The Association between Single Nucleotide Polymorphisms, including miR-499a Genetic Variants, and Dyslipidemia in Subjects Treated with Pharmacological or Phytochemical Lipid-Lowering Agents
Abstract
:1. Introduction
2. Results
2.1. Associations of Selected SNPs with the Baseline Lipid Profile
2.2. Associations of Selected SNPs with Lipid Profile in Subjects under Lipid-Lowering Therapy
2.3. Bioinformatic Prediction of miR-499a-5p Target Genes Related to Lipid Metabolism
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Data Collection
4.3. DNA Extraction and Genotyping
4.4. Statistical Analysis and Quality Control of Genotyping Data
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pirillo, A.; Casula, M.; Olmastroni, E.; Norata, G.D.; Catapano, A.L. Global epidemiology of dyslipidaemias. Nat. Rev. Cardiol. 2021, 18, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Michos, E.D.; McEvoy, J.W.; Blumenthal, R.S. Lipid Management for the Prevention of Atherosclerotic Cardiovascular Disease. N. Engl. J. Med. 2019, 381, 1557–1567. [Google Scholar] [CrossRef]
- Jarauta, E.; Bea-Sanz, A.M.; Marco-Benedi, V.; Lamiquiz-Moneo, I. Genetics of Hypercholesterolemia: Comparison between Familial Hypercholesterolemia and Hypercholesterolemia Nonrelated to LDL Receptor. Front. Genet. 2020, 11, 554931. [Google Scholar] [CrossRef] [PubMed]
- Trinder, M.; Francis, G.A.; Brunham, L.R. Association of Monogenic vs Polygenic Hypercholesterolemia with Risk of Atherosclerotic Cardiovascular Disease. JAMA Cardiol. 2020, 5, 390–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giral, H.; Landmesser, U.; Kratzer, A. Into the Wild: GWAS Exploration of Non-coding RNAs. Front. Cardiovasc. Med. 2018, 5, 181. [Google Scholar] [CrossRef]
- Li, Q.; Chen, L.; Chen, D.; Wu, X.; Chen, M. Influence of microRNA-related polymorphisms on clinical outcomes in coronary artery disease. Am. J. Transl. Res. 2015, 7, 393–400. [Google Scholar]
- Chen, L.B.; Zheng, H.K.; Zhang, L.; An, Z.; Wang, X.P.; Shan, R.T.; Zhang, W.Q. A single nucleotide polymorphism located in microRNA-499a causes loss of function resulting in increased expression of osbpl1a and reduced serum HDL level. Oncol. Rep. 2017, 38, 3515–3521. [Google Scholar] [CrossRef] [Green Version]
- Qiu, X.Y.; Lu, S.T.; Fan, M.K.; Geng, H.H.; Han, Z.Y.; Gao, S.P.; Pan, H.Y.; Huang, R.; Pan, M. Effects of Polymorphisms in Pre-miRNA on Inflammatory Markers in Atrial Fibrillation in Han Chinese. Clin. Lab. 2019, 65. [Google Scholar] [CrossRef]
- Ciccacci, C.; Latini, A.; Greco, C.; Politi, C.; D’Amato, C.; Lauro, D.; Novelli, G.; Borgiani, P.; Spallone, V. Association between a MIR499A polymorphism and diabetic neuropathy in type 2 diabetes. J. Diabetes Complicat. 2018, 32, 11–17. [Google Scholar] [CrossRef]
