transition checkpoint and chromosomes segregation [75,76] Phosphorylation of the central element of the synaptonemal complex [74] Cohesion of sister chromatids [77] Chromosomes alignment [78] |
| 117524 | Ccnf | 1.40 × 104 | P1 | Down (−1.78) | Down (−1.76) | Down (−1.67) | Dividing cells [79], spermatozoa [80] | Cell cycle [79] Potentially involved in proliferation and sperm motility [80] |
| 498709 | Cks2 | 1.40 × 104 | P4 | Down (−1.36) | Down (−1.36) | Down (−1.56) | Meiotic cells [81] | Regulation of meiotic cell cycle, contribution to the control of the first metaphase/anaphase transition during mammalian meiosis [33,81,82] |
| 81639 | Alox15 | 1.70 × 104 | P1 | Down (−7.34) | Down (−6.72) | Down (−5.39) | Spermatozoa [83] | Contribution to spermiogenesis with formation and resorption of the cytoplasmic droplet [83] Constituent of the oxidative stress pathway [84,85,86] |
| 171576 | Bub1b | 1.70 × 104 | P1 | Down (−1.26) | Down (−1.42) | Down (−1.49) | Mitotic and meiotic cells [87] | Regulation of the SAC and chromosomal alignment, interaction with Plk1 [78,88,89,90] |
| 114494 | Ccna2 | 1.70 × 104 | P1 | Down (−1.54) | Down (−1.61) | Down (−1.68) | Spermatogonial stem cells (SSC), spermatogonia, spermatocytes (preleptotene), Sertoli cells [91,92,93] | Formation of the SSC pool [94] Regulation of microtubule dynamics during the rapid formation of the metaphase II spindle (by similarity with oocytes) [95] |
| 303730 | Cbx2 | 1.80 × 104 | P1 | Down (−1.12) | Down (−1.22) | Down (−1.12) | Germ cells [96] | Establishment of the synaptonemal complex [96] |
| Patterns 5–7 | ||||||||
| ID | Gene | Adjusted F-Value | Pattern | Fold Change (Log(2)) | Cell Type (Testis) | Function in Spermatogenesis | ||
| D16 | D22 | D28 | ||||||
| 81687 | Mmp9 | 4.10 × 105 | P7 | Up (8.06) | Up (8.32) | Up (7.94) | Early SSC [97], gonocytes and Sertoli cells [98,99,100], spermatocytes and spermatids [101] | Cell junctions and BTB maintenance [101,102,103,104] Sperm motility [105] |
| 25696 | Vldlr | 4.10 × 105 | P7 | Up (2.28) | Up (2.90) | Up (2.48) | Spermatocytes (pachytene) and Leydig cells [106] | Regulation of meiosis [106,107] |
| 252917 | Nr1d1 | 4.70 × 105 | P7 | Up (1.31) | Up (1.43) | Up (1.80) | Leydig cells [108] and germ cells [109] | Regulation of meiotic entry, Stra8/Nr1d1 balance [109] Stimulation of testosterone production and steroidogenic gene expression [108] |
| 24484 | Igfbp3 | 5.20 × 105 | P5 | Up (3.62) | Up (3.69) | Up (3.70) | Sertoli cells [110], Leydig cells [111] | Regulation of testicular cell homeostasis via apoptosis [112], inhibition of IGF1 stimulation of steroidogenesis [111] |
| 245920 | Cxcl10 | 9.70 × 105 | P5 | Up (2.35) | Up (2.78) | Up (2.72) | Leydig cells, T cells, and macrophages [113,114], peritubular and Sertoli cells during inflammation [115] | Role in inflammatory process, induction of germ cell apoptosis [115,116] |
| 313210 | Abca1 | 1.10 × 104 | P5 | Up (1.47) | Up (1.27) | Up (1.69) | Leydig and Sertoli cells, round spermatids, spermatozoa [117,118] | Cholesterol transporter [118] Modulation of Sertoli cell phagocytosis [117] Impact on fertility [119] |
| 114031 | Fstl3 | 1.10 × 104 | P5 | Up (1.94) | Up (2.04) | Up (2.08) | Leydig cells, spermatogonia, mature spermatids [120] | Regulation of gonadal development via interaction with activin [120,121,122] |
