Nuclear mRNA Export and Aging
Abstract
:1. Introduction
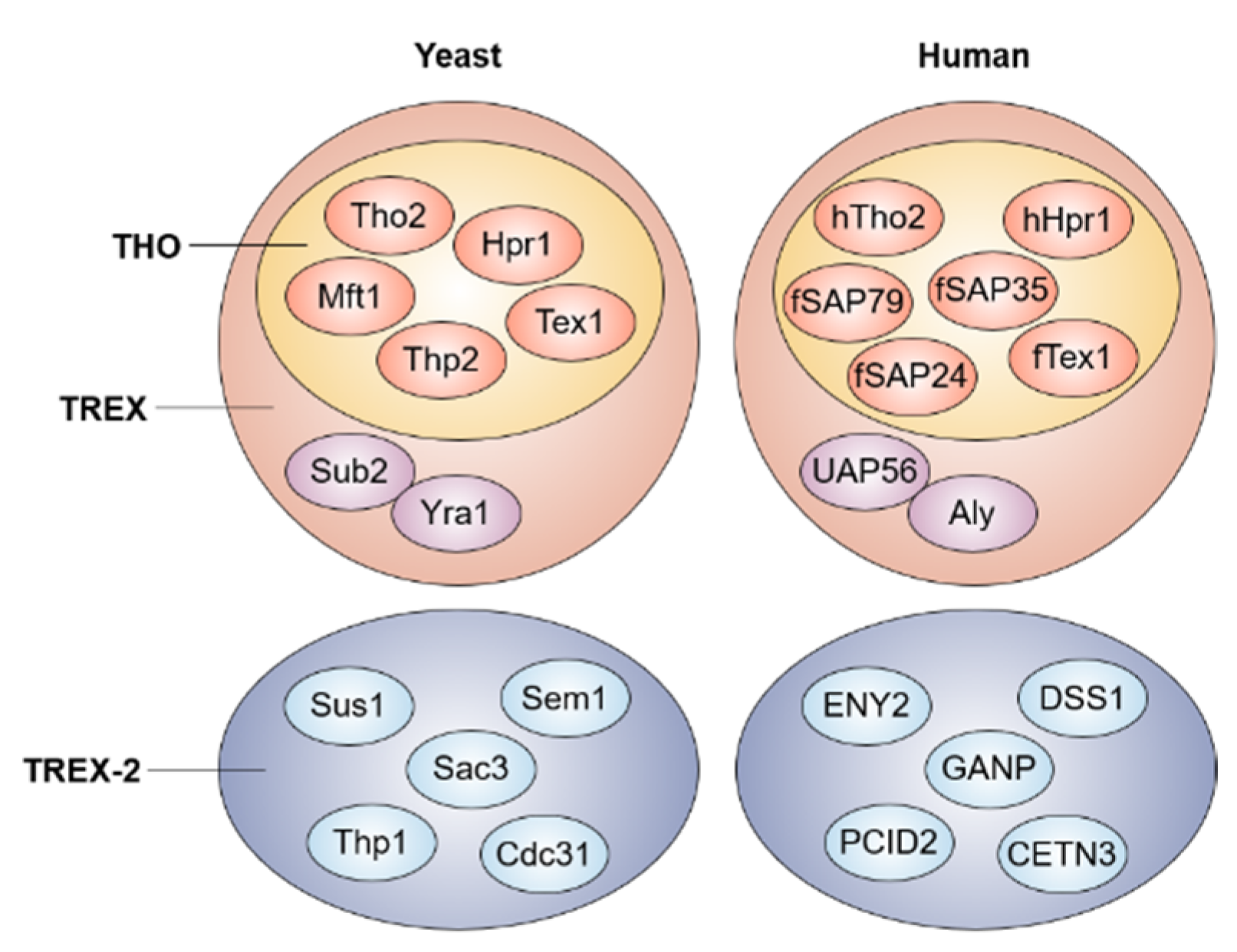
2. Nuclear mRNA Export Pathway
3. NPC and mRNA Export
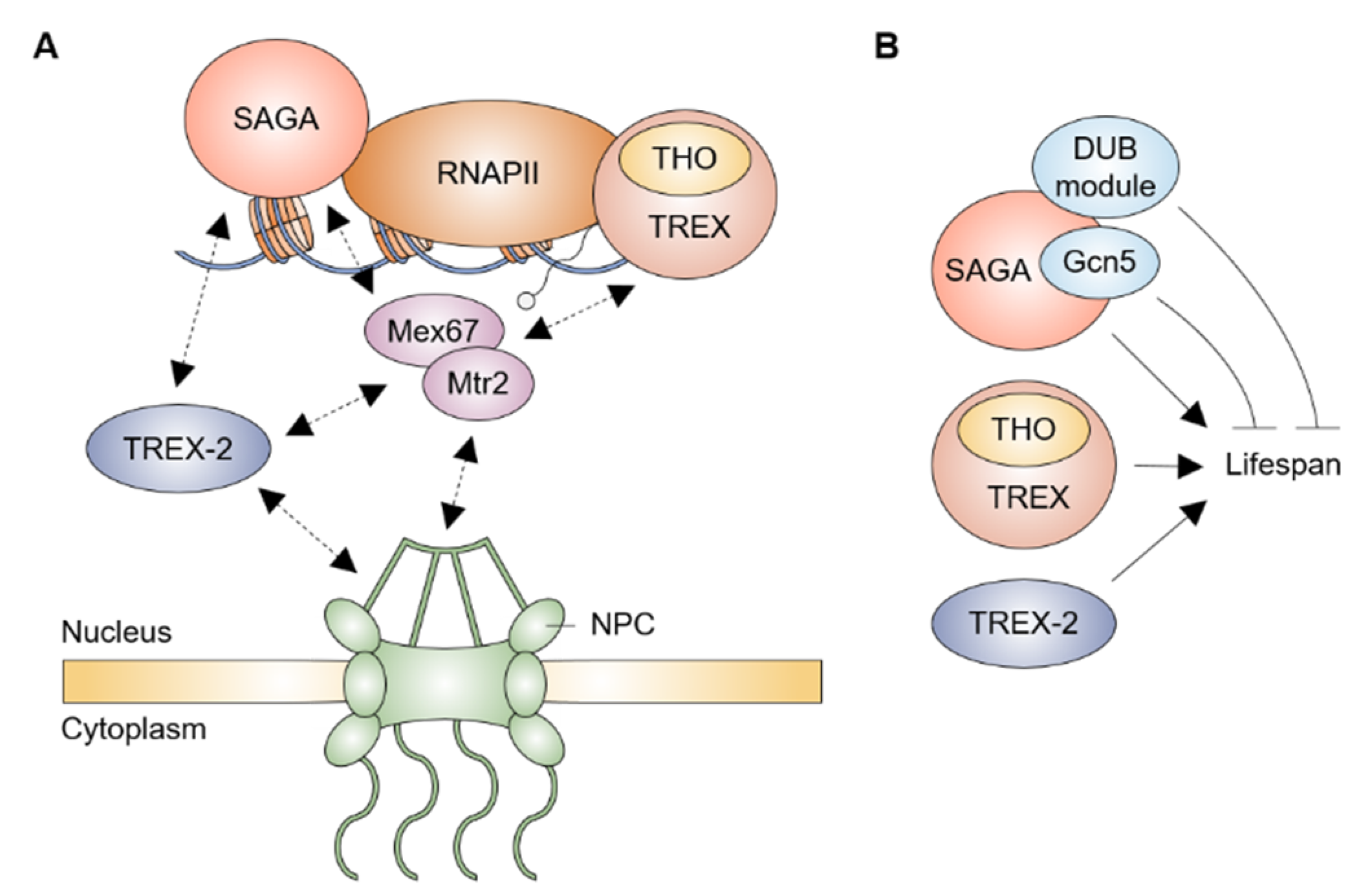
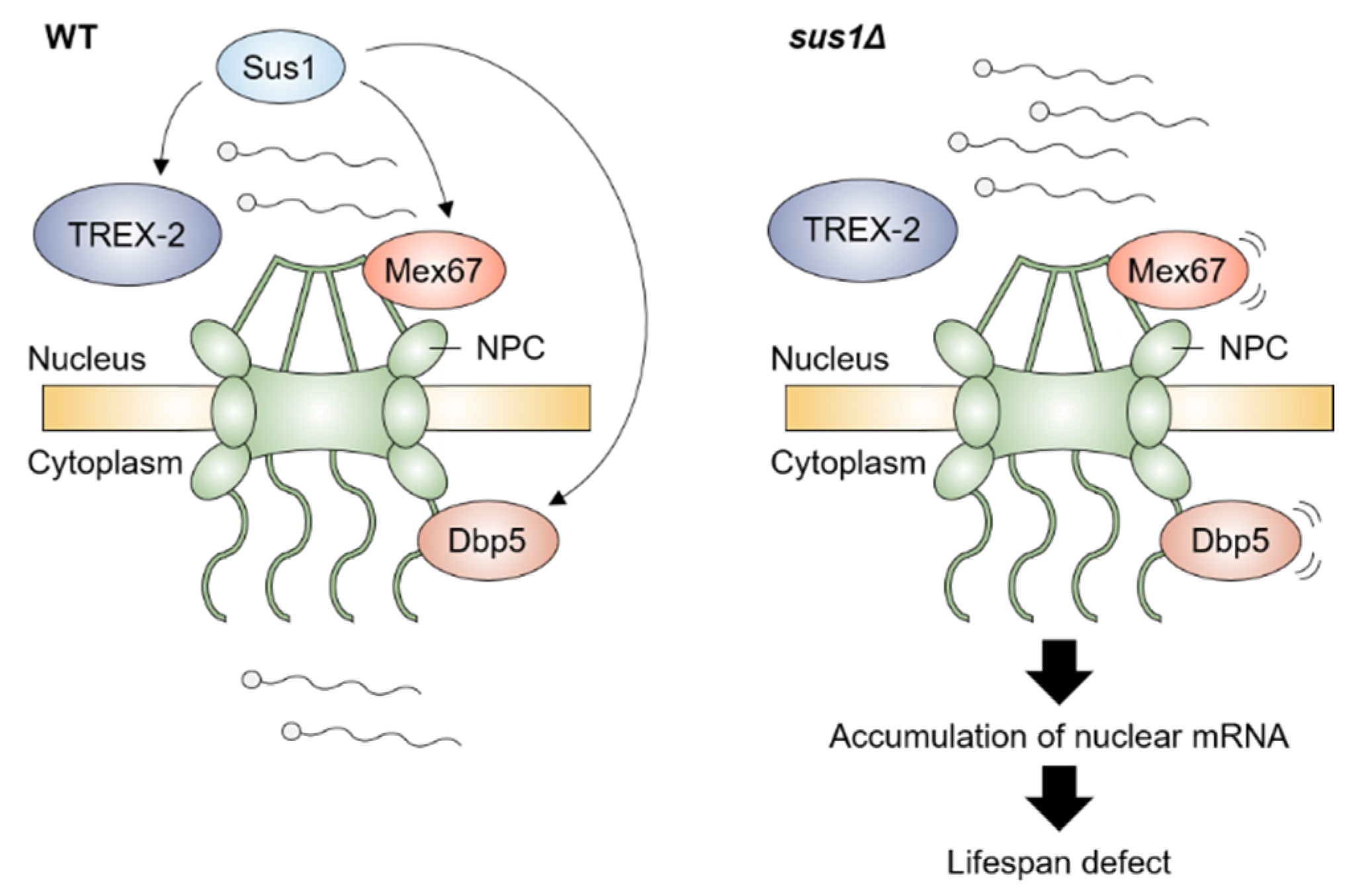
4. mRNA Export Factors and Aging
5. NPC and Aging
6. MRNA Turnover and Aging
7. MRNA Export and Age-Related Neurodegenerative Disorders
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kornberg, R.D. The molecular basis of eukaryotic transcription. Proc. Natl. Acad. Sci. USA 2007, 104, 12955–12961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maniatis, T.; Reed, R. An extensive network of coupling among gene expression machines. Nature 2002, 416, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Partridge, L.; Longo, V.D. Extending Healthy Life Span—From Yeast to Humans. Science 2010, 328, 321–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, R.W.; Lu, R.; Danthi, P.S.; Bravo, J.I.; Goumba, A.; Sampathkumar, N.K.; Benayoun, B.A. Multi-level remodeling of transcriptional landscapes in aging and longevity. BMB Rep. 2019, 52, 86–108. [Google Scholar] [CrossRef] [PubMed]
- Stegeman, R.; Weake, V.M. Transcriptional Signatures of Aging. J. Mol. Biol. 2017, 429, 2427–2437. [Google Scholar] [CrossRef] [PubMed]
- Callegari, A.J. Does transcription-associated DNA damage limit lifespan? DNA Repair 2016, 41, 1–7. [Google Scholar] [CrossRef]
- Verheijen, B.M.; Van Leeuwen, F.W. Commentary: The landscape of transcription errors in eukaryotic cells. Front. Genet. 2017, 8, 219. [Google Scholar] [CrossRef] [Green Version]
- Van Leeuwen, F.W.; Hol, E.M.; Burbach, J.P. Mutations in RNA: A first example of molecular misreading in Alzheimer’s disease. Trends Neurosci. 1998, 21, 331–335. [Google Scholar] [CrossRef]
- van Leeuwen, F.W.; de Kleijn, D.P.V.; Van Den Hurk, H.H.; Neubauer, A.; Sonnemans, M.A.F.; Sluijs, J.A.; Köycü, S.; Ramdjielal, R.D.J.; Salehi, A.; Martens, G.J.M.; et al. Frameshift Mutants of β Amyloid Precursor Protein and Ubiquitin-B in Alzheimer’s and Down Patients. Science 1998, 279, 242–247. [Google Scholar] [CrossRef]
- Saxowsky, T.T.; Meadows, K.L.; Klungland, A.; Doetsch, P.W. 8-Oxoguanine-mediated transcriptional mutagenesis causes Ras activation in mammalian cells. Proc. Natl. Acad. Sci. USA 2008, 105, 18877–18882. [Google Scholar] [CrossRef] [Green Version]
- Vermulst, M.; Denney, A.S.; Lang, M.; Hung, C.-W.; Moore, S.; Moseley, M.A.; Thompson, W.J.; Madden, V.; Gauer, J.; Wolfe, K.J.; et al. Transcription errors induce proteotoxic stress and shorten cellular lifespan. Nat. Commun. 2015, 6, 8065. [Google Scholar] [CrossRef] [PubMed]
- Björk, P.; Wieslander, L. Mechanisms of mRNA export. Semin. Cell Dev. Biol. 2014, 32, 47–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmid, M.; Jensen, T.H. Quality control of mRNP in the nucleus. Chromosoma 2008, 117, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Lykke-Andersen, S.; Nasser, T.; Saguez, C.; Bertrand, E.; Jensen, T.H.; Moore, C. Assembly of an Export-Competent mRNP Is Needed for Efficient Release of the 3′-End Processing Complex after Polyadenylation. Mol. Cell. Biol. 2009, 29, 5327–5338. [Google Scholar] [CrossRef] [Green Version]
- Strässer, K.; Hurt, E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 2000, 19, 410–420. [Google Scholar] [CrossRef] [Green Version]
- Batisse, J.; Batisse, C.; Budd, A.; Böttcher, B.; Hurt, E. Purification of Nuclear Poly(A)-binding Protein Nab2 Reveals Association with the Yeast Transcriptome and a Messenger Ribonucleoprotein Core Structure. J. Biol. Chem. 2009, 284, 34911–34917. [Google Scholar] [CrossRef] [Green Version]