- Qiu, H.; Chen, Z.; Lv, L.; Tang, W.; Hu, R. Associations between microRNA Polymorphisms and Development of Coronary Artery Disease: A Case-Control Study. DNA Cell Biol. 2020, 39, 25–36. [Google Scholar] [CrossRef]
- Liu, F.; Wang, S.; Luo, Z. Associations of the miRNA-146a rs2910164 and the miRNA-499a rs3746444 Polymorphisms with Plasma Lipid Levels: A Meta-Analysis. Front. Genet. 2021, 12, 746686. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, F.; Antonicelli, R.; Lorenzi, M.; D’Alessandra, Y.; Lazzarini, R.; Santini, G.; Spazzafumo, L.; Lisa, R.; La Sala, L.; Galeazzi, R.; et al. Diagnostic potential of circulating miR-499-5p in elderly patients with acute non ST-elevation myocardial infarction. Int. J. Cardiol. 2013, 167, 531–536. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, F.; Antonicelli, R.; Spazzafumo, L.; Santini, G.; Rippo, M.R.; Galeazzi, R.; Giovagnetti, S.; D’Alessandra, Y.; Marcheselli, F.; Capogrossi, M.C.; et al. Admission levels of circulating miR-499-5p and risk of death in elderly patients after acute non-ST elevation myocardial infarction. Int. J. Cardiol. 2014, 172, e276–e278. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Huang, W.; Fan, X.; Liu, F.; Luo, L.; Yuan, H.; Jiang, Y.; Xiao, H.; Zhou, Z.; Deng, C.; et al. miR-499 released during myocardial infarction causes endothelial injury by targeting alpha7-nAchR. J. Cell. Mol. Med. 2019, 23, 6085–6097. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Yu, H.; Yan, P.; Zhou, X.; Wang, Y.; Yao, Y. Circulating MicroRNA-499 as a Diagnostic Biomarker for Acute Myocardial Infarction: A Meta-analysis. Dis. Markers 2019, 2019, 6121696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Hong, H.; Chen, L.; Shi, X.; Chen, Y.; Weng, Q. Association of microRNA polymorphisms with the risk of myocardial infarction in a Chinese population. Tohoku J. Exp. Med. 2014, 233, 89–94. [Google Scholar] [CrossRef] [Green Version]
- Gurau, F.; Baldoni, S.; Prattichizzo, F.; Espinosa, E.; Amenta, F.; Procopio, A.D.; Albertini, M.C.; Bonafe, M.; Olivieri, F. Anti-senescence compounds: A potential nutraceutical approach to healthy aging. Ageing Res. Rev. 2018, 46, 14–31. [Google Scholar] [CrossRef]
- Matacchione, G.; Gurău, F.; Baldoni, S.; Prattichizzo, F.; Silvestrini, A.; Giuliani, A.; Pugnaloni, A.; Espinosa, E.; Amenta, F.; Bonafè, M.; et al. Pleiotropic effects of polyphenols on glucose and lipid metabolism: Focus on clinical trials. Ageing Res. Rev. 2020, 61, 101074. [Google Scholar] [CrossRef]
- Bonfigli, A.R.; Protic, O.; Olivieri, F.; Montesanto, A.; Malatesta, G.; Di Pillo, R.; Antonicelli, R. Effects of a novel nutraceutical combination (BruMeChol) in subjects with mild hypercholesterolemia: Study protocol of a randomized, double-blind, controlled trial. Trials 2020, 21, 616. [Google Scholar] [CrossRef]