| 84426 | Wnt4 | 1.10 × 104 | P7 | Up (3.34) | Up (3.90) | Up (4.18) | SSC [123] and Leydig cells [124] | Cell activity and apoptosis [123] |
| 60350 | Cd14 | 1.40 × 104 | P5 | Up (2.28) | Up (2.53) | Up (2.85) | Putative SSC and early spermatogonia [125,126] Macrophages [127] | Inflammatory response signaling pathway [128] Immunoregulation of the testicular environment [129] |
| 25125 | Stat3 | 1.40 × 104 | P5 | Up (1.29) | Up (1.60) | Up (1.71) | Gonocytes, pro-spermatogonia, round spermatids [130] Sertoli cells (Rete testis) [131] | Cell cycle regulation of G1 to S phase transition [132] Promotion of SSC differentiation [133,134] Cell viability [135] Assembly of the meiotic spindle (by similarity with oocytes) [136] |
| 25425 | Ctsh | 1.40 × 104 | P7 | Up (1.25) | Up (1.25) | Up (1.38) | Germ cells, Sertoli and Leydig cells [137,138] | Germ cells, Sertoli and Leydig cells [137,138] |
| 89808 | Cx3cl1 | 1.60 × 104 | P5 | Up (1.51) | Up (2.04) | Up (2.42) | Interstitial tissue [139], Sertoli and Leydig cells, spermatogonia, spermatocytes and peritubular cells | Inflammation process [139] Stimulation of macrophage recruitment [140] |
| 25589 | Kdr | 1.70 × 104 | P5 | Up (1.57) | Up (1.74) | Up (1.91) | Spermatids, Sertoli and Leydig cells, lamina propria and blood vessels [141,142] | Participation to germ cell survival and enhancement of vascularization [143,144] |
| 78968 | Srebf1 | 1.70 × 104 | P5 | Up (1.22) | Up (1.46) | Up (1.74) | Germ cells [145] | Regulation of cholesterol transport [146] Glucose homeostasis and fat metabolism [147] |
| 24825 | Tf | 1.70 × 104 | P7 | Up (3.25) | Up (3.52) | Up (2.95) | Sertoli cells, spermatocytes and early spermatids [148,149,150,151] | Regulation of sperm yield [152] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saulnier, J.; Chalmel, F.; Delessard, M.; Moutard, L.; Pereira, T.; Fraissinet, F.; Dumont, L.; Rives-Feraille, A.; Rondanino, C.; Rives, N. Understanding the Underlying Molecular Mechanisms of Meiotic Arrest during In Vitro Spermatogenesis in Rat Prepubertal Testicular Tissue. Int. J. Mol. Sci. 2022, 23, 5893. https://doi.org/10.3390/ijms23115893
Saulnier J, Chalmel F, Delessard M, Moutard L, Pereira T, Fraissinet F, Dumont L, Rives-Feraille A, Rondanino C, Rives N. Understanding the Underlying Molecular Mechanisms of Meiotic Arrest during In Vitro Spermatogenesis in Rat Prepubertal Testicular Tissue. International Journal of Molecular Sciences. 2022; 23(11):5893. https://doi.org/10.3390/ijms23115893
Chicago/Turabian StyleSaulnier, Justine, Frédéric Chalmel, Marion Delessard, Laura Moutard, Tony Pereira, François Fraissinet, Ludovic Dumont, Aurélie Rives-Feraille, Christine Rondanino, and Nathalie Rives. 2022. "Understanding the Underlying Molecular Mechanisms of Meiotic Arrest during In Vitro Spermatogenesis in Rat Prepubertal Testicular Tissue" International Journal of Molecular Sciences 23, no. 11: 5893. https://doi.org/10.3390/ijms23115893
APA StyleSaulnier, J., Chalmel, F., Delessard, M., Moutard, L., Pereira, T., Fraissinet, F., Dumont, L., Rives-Feraille, A., Rondanino, C., & Rives, N. (2022). Understanding the Underlying Molecular Mechanisms of Meiotic Arrest during In Vitro Spermatogenesis in Rat Prepubertal Testicular Tissue. International Journal of Molecular Sciences, 23(11), 5893. https://doi.org/10.3390/ijms23115893