- Green, D.M.; Marfatia, K.A.; Crafton, E.B.; Zhang, X.; Cheng, X.; Corbett, A.H. Nab2p Is Required for Poly(A) RNA Export in Saccharomyces cerevisiae and Is Regulated by Arginine Methylation via Hmt1p. J. Biol. Chem. 2002, 277, 7752–7760. [Google Scholar] [CrossRef] [Green Version]
- Hector, R.E.; Nykamp, K.R.; Dheur, S.; Anderson, J.T.; Non, P.J.; Urbinati, C.R.; Wilson, S.; Minvielle-Sebastia, L.; Swanson, M.S. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J. 2002, 21, 1800–1810. [Google Scholar] [CrossRef] [Green Version]
- Strässer, K.; Hurt, E. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature 2001, 413, 648–652. [Google Scholar] [CrossRef]
- Strässer, K.; Masuda, S.; Mason, P.; Pfannstiel, J.; Oppizzi, M.; Rodriguez-Navarro, S.; Rondón, A.G.; Aguilera, A.; Struhl, K.; Reed, R.; et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 2002, 417, 304–308. [Google Scholar] [CrossRef]
- Johnson, S.A.; Kim, H.; Erickson, B.; Bentley, D.L. The export factor Yra1 modulates mRNA 3′ end processing. Nat. Struct. Mol. Biol. 2011, 18, 1164–1171. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.A.; Cubberley, G.; Bentley, D.L. Cotranscriptional Recruitment of the mRNA Export Factor Yra1 by Direct Interaction with the 3′ End Processing Factor Pcf11. Mol. Cell 2009, 33, 215–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saguez, C.; Schmid, M.; Olesen, J.R.; Ghazy, M.A.E.-H.; Qu, X.; Poulsen, M.B.; Nasser, T.; Moore, C.; Jensen, T.H. Nuclear mRNA Surveillance in THO/sub2 Mutants Is Triggered by Inefficient Polyadenylation. Mol. Cell 2008, 31, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Chávez, S.; Beilharz, T.; Rondón, A.G.; Erdjument-Bromage, H.; Tempst, P.; Svejstrup, J.; Lithgow, T.; Aguilera, A. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J. 2000, 19, 5824–5834. [Google Scholar] [CrossRef] [Green Version]
- Hur, J.K.; Chung, Y.D. A novel model of THO/TREX loading onto target RNAs in metazoan gene expression. BMB Rep. 2016, 49, 355–356. [Google Scholar] [CrossRef] [Green Version]
- Zuckerman, B.; Ron, M.; Mikl, M.; Segal, E.; Ulitsky, I. Gene Architecture and Sequence Composition Underpin Selective Dependency of Nuclear Export of Long RNAs on NXF1 and the TREX Complex. Mol. Cell 2020, 79, 251–267.e6. [Google Scholar] [CrossRef]
- Müller-McNicoll, M.; Botti, V.; Domingues, A.M.D.J.; Brandl, H.; Schwich, O.D.; Steiner, M.C.; Curk, T.; Poser, I.; Zarnack, K.; Neugebauer, K.M. SR proteins are NXF1 adaptors that link alternative RNA processing to mRNA export. Genes Dev. 2016, 30, 553–566. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Navarro, S. Insights into SAGA function during gene expression. EMBO Rep. 2009, 10, 843–850. [Google Scholar] [CrossRef]
- Ellisdon, A.; Dimitrova, L.; Hurt, E.; Stewart, M. Structural basis for the assembly and nucleic acid binding of the TREX-2 transcription-export complex. Nat. Struct. Mol. Biol. 2012, 19, 328–336. [Google Scholar] [CrossRef] [Green Version]
- Fischer, T.; Strässer, K.; Racz, A.; Rodriguez-Navarro, S.; Oppizzi, M.; Ihrig, P.; Lechner, J.; Hurt, E. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 2002, 21, 5843–5852. [Google Scholar] [CrossRef] [Green Version]