- Ding, W.; Li, M.; Sun, T.; Han, D.; Guo, X.; Chen, X.; Wan, Q.; Zhang, X.; Wang, J. A polymorphism rs3746444 within the pre-miR-499 alters the maturation of miR-499-5p and its antiapoptotic function. J. Cell. Mol. Med. 2018, 22, 5418–5428. [Google Scholar] [CrossRef] [Green Version]
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Vlachos, I.S.; Vergoulis, T.; Reczko, M.; Filippidis, C.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-microT web server v5.0: Service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013, 41, W169–W173. [Google Scholar] [CrossRef] [Green Version]
- Belinky, F.; Nativ, N.; Stelzer, G.; Zimmerman, S.; Iny Stein, T.; Safran, M.; Lancet, D. PathCards: Multi-source consolidation of human biological pathways. Database 2015, 2015, bav006. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Kumar, S.; Ahmad, M.K.; Singh, R.; Kant Kumar, S.; Pradhan, A.; Chandra, S.; Kumar, S. Promoter variants of TNF-alpha rs1800629 and IL-10 rs1800871 are independently associated with the susceptibility of coronary artery disease in north Indian. Cytokine 2018, 110, 131–136. [Google Scholar] [CrossRef]
- Leonska-Duniec, A.; Ficek, K.; Switala, K.; Cieszczyk, P. Association of the TNF-alpha -308G/A polymorphism with lipid profile changes in response to aerobic training program. Biol. Sport 2019, 36, 291–296. [Google Scholar] [CrossRef]
- Ghareeb, D.; Abdelazem, A.S.; Hussein, E.M.; Al-Karamany, A.S. Association of TNF-alpha-308 G>A (rs1800629) polymorphism with susceptibility of metabolic syndrome. J. Diabetes Metab. Disord. 2021, 20, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Gusti, A.M.T.; Qusti, S.Y.; Alshammari, E.M.; Toraih, E.A.; Fawzy, M.S. Antioxidants-Related Superoxide Dismutase (SOD), Catalase (CAT), Glutathione Peroxidase (GPX), Glutathione-S-Transferase (GST), and Nitric Oxide Synthase (NOS) Gene Variants Analysis in an Obese Population: A Preliminary Case-Control Study. Antioxidants 2021, 10, 595. [Google Scholar] [CrossRef] [PubMed]
- Stathopoulou, M.G.; Bonnefond, A.; Ndiaye, N.C.; Azimi-Nezhad, M.; El Shamieh, S.; Saleh, A.; Rancier, M.; Siest, G.; Lamont, J.; Fitzgerald, P.; et al. A common variant highly associated with plasma VEGFA levels also contributes to the variation of both LDL-C and HDL-C. J. Lipid Res. 2013, 54, 535–541. [Google Scholar] [CrossRef] [Green Version]
- Sandhofer, A.; Tatarczyk, T.; Kirchmair, R.; Iglseder, B.; Paulweber, B.; Patsch, J.R.; Schratzberger, P. Are plasma VEGF and its soluble receptor sFlt-1 atherogenic risk factors? Cross-sectional data from the SAPHIR study. Atherosclerosis 2009, 206, 265–269. [Google Scholar] [CrossRef]
- Debette, S.; Visvikis-Siest, S.; Chen, M.H.; Ndiaye, N.C.; Song, C.; Destefano, A.; Safa, R.; Azimi Nezhad, M.; Sawyer, D.; Marteau, J.B.; et al. Identification of cis- and trans-acting genetic variants explaining up to half the variation in circulating vascular endothelial growth factor levels. Circ. Res. 2011, 109, 554–563. [Google Scholar] [CrossRef] [Green Version]
- Kaess, B.M.; Preis, S.R.; Beiser, A.; Sawyer, D.B.; Chen, T.C.; Seshadri, S.; Vasan, R.S. Circulating vascular endothelial growth factor and the risk of cardiovascular events. Heart 2016, 102, 1898–1901. [Google Scholar] [CrossRef]
- Celletti, F.L.; Waugh, J.M.; Amabile, P.G.; Brendolan, A.; Hilfiker, P.R.; Dake, M.D. Vascular endothelial growth factor enhances atherosclerotic plaque progression. Nat. Med. 2001, 7, 425–429. [Google Scholar] [CrossRef] [PubMed]
- Chmurzynska, A.; Muzsik, A.; Krzyzanowska-Jankowska, P.; Walkowiak, J.; Bajerska, J. The Effect of Habitual Fat Intake, IL6 Polymorphism, and Different Diet Strategies on Inflammation in Postmenopausal Women with Central Obesity. Nutrients 2019, 11, 1557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becker, M.L.; Elens, L.L.; Visser, L.E.; Hofman, A.; Uitterlinden, A.G.; van Schaik, R.H.; Stricker, B.H. Genetic variation in the ABCC2 gene is associated with dose decreases or switches to other cholesterol-lowering drugs during simvastatin and atorvastatin therapy. Pharm. J. 2013, 13, 251–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Y.; Wang, J.; Liu, Y.; Luo, X.; Yao, Z.; Wang, X.; Zhang, Y.; Xu, C.; Zhao, X. MassARRAY multigene screening combined with LDL-C and sdLDL-C detection for more favorable outcomes in type 2 diabetes mellitus therapy. BMC Med. Genom. 2021, 14, 83. [Google Scholar] [CrossRef]
- Kee, P.S.; Chin, P.K.L.; Kennedy, M.A.; Maggo, S.D.S. Pharmacogenetics of Statin-Induced Myotoxicity. Front. Genet. 2020, 11, 575678. [Google Scholar] [CrossRef]
- Abdullah, M.M.H.; Vazquez-Vidal, I.; Baer, D.J.; House, J.D.; Jones, P.J.H.; Desmarchelier, C. Common Genetic Variations Involved in the Inter-Individual Variability of Circulating Cholesterol Concentrations in Response to Diets: A Narrative Review of Recent Evidence. Nutrients 2021, 13, 695. [Google Scholar] [CrossRef]
- Piko, P.; Fiatal, S.; Kosa, Z.; Sandor, J.; Adany, R. Generalizability and applicability of results obtained from populations of European descent regarding the effect direction and size of HDL-C level-associated genetic variants to the Hungarian general and Roma populations. Gene 2019, 686, 187–193. [Google Scholar] [CrossRef]