- Jani, D.; Lutz, S.; Marshall, N.J.; Fischer, T.; Köhler, A.; Ellisdon, A.M.; Hurt, E.; Stewart, M. Sus1, Cdc31, and the Sac3 CID Region Form a Conserved Interaction Platform that Promotes Nuclear Pore Association and mRNA Export. Mol. Cell 2009, 33, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Jani, D.; Valkov, E.; Stewart, M. Structural basis for binding the TREX2 complex to nuclear pores, GAL1 localisation and mRNA export. Nucleic Acids Res. 2014, 42, 6686–6697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, E.P.; Stern, C.A.; Fahrenkrog, B.; Krebber, H.; Moy, T.I.; Aebi, U.; Silver, P.A. Sac3 Is an mRNA Export Factor That Localizes to Cytoplasmic Fibrils of Nuclear Pore Complex. Mol. Biol. Cell 2003, 14, 836–847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Molinero, V.; García-Martínez, J.; Reja, R.; Furió-Tarí, P.; Antúnez, O.; Vinayachandran, V.; Conesa, A.; Pugh, B.F.; Pérez-Ortín, J.E.; Rodríguez-Navarro, S. The SAGA/TREX-2 subunit Sus1 binds widely to transcribed genes and affects mRNA turnover globally. Epigenetics Chromatin 2018, 11, 13. [Google Scholar] [CrossRef]
- Köhler, A.; Garcia, P.P.; Llopis, A.; Zapater, M.; Posas, F.; Hurt, E.; Rodríguez-Navarro, S. The mRNA Export Factor Sus1 Is Involved in Spt/Ada/Gcn5 Acetyltransferase-mediated H2B Deubiquitinylation through Its Interaction with Ubp8 and Sgf11. Mol. Biol. Cell 2006, 17, 4228–4236. [Google Scholar] [CrossRef]
- Rodriguez-Navarro, S.; Fischer, T.; Luo, M.-J.; Antúnez, O.; Brettschneider, S.; Lechner, J.; Pérez-Ortín, J.E.; Reed, R.; Hurt, E. Sus1, a Functional Component of the SAGA Histone Acetylase Complex and the Nuclear Pore-Associated mRNA Export Machinery. Cell 2004, 116, 75–86. [Google Scholar] [CrossRef] [Green Version]
- Schneider, M.; Hellerschmied, D.; Schubert, T.; Amlacher, S.; Vinayachandran, V.; Reja, R.; Pugh, B.F.; Clausen, T.; Köhler, A. The Nuclear Pore-Associated TREX-2 Complex Employs Mediator to Regulate Gene Expression. Cell 2015, 162, 1016–1028. [Google Scholar] [CrossRef] [Green Version]
- Cabal, G.; Genovesio, A.; Rodriguez-Navarro, S.; Zimmer, C.; Gadal, O.; Lesne, A.; Buc, H.; Feuerbach-Fournier, F.; Olivo-Marin, J.-C.; Hurt, E.C.; et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature 2006, 441, 770–773. [Google Scholar] [CrossRef]
- Chekanova, J.A.; Abruzzi, K.C.; Rosbash, M.; Belostotsky, D.A. Sus1, Sac3, and Thp1 mediate post-transcriptional tethering of active genes to the nuclear rim as well as to non-nascent mRNP. RNA 2008, 14, 66–77. [Google Scholar] [CrossRef] [Green Version]
- Kurshakova, M.M.; Krasnov, A.; Kopytova, D.V.; Shidlovskii, Y.V.; Nikolenko, J.V.; Nabirochkina, E.N.; Spehner, D.; Schultz, P.; Tora, L.; Georgieva, S.G. SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. EMBO J. 2007, 26, 4956–4965. [Google Scholar] [CrossRef]
- Faza, M.B.; Kemmler, S.; Jimeno, S.; González-Aguilera, C.; Aguilera, A.; Hurt, E.; Panse, V.G. Sem1 is a functional component of the nuclear pore complex–associated messenger RNA export machinery. J. Cell Biol. 2009, 184, 833–846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wickramasinghe, V.O.; McMurtrie, P.I.; Mills, A.D.; Takei, Y.; Penrhyn-Lowe, S.; Amagase, Y.; Main, S.; Marr, J.; Stewart, M.; Laskey, R.A. mRNA Export from Mammalian Cell Nuclei Is Dependent on GANP. Curr. Biol. 2010, 20, 25–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jani, D.; Lutz, S.; Hurt, E.; Laskey, R.A.; Stewart, M.; Wickramasinghe, V.O. Functional and structural characterization of the mammalian TREX-2 complex that links transcription with nuclear messenger RNA export. Nucleic Acids Res. 2012, 40, 4562–4573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wente, S.R.; Rout, M.P. The Nuclear Pore Complex and Nuclear Transport. Cold Spring Harb. Perspect. Biol. 2010, 2, a000562. [Google Scholar] [CrossRef] [PubMed]