- Wang, J.; Wang, L.J.; Zhong, Y.; Gu, P.; Shao, J.Q.; Jiang, S.S.; Gong, J.B. CETP gene polymorphisms and risk of coronary atherosclerosis in a Chinese population. Lipids Health Dis. 2013, 12, 176. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Zhang, Y.; Yan, W.; Wang, W.; Zhao, X.; Ma, X.; Gao, X.; Zhang, S. Association between three functional microRNA polymorphisms (miR-499 rs3746444, miR-196a rs11614913 and miR-146a rs2910164) and breast cancer risk: A meta-analysis. Oncotarget 2017, 8, 393–407. [Google Scholar] [CrossRef] [Green Version]
- Ahmed Ali, M.; Gamil Shaker, O.; Mohamed Eid, H.; Elsayed Mahmoud, E.; Mahmoud Ezzat, E.; Nady Gaber, S. Relationship between miR-155 and miR-146a polymorphisms and susceptibility to multiple sclerosis in an Egyptian cohort. Biomed. Rep. 2020, 12, 276–284. [Google Scholar] [CrossRef] [Green Version]
- Hu, Z.; Liang, J.; Wang, Z.; Tian, T.; Zhou, X.; Chen, J.; Miao, R.; Wang, Y.; Wang, X.; Shen, H. Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum. Mutat. 2009, 30, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Chistiakov, D.A.; Orekhov, A.N.; Bobryshev, Y.V. Cardiac-specific miRNA in cardiogenesis, heart function, and cardiac pathology (with focus on myocardial infarction). J. Mol. Cell. Cardiol. 2016, 94, 107–121. [Google Scholar] [CrossRef]
- Mach, F.; Baigent, C.; Catapano, A.L.; Koskinas, K.C.; Casula, M.; Badimon, L.; Chapman, M.J.; De Backer, G.G.; Delgado, V.; Ference, B.A.; et al. 2019 ESC/EAS Guidelines for the management of dyslipidaemias: Lipid modification to reduce cardiovascular risk. Eur. Heart J. 2020, 41, 111–188. [Google Scholar] [CrossRef] [PubMed]
- Ference, B.A.; Robinson, J.G.; Brook, R.D.; Catapano, A.L.; Chapman, M.J.; Neff, D.R.; Voros, S.; Giugliano, R.P.; Davey Smith, G.; Fazio, S.; et al. Variation in PCSK9 and HMGCR and Risk of Cardiovascular Disease and Diabetes. N. Engl. J. Med. 2016, 375, 2144–2153. [Google Scholar] [CrossRef] [Green Version]
- Kotur-Stevuljevic, J.; Vekic, J.; Stefanovic, A.; Zeljkovic, A.; Ninic, A.; Ivanisevic, J.; Miljkovic, M.; Sopic, M.; Munjas, J.; Mihajlovic, M.; et al. Paraoxonase 1 and atherosclerosis-related diseases. Biofactors 2020, 46, 193–205. [Google Scholar] [CrossRef]
- Rizzi, F.; Conti, C.; Dogliotti, E.; Terranegra, A.; Salvi, E.; Braga, D.; Ricca, F.; Lupoli, S.; Mingione, A.; Pivari, F.; et al. Interaction between polyphenols intake and PON1 gene variants on markers of cardiovascular disease: A nutrigenetic observational study. J. Transl. Med. 2016, 14, 186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.H.; Suchindran, S.; Shah, S.H.; Kraus, W.E.; Ginsburg, G.S.; Voora, D. SLCO1B1 genetic variants, long-term low-density lipoprotein cholesterol levels and clinical events in patients following cardiac catheterization. Pharmacogenomics 2015, 16, 449–458. [Google Scholar] [CrossRef] [Green Version]
- Bowden, K.L.; Dubland, J.A.; Chan, T.; Xu, Y.H.; Grabowski, G.A.; Du, H.; Francis, G.A. LAL (Lysosomal Acid Lipase) Promotes Reverse Cholesterol Transport In Vitro and In Vivo. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 1191–1201. [Google Scholar] [CrossRef] [Green Version]