- Aitchison, J.D.; Rout, M. The Yeast Nuclear Pore Complex and Transport Through It. Genetics 2012, 190, 855–883. [Google Scholar] [CrossRef] [Green Version]
- Terry, L.J.; Wente, S.R. Flexible Gates: Dynamic Topologies and Functions for FG Nucleoporins in Nucleocytoplasmic Transport. Eukaryot. Cell 2009, 8, 1814–1827. [Google Scholar] [CrossRef] [Green Version]
- Terry, L.J.; Wente, S.R. Nuclear mRNA export requires specific FG nucleoporins for translocation through the nuclear pore complex. J. Cell Biol. 2007, 178, 1121–1132. [Google Scholar] [CrossRef]
- Siebrasse, J.P.; Kaminski, T.; Kubitscheck, U. Nuclear export of single native mRNA molecules observed by light sheet fluorescence microscopy. Proc. Natl. Acad. Sci. USA 2012, 109, 9426–9431. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Liu, Z.; Michelotti, N.; Pitchiaya, S.; Veerapaneni, R.; Androsavich, J.R.; Walter, N.G.; Yang, W. High-resolution three-dimensional mapping of mRNA export through the nuclear pore. Nat. Commun. 2013, 4, 2414. [Google Scholar] [CrossRef] [Green Version]
- Mor, A.; Suliman, S.; Ben-Yishay, R.; Yunger, S.; Brody, Y.; Shav-Tal, Y. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat. Cell Biol. 2010, 12, 543–552. [Google Scholar] [CrossRef]
- Grünwald, D.; Singer, R.H. In vivo imaging of labelled endogenous β-actin mRNA during nucleocytoplasmic transport. Nature 2010, 467, 604–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alic, N.; Giannakou, M.E.; Papatheodorou, I.; Hoddinott, M.P.; Andrews, T.D.; Bolukbasi, E.; Partridge, L. Interplay of dFOXO and Two ETS-Family Transcription Factors Determines Lifespan in Drosophila melanogaster. PLoS Genet. 2014, 10, e1004619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, L.; McCormick, M.A.; Kennedy, B.K.; Tu, B.P. Integration of Multiple Nutrient Cues and Regulation of Lifespan by Ribosomal Transcription Factor Ifh1. Cell Rep. 2013, 4, 1063–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghazi, A.; Henis-Korenblit, S.; Kenyon, C. A Transcription Elongation Factor That Links Signals from the Reproductive System to Lifespan Extension in Caenorhabditis elegans. PLoS Genet. 2009, 5, e1000639. [Google Scholar] [CrossRef] [Green Version]
- Mittal, N.; Guimaraes, J.C.; Gross, T.; Schmidt, A.; Vina-Vilaseca, A.; Nedialkova, D.D.; Aeschimann, F.; Leidel, S.; Spang, A.; Zavolan, M. The Gcn4 transcription factor reduces protein synthesis capacity and extends yeast lifespan. Nat. Commun. 2017, 8, 457. [Google Scholar] [CrossRef] [Green Version]
- Oh, S.W.; Mukhopadhyay, A.; Svrzikapa, N.; Jiang, F.; Davis, R.J.; Tissenbaum, H.A. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc. Natl. Acad. Sci. USA 2005, 102, 4494–4499. [Google Scholar] [CrossRef] [Green Version]
- Postnikoff, S.D.L.; Malo, M.E.; Wong, B.; Harkness, T.A.A. The Yeast Forkhead Transcription Factors Fkh1 and Fkh2 Regulate Lifespan and Stress Response Together with the Anaphase-Promoting Complex. PLoS Genet. 2012, 8, e1002583. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Lopez, M.; Gonzalez, S.; Hillson, O.; Tunnacliffe, E.; Codlin, S.; Tallada, V.A.; Bähler, J.; Rallis, C.; Rodriguez-Lopez, M.; Gonzalez, S.; et al. The GATA Transcription Factor Gaf1 Represses tRNAs, Inhibits Growth, and Extends Chronological Lifespan Downstream of Fission Yeast TORC1. Cell Rep. 2020, 30, 3240–3249.e4. [Google Scholar] [CrossRef] [Green Version]
- Ryu, H.-Y.; Rhie, B.-H.; Ahn, S.H. Loss of the Set2 histone methyltransferase increases cellular lifespan in yeast cells. Biochem. Biophys. Res. Commun. 2014, 446, 113–118. [Google Scholar] [CrossRef]