- Soll, D.; Spira, D.; Hollstein, T.; Haberbosch, L.; Demuth, I.; Steinhagen-Thiessen, E.; Bobbert, T.; Spranger, J.; Kassner, U. Clinical outcome of a patient with lysosomal acid lipase deficiency and first results after initiation of treatment with Sebelipase alfa: A case report. Mol. Genet. Metab. Rep. 2019, 20, 100479. [Google Scholar] [CrossRef]
- Motazacker, M.M.; Pirhonen, J.; van Capelleveen, J.C.; Weber-Boyvat, M.; Kuivenhoven, J.A.; Shah, S.; Hovingh, G.K.; Metso, J.; Li, S.; Ikonen, E.; et al. A loss-of-function variant in OSBPL1A predisposes to low plasma HDL cholesterol levels and impaired cholesterol efflux capacity. Atherosclerosis 2016, 249, 140–147. [Google Scholar] [CrossRef] [Green Version]
- Conroy, R.M.; Pyorala, K.; Fitzgerald, A.P.; Sans, S.; Menotti, A.; De Backer, G.; De Bacquer, D.; Ducimetiere, P.; Jousilahti, P.; Keil, U.; et al. Estimation of ten-year risk of fatal cardiovascular disease in Europe: The SCORE project. Eur. Heart J. 2003, 24, 987–1003. [Google Scholar] [CrossRef]
| Patients Affected by Moderate Dyslipidemia Not on Lipid-Lowering Therapy (n = 125) | Patients with Dyslipidemia Treated with Lipid-Lowering Therapy (n = 302) | p | |
|---|---|---|---|
| Age (years) | 58.0 (8.7) | 69.3 (7.9) | <0.001 |
| Sex (males, %) | 53 (42%) | 140 (46%) | 0.455 |
| BMI (Kg/m2) | 25.6 (4.2) | 27.1 (4.4) | 0.001 |
| Weight (Kg) | 72.0 (14.8) | 73.1 (14.3) | 0.474 |
| Glucose (mg/dL) | 92.9 (9.3) | 121.0 (44.3) | <0.001 |
| Total-cholesterol (mg/dL) | 228.14 (21.2) | 194.9 (42.6) | <0.001 |
| HDL-cholesterol (mg/dL) | 65.3 (16.4) | 56.5 (15.2) | <0.001 |
| LDL-cholesterol (mg/dL) | 157.2 (20.9) | 110.3 (32.1) | <0.001 |
| Total cholesterol/HDL | 3.7 (0.9) | 3.62 (1.0) | 0.439 |
| Triglycerides (mg/dL) | 111.6 (52.5) | 118.8 (59.2) | 0.238 |
| hs-CRP (mg/L) | 0.2 (0.4) | 3.1 (9.0) | <0.001 |
| Creatine kinase (U/L) | 114.4 (72.9) | 123.4 (73.9) | 0.251 |
| Lp(a) (mg/dL) | 276.4 (324.7) | 269.0 (272.5) | 0.810 |
| Myoglobin (mg/dL) | 34.7 (21.3) | 43.5 (22.8) | <0.001 |
| Monocytes (n/mm3) | 0.4 (0.1) | 0.4 (0.1) | 0.999 |
| Neutrophils (n/mm3) | 3.3 (1.1) | 3.6 (1.2) | 0.016 |
| Lymphocytes (n/mm3) | 2.1 (0.6) | 2.0 (0.5) | 0.077 |
| Creatinine (mg/dL) | 0.9 (0.2) | 0.9 (0.3) | 0.999 |
| SNP ID | Chr. | Position | Locus | Functional Implication | Alleles | Type |
|---|---|---|---|---|---|---|
| rs3746444 | 20 | 34,990,448 | MYH7B MIR-499 | - | A/G | Intronic MYH7B, miR-499 |
| rs366631 | 1 | 109,709,850 | GSTM5 | DM | A | upstream_transcript_variant |
| rs1800896 | 1 | 206,773,552 | IL-10 | I/C | T/C | intron_variant |
| rs4693570 | 4 | 83,170,698 | 100 kb downstream of COQ2 | LM | T/C | Intergenic variant |