- Yamamoto, R.; Tatar, M. Insulin receptor substrate chico acts with the transcription factor FOXO to extend Drosophila lifespan. Aging Cell 2011, 10, 729–732. [Google Scholar] [CrossRef] [Green Version]
- Samara, N.L.; Wolberger, C. A new chapter in the transcription SAGA. Curr. Opin. Struct. Biol. 2011, 21, 767–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, B.; Zhong, D.; Zhu, J.; An, Y.; Gao, M.; Zhu, S.; Dang, W.; Wang, X.; Yang, B.; Xie, Z. Inhibition of histone acetyltransferase GCN5 extends lifespan in both yeast and human cell lines. Aging Cell 2020, 19, e13129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horiuchi, J.; Silverman, N.; Marcus, G.A.; Guarente, L. ADA3, a putative transcriptional adaptor, consists of two separable domains and interacts with ADA2 and GCN5 in a trimeric complex. Mol. Cell. Biol. 1995, 15, 1203–1209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, S.; Ahn, H.; Duan, R.; Liu, Y.; Ryu, H.-Y.; Ahn, S.H. The Spt7 subunit of the SAGA complex is required for the regulation of lifespan in both dividing and nondividing yeast cells. Mech. Ageing Dev. 2021, 196, 111480. [Google Scholar] [CrossRef] [PubMed]
- McCormick, M.A.; Mason, A.G.; Guyenet, S.J.; Dang, W.; Garza, R.M.; Ting, M.K.; Moller, R.M.; Berger, S.L.; Kaeberlein, M.; Pillus, L.; et al. The SAGA Histone Deubiquitinase Module Controls Yeast Replicative Lifespan via Sir2 Interaction. Cell Rep. 2014, 8, 477–486. [Google Scholar] [CrossRef] [Green Version]
- Denoth-Lippuner, A.; Krzyzanowski, M.K.; Stober, C.; Barral, Y. Role of SAGA in the asymmetric segregation of DNA circles during yeast ageing. eLife 2014, 3, e03790. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Cho, B.; Moon, S.; Chung, Y.D. The THO complex is required for stress tolerance and longevity in Drosophila. Genes Genom. 2011, 33, 291–297. [Google Scholar] [CrossRef]
- Kim, S.Y.; Yang, E.J.; Lee, S.B.; Lee, Y.-S.; Cho, K.A.; Park, S.C. Global transcriptional downregulation of TREX and nuclear trafficking machinery as pan-senescence phenomena: Evidence from human cells and tissues. Exp. Mol. Med. 2020, 52, 1351–1359. [Google Scholar] [CrossRef]
- Lim, S.; Liu, Y.; Rhie, B.-H.; Kim, C.; Ryu, H.-Y.; Ahn, S.H. Sus1 maintains a normal lifespan through regulation of TREX-2 complex-mediated mRNA export. bioRxiv 2022. [Google Scholar] [CrossRef]
- D’Angelo, M.A.; Raices, M.; Panowski, S.H.; Hetzer, M.W. Age-Dependent Deterioration of Nuclear Pore Complexes Causes a Loss of Nuclear Integrity in Postmitotic Cells. Cell 2009, 136, 284–295. [Google Scholar] [CrossRef] [Green Version]
- Savas, J.N.; Toyama, B.H.; Xu, T.; Yates, J.R., 3rd; Hetzer, M.W. Extremely Long-Lived Nuclear Pore Proteins in the Rat Brain. Science 2012, 335, 942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lord, C.; Timney, B.L.; Rout, M.; Wente, S.R. Altering nuclear pore complex function impacts longevity and mitochondrial function in S. cerevisiae. J. Cell Biol. 2015, 208, 729–744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lord, C.L.; Ospovat, O.; Wente, S.R. Nup100 regulates Saccharomyces cerevisiae replicative life span by mediating the nuclear export of specific tRNAs. RNA 2017, 23, 365–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, U.H.; Hetzer, M.W. Nuclear Periphery Takes Center Stage: The Role of Nuclear Pore Complexes in Cell Identity and Aging. Neuron 2020, 106, 899–911. [Google Scholar] [CrossRef] [PubMed]
- Ryu, S.J.; Cho, K.A.; Oh, Y.S.; Park, S.C. Role of Src-specific phosphorylation site on focal adhesion kinase for senescence-associated apoptosis resistance. Apoptosis 2006, 11, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.A.; Ryu, S.J.; Park, J.S.; Jang, I.S.; Ahn, J.S.; Kim, K.T.; Park, S.C. Senescent Phenotype Can Be Reversed by Reduction of Caveolin Status. J. Biol. Chem. 2003, 278, 27789–27795. [Google Scholar] [CrossRef] [Green Version]
- Park, W.-Y.; Park, J.-S.; Cho, K.-A.; Kim, D.-I.; Ko, Y.-G.; Seo, J.-S.; Park, S.C. Up-regulation of Caveolin Attenuates Epidermal Growth Factor Signaling in Senescent Cells. J. Biol. Chem. 2000, 275, 20847–20852. [Google Scholar] [CrossRef] [Green Version]
- Seluanov, A.; Gorbunova, V.; Falcovitz, A.; Sigal, A.; Milyavsky, M.; Zurer, I.; Shohat, G.; Goldfinger, N.; Rotter, V. Change of the Death Pathway in Senescent Human Fibroblasts in Response to DNA Damage Is Caused by an Inability To Stabilize p53. Mol. Cell. Biol. 2001, 21, 1552–1564. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.Y.; Ryu, S.J.; Ahn, H.J.; Choi, H.R.; Kang, H.T.; Park, S.C. Senescence-related functional nuclear barrier by down-regulation of nucleo-cytoplasmic trafficking gene expression. Biochem. Biophys. Res. Commun. 2010, 391, 28–32. [Google Scholar] [CrossRef]
- Janssens, G.; Meinema, A.C.; González, J.; Wolters, J.C.; Schmidt, A.; Guryev, V.; Bischoff, R.; Wit, E.C.; Veenhoff, L.M.; Heinemann, M. Protein biogenesis machinery is a driver of replicative aging in yeast. eLife 2015, 4, e08527. [Google Scholar] [CrossRef]
- Rempel, I.L.; Crane, M.M.; Thaller, D.J.; Mishra, A.; Jansen, D.P.; Janssens, G.; Popken, P.; Akşit, A.; Kaeberlein, M.; Van Der Giessen, E.; et al. Age-dependent deterioration of nuclear pore assembly in mitotic cells decreases transport dynamics. eLife 2019, 8, e48186. [Google Scholar] [CrossRef] [PubMed]
- Maeshima, K.; Yahata, K.; Sasaki, Y.; Nakatomi, R.; Tachibana, T.; Hashikawa, T.; Imamoto, F.; Imamoto, N. Cell-cycle-dependent dynamics of nuclear pores: Pore-free islands and lamins. J. Cell Sci. 2006, 119, 4442–4451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hieronymus, H.; Yu, M.C.; Silver, P.A. Genome-wide mRNA surveillance is coupled to mRNA export. Genes Dev. 2004, 18, 2652–2662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Son, H.G.; Lee, S.-J.V. Longevity regulation by NMD-mediated mRNA quality control. BMB Rep. 2017, 50, 160–161. [Google Scholar] [CrossRef] [Green Version]
- Mullani, N.; Porozhan, Y.; Mangelinck, A.; Rachez, C.; Costallat, M.; Batsché, E.; Goodhardt, M.; Cenci, G.; Mann, C.; Muchardt, C. Reduced RNA turnover as a driver of cellular senescence. Life Sci. Alliance 2021, 4, e202000809. [Google Scholar] [CrossRef]
- Camblong, J.; Iglesias, N.; Fickentscher, C.; Dieppois, G.; Stutz, F. Antisense RNA Stabilization Induces Transcriptional Gene Silencing via Histone Deacetylation in S. cerevisiae. Cell 2007, 131, 706–717. [Google Scholar] [CrossRef]
- Vinciguerra, P.; Iglesias, N.; Camblong, J.; Zenklusen, D.; Stutz, F. Perinuclear Mlp proteins downregulate gene expression in response to a defect in mRNA export. EMBO J. 2005, 24, 813–823. [Google Scholar] [CrossRef]
- Niño, C.A.; Hérissant, L.; Babour, A.; Dargemont, C. mRNA Nuclear Export in Yeast. Chem. Rev. 2013, 113, 8523–8545. [Google Scholar] [CrossRef]
- Cookson, M.R. Aging-RNA in development and disease. Wiley Interdiscip. Rev. RNA 2012, 3, 133–143. [Google Scholar] [CrossRef] [Green Version]
- Sakuma, S.; D’Angelo, M.A. The roles of the nuclear pore complex in cellular dysfunction, aging and disease. Semin. Cell Dev. Biol. 2017, 68, 72–84. [Google Scholar] [CrossRef]