| rs17238540 | 5 | 74,655,498 | HMGCR | LM | T/G | intron_variant |
| rs3761740 | 5 | 75,336,308 | HMGCR | LM | C/A | upstream_transcript_variant |
| rs17238484 | 5 | 75,352,671 | HMGCR | LM | G/T | intron_variant |
| rs12916 | 5 | 75,360,714 | HMGCR | LM | T/C | 3_prime_UTR_variant |
| rs1800629 | 6 | 31,575,254 | TNFA | I/C | G/A | upstream_transcript_variant |
| rs699947 | 6 | 43,768,652 | VEGFA | I/C | C/A | upstream_transcript_variant |
| rs2010963 | 6 | 43,770,613 | VEGFA | I/C | G/C | upstream_transcript_variant |
| rs3025039 | 6 | 43,784,799 | VEGFA | I/C | C/T | 3_prime_UTR_variant |
| rs4880 | 6 | 159,692,840 | SOD2 | I/C | A/G | missense_variant |
| rs1800795 | 7 | 22,727,026 | IL-6 | I/C | G/C | intron_variant |
| rs662 | 7 | 95,308,134 | PON1 | LM | T/C | missense_variant |
| rs705379 | 7 | 95,324,583 | PON1 | LM | A/G | upstream_transcript_variant |
| rs35599367 | 7 | 99,366,316 | CYP3A4 | DM | G/A | intron_variant |
| rs776746 | 7 | 99,672,916 | CYP3A5 | DM | C/T | intron_variant |
| rs2740574 | 7 | 99,784,473 | CYP3A4 | DM | T/C | upstream_transcript_variant |
| rs328 | 8 | 19,962,213 | LPL | LM | C/G | stop_gained, coding_sequence_variant |
| rs4636297 | 9 | 136,670,698 | EGFL7 | I/C | G/A | downstream_transcript_variant |
| rs1799853 | 10 | 94,942,290 | CYP2C9 | DM | C/T | missense_variant |
| rs72558195 | 10 | 95,064,886 | CYP2C8 | DM | G/A | stop_gained, missense_variant |
| rs1057910 | 10 | 96,741,053 | CYP2C9 | DM | A/C | missense_variant |
| rs717620 | 10 | 99,782,821 | ABCC2 | DM | C/T | upstream_transcript_variant |
| rs17222723 | 10 | 99,836,239 | ABCC2 | DM | T/A | missense_variant |
| rs8187710 | 10 | 99,851,537 | ABCC2 | DM | G/A | missense_variant |
| rs1695 | 11 | 67,585,218 | GSTP1 | DM | A/G | missense_variant |
| rs1799837 | 11 | 116,837,537 | APOA1 | LM | C/T | 5_prime_UTR_variant |
| rs4149056 | 12 | 21,331,549 | SLCO1B1 | DM | T/C | missense_variant |
| rs4363657 | 12 | 21,368,722 | SLCO1B1 | DM | T/C | intron_variant |
| rs708272 | 16 | 56,962,376 | CETP | LM | G/A | intron_variant |
| rs1532624 | 16 | 56,971,567 | CETP | LM | C/A | intron_variant |
| rs2228314 | 22 | 41,880,738 | SREBF2 | LM | G/C | missense_variant |
| p-Values | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP ID | Gene | Alleles | MAF | HWE | Call Rate | TC | LDL-C | HDL-C | TRIG | TC:HDL-C Ratio | Non-HDL-C |
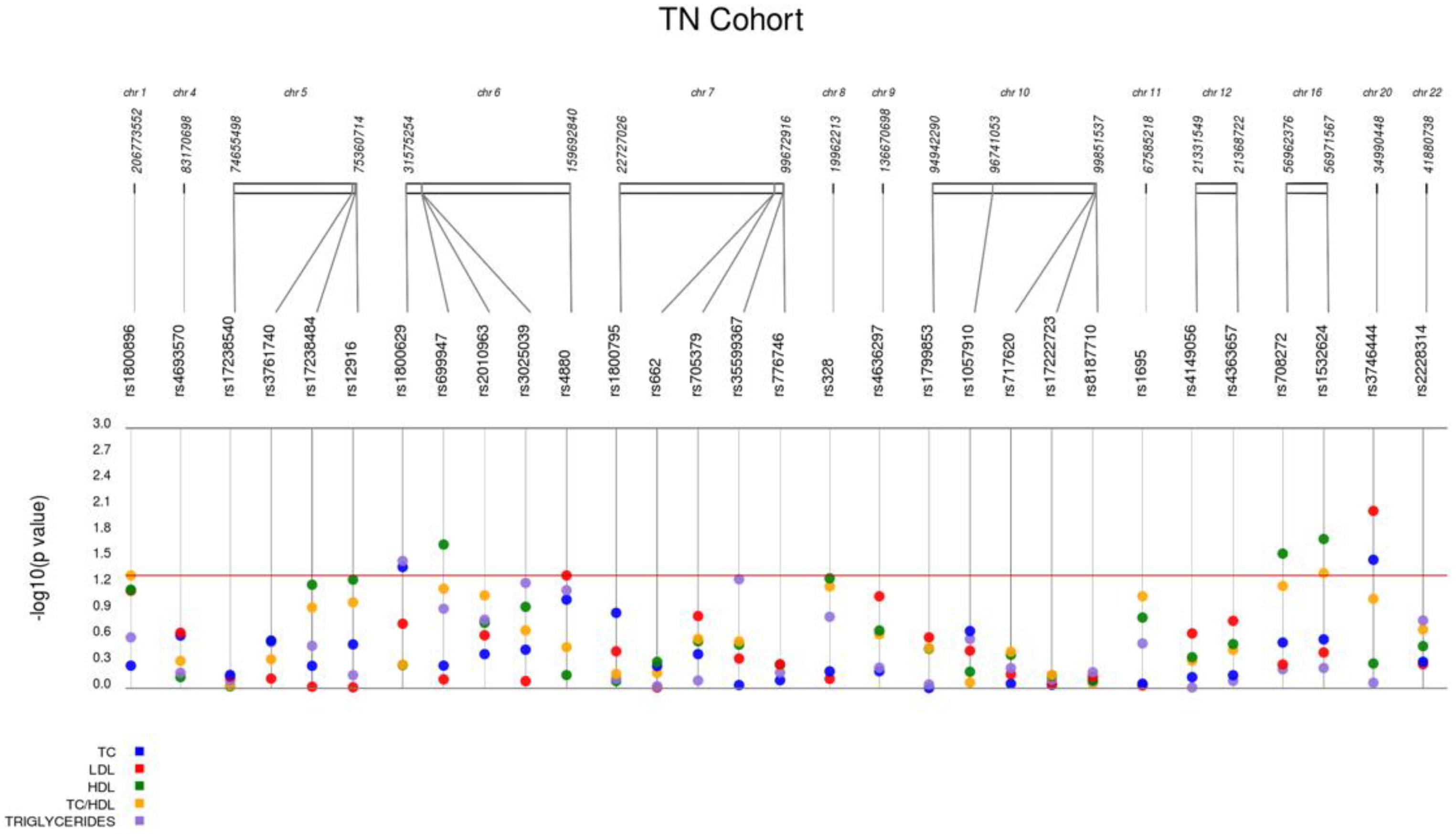
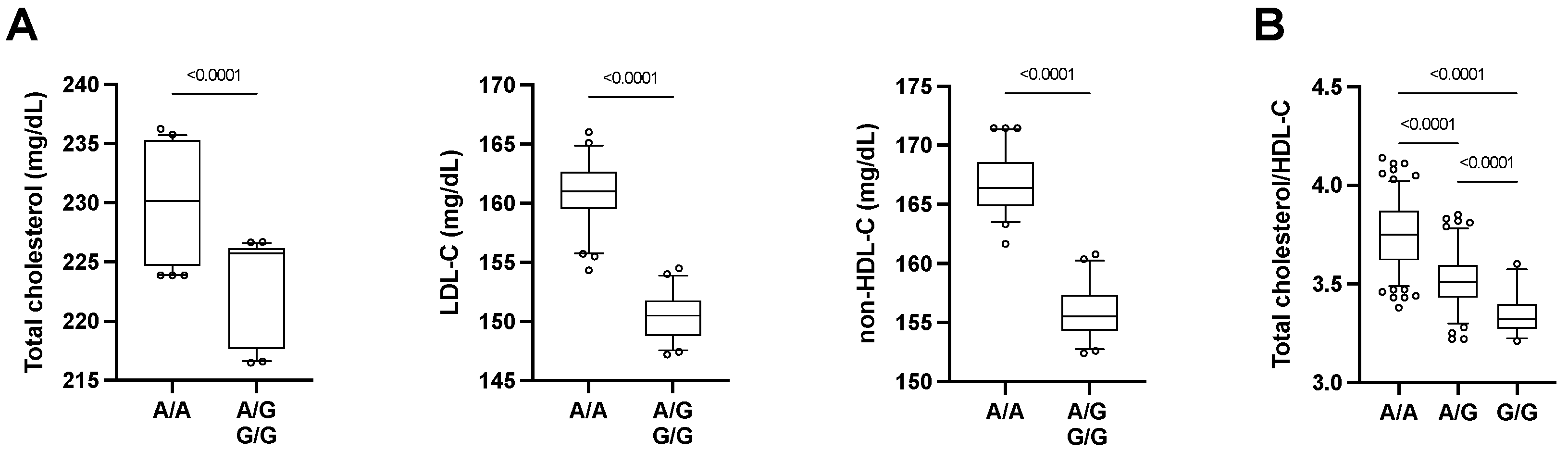
| rs3746444 | MYH7B miR-499 | A/G | 24.1 | 0.210 | 92.80 | 0.033 ↓ | 0.009 ↓ | 0.519 | 0.863 | 0.088 | 0.009 ↓ |
| rs1800629 | TNFA | G/A | 10.3 | 1.000 | 93.60 | 0.040 ↑ | 0.180 | 0.541 | 0.034 ↑ | 0.530 | 0.110 |
| rs708272 | CETP | G/A | 40.0 | 1.000 | 92.00 | 0.297 | 0.532 | 0.028 ↑ | 0.604 | 0.068 | 0.570 |