- Woerner, A.C.; Frottin, F.; Hornburg, D.; Feng, L.R.; Meissner, F.; Patra, M.; Tatzelt, J.; Mann, M.; Winklhofer, K.F.; Hartl, F.U.; et al. Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science 2016, 351, 173–176. [Google Scholar] [CrossRef] [PubMed]
- Freibaum, B.D.; Lu, Y.; Lopez-Gonzalez, R.; Kim, N.C.; Almeida, S.; Lee, K.-H.; Badders, N.; Valentine, M.; Miller, B.L.; Wong, P.C.; et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 2015, 525, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Maeder, C.I.; Kim, J.-I.; Liang, X.; Kaganovsky, K.; Shen, A.; Li, Q.; Li, Z.; Wang, S.; Xu, X.S.; Li, J.B.; et al. The THO Complex Coordinates Transcripts for Synapse Development and Dopamine Neuron Survival. Cell 2018, 174, 1436–1449.e20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boehringer, A.; Garcia-Mansfield, K.; Singh, G.; Bakkar, N.; Pirrotte, P.; Bowser, R. ALS Associated Mutations in Matrin 3 Alter Protein-Protein Interactions and Impede mRNA Nuclear Export. Sci. Rep. 2017, 7, 14529. [Google Scholar] [CrossRef]
- Tollervey, J.R.; Curk, T.; Rogelj, B.; Briese, M.; Cereda, M.; Kayikci, M.; König, J.; Hortobágyi, T.; Nishimura, A.L.; Župunski, V.; et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 2011, 14, 452–458. [Google Scholar] [CrossRef] [Green Version]
- Chou, C.-C.; Zhang, Y.; Umoh, M.E.; Vaughan, S.W.; Lorenzini, I.; Liu, F.; Sayegh, M.; Donlin-Asp, P.; Chen, Y.H.; Duong, D.; et al. TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat. Neurosci. 2018, 21, 228–239. [Google Scholar] [CrossRef] [Green Version]
- Kapeli, K.; Pratt, G.A.; Vu, A.Q.; Hutt, K.R.; Martinez, F.J.; Sundararaman, B.; Batra, R.; Freese, P.D.; Lambert, N.J.; Huelga, S.C.; et al. Distinct and shared functions of ALS-associated proteins TDP-43, FUS and TAF15 revealed by multisystem analyses. Nat. Commun. 2016, 7, 12143. [Google Scholar] [CrossRef] [Green Version]
- Fujii, R.; Takumi, T. TLS facilitates transport of mRNA encoding an actin-stabilizing protein to dendritic spines. J. Cell Sci. 2005, 118, 5755–5765. [Google Scholar] [CrossRef] [Green Version]
- Fujii, R.; Okabe, S.; Urushido, T.; Inoue, K.; Yoshimura, A.; Tachibana, T.; Nishikawa, T.; Hicks, G.; Takumi, T. The RNA Binding Protein TLS Is Translocated to Dendritic Spines by mGluR5 Activation and Regulates Spine Morphology. Curr. Biol. 2005, 15, 587–593. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Donnelly, C.J.; Haeusler, A.R.; Grima, J.C.; Machamer, J.B.; Steinwald, P.; Daley, E.; Miller, S.J.; Cunningham, K.; Vidensky, S.; et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 2015, 525, 56–61. [Google Scholar] [CrossRef] [Green Version]
- Kinoshita, Y.; Ito, H.; Hirano, A.; Fujita, K.; Wate, R.; Nakamura, M.; Kaneko, S.; Nakano, S.; Kusaka, H. Nuclear Contour Irregularity and Abnormal Transporter Protein Distribution in Anterior Horn Cells in Amyotrophic Lateral Sclerosis. J. Neuropathol. Exp. Neurol. 2009, 68, 1184–1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaneb, H.M.; Folkmann, A.W.; Belzil, V.V.; Jao, L.-E.; Leblond, C.S.; Girard, S.L.; Daoud, H.; Noreau, A.; Rochefort, D.; Hince, P.; et al. Deleterious mutations in the essential mRNA metabolism factor, hGle1, in amyotrophic lateral sclerosis. Hum. Mol. Genet. 2015, 24, 1363–1373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, K.-I.; Yoon, D.; Qiu, S.; Danziger, Z.; Grill, W.M.; Wetsel, W.C.; Ferreira, P.A. Loss of Ranbp2 in motor neurons causes the disruption of nucleocytoplasmic and chemokine signaling and proteostasis of hnRNPH3 and Mmp28, and the development of amyotrophic lateral sclerosis (ALS)-like syndromes. Dis. Model. Mech. 2017, 10, 559–579. [Google Scholar] [CrossRef] [Green Version]