| rs1532624 | CETP | C/A | 41.5 | 0.707 | 93.60 | 0.273 | 0.387 | 0.019 ↑ | 0.584 | 0.049 ↓ | 0.540 |
| rs4880 | SOD2 | A/G | 46.6 | 1.000 | 93.60 | 0.095 | 0.049 ↓ | 0.704 | 0.074 | 0.340 | 0.051 |
| rs699947 | VEGFA | C/A | 38.5 | 0.845 | 93.60 | 0.548 | 0.790 | 0.022 ↑ | 0.121 | 0.064 | 0.280 |
| p-Values | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP ID | Gene | Alleles | MAF | HWE | Call Rate | TC | LDL-C | HDL-C | TRIG | TC:HDL-C Ratio | Non-HDL-C |
| rs3746444 | MYH7B miR-499a | A/G | 28.5 | 0.32 | 100 | 0.850 | 0.970 | 0.079 | 0.310 | 0.016 ↓ | 0.380 |
| rs1532624 | CETP | C/A | 40.8 | 0.63 | 99.34 | 0.490 | 0.220 | 0.035 ↑ | 0.089 | 0.160 | 0.930 |
| rs708272 | CETP | G/A | 39.1 | 0.72 | 100 | 0.540 | 0.260 | 0.049 ↑ | 0.100 | 0.170 | 0.910 |
| rs1057910 | CYP2C9 | A/C | 8.1 | 0.71 | 100 | 0.020 ↑ | 0.790 | 0.990 | 0.016 ↑ | 0.028 ↑ | 0.015 ↑ |
| rs1800795 | IL-6 | G/C | 28.2 | 0.89 | 99.67 | 0.340 | 0.020 ↓ | 0.190 | 0.510 | 0.190 | 0.130 |
| rs717620 | ABCC2 | C/T | 16.6 | 0.41 | 100 | 0.049 ↓ | 0.0082 ↓ | 0.210 | 0.230 | 0.016 ↓ | 0.011 ↓ |
| rs705379 | PON1 | A/G | 46.8 | 0.35 | 99.01 | 0.150 | 0.024 ↓ | 0.860 | 1.000 | 0.580 | 0.160 |
| rs1800896 | IL-10 | T/C | 37.3 | 0.33 | 100 | 0.790 | 0.990 | 0.270 | 0.0011 ↑ | 0.093 | 0.480 |
| rs2010963 | VEGFA | G/C | 38.4 | 0.9 | 100 | 0.670 | 0.960 | 0.190 | 0.043 ↑ | 0.160 | 0.340 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giuliani, A.; Montesanto, A.; Matacchione, G.; Graciotti, L.; Ramini, D.; Protic, O.; Galeazzi, R.; Antonicelli, R.; Tortato, E.; Bonfigli, A.R.; et al. The Association between Single Nucleotide Polymorphisms, including miR-499a Genetic Variants, and Dyslipidemia in Subjects Treated with Pharmacological or Phytochemical Lipid-Lowering Agents. Int. J. Mol. Sci. 2022, 23, 5617. https://doi.org/10.3390/ijms23105617
Giuliani A, Montesanto A, Matacchione G, Graciotti L, Ramini D, Protic O, Galeazzi R, Antonicelli R, Tortato E, Bonfigli AR, et al. The Association between Single Nucleotide Polymorphisms, including miR-499a Genetic Variants, and Dyslipidemia in Subjects Treated with Pharmacological or Phytochemical Lipid-Lowering Agents. International Journal of Molecular Sciences. 2022; 23(10):5617. https://doi.org/10.3390/ijms23105617
Chicago/Turabian StyleGiuliani, Angelica, Alberto Montesanto, Giulia Matacchione, Laura Graciotti, Deborah Ramini, Olga Protic, Roberta Galeazzi, Roberto Antonicelli, Elena Tortato, Anna Rita Bonfigli, and et al. 2022. "The Association between Single Nucleotide Polymorphisms, including miR-499a Genetic Variants, and Dyslipidemia in Subjects Treated with Pharmacological or Phytochemical Lipid-Lowering Agents" International Journal of Molecular Sciences 23, no. 10: 5617. https://doi.org/10.3390/ijms23105617