- Gasset-Rosa, F.; Chillon-Marinas, C.; Goginashvili, A.; Atwal, R.S.; Artates, J.W.; Tabet, R.; Wheeler, V.C.; Bang, A.G.; Cleveland, D.W.; Lagier-Tourenne, C. Polyglutamine-Expanded Huntingtin Exacerbates Age-Related Disruption of Nuclear Integrity and Nucleocytoplasmic Transport. Neuron 2017, 94, 48–57.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grima, J.C.; Daigle, J.G.; Arbez, N.; Cunningham, K.C.; Zhang, K.; Ochaba, J.; Geater, C.; Morozko, E.; Stocksdale, J.; Glatzer, J.C.; et al. Mutant Huntingtin Disrupts the Nuclear Pore Complex. Neuron 2017, 94, 93.e6–107.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheffield, L.G.; Miskiewicz, H.B.; Tannenbaum, L.B.; Mirra, S.S. Nuclear Pore Complex Proteins in Alzheimer Disease. J. Neuropathol. Exp. Neurol. 2006, 65, 45–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Um, J.W.; Min, D.S.; Rhim, H.; Kim, J.; Paik, S.R.; Chung, K.C. Parkin Ubiquitinates and Promotes the Degradation of RanBP2. J. Biol. Chem. 2006, 281, 3595–3603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mata, I.F.; Lockhart, P.J.; Farrer, M.J. Parkin genetics: One model for Parkinson’s disease. Hum. Mol. Genet. 2004, 13, R127–R133. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Hassinger, L.; Thomson, T.; Ding, B.; Ashley, J.; Hassinger, W.; Budnik, V. Lamin Mutations Accelerate Aging via Defective Export of Mitochondrial mRNAs through Nuclear Envelope Budding. Curr. Biol. 2016, 26, 2052–2059. [Google Scholar] [CrossRef] [Green Version]
- Thakur, M.; Oka, T.; Natori, Y. Gene expression and aging. Mech. Ageing Dev. 1993, 66, 283–298. [Google Scholar] [CrossRef]
- Lennartsson, A.; Ekwall, K. Histone modification patterns and epigenetic codes. Biochim. Biophys. Acta 2009, 1790, 863–868. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.-Y.; Hochstrasser, M. Histone sumoylation and chromatin dynamics. Nucleic Acids Res. 2021, 49, 6043–6052. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.; Su, D.; Wilson-Eisele, N.R.; Zhao, D.; López-Giráldez, F.; Hochstrasser, M. The Ulp2 SUMO protease promotes transcription elongation through regulation of histone sumoylation. EMBO J. 2019, 38, e102003. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.-Y.; Zhao, D.; Li, J.; Su, D.; Hochstrasser, M. Histone sumoylation promotes Set3 histone-deacetylase complex-mediated transcriptional regulation. Nucleic Acids Res. 2020, 48, 12151–12168. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Ryoo, Z.Y.; Cho, D.-H.; Lee, H.-S.; Ryu, H.-Y. Trans-tail regulation-mediated suppression of cryptic transcription. Exp. Mol. Med. 2021, 53, 1683–1688. [Google Scholar] [CrossRef] [PubMed]
- Oliete-Calvo, P.; Serrano-Quílez, J.; Nuño-Cabanes, C.; Pérez-Martínez, M.E.; Soares, L.M.; Dichtl, B.; Buratowski, S.; Pérez-Ortín, J.E.; Rodríguez-Navarro, S. A role for Mog1 in H2Bub1 and H3K4me3 regulation affecting RNAPII transcription and mRNA export. EMBO Rep. 2018, 19, e45992. [Google Scholar] [CrossRef]
- Yoh, S.M.; Lucas, J.S.; Jones, K.A. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev. 2008, 22, 3422–3434. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Yuan, Q.; Xie, L. Histone Modifications in Aging: The Underlying Mechanisms and Implications. Curr. Stem Cell Res. Ther. 2018, 13, 125–135. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, H.-S.; Lee, J.; Lee, H.-S.; Ahn, S.H.; Ryu, H.-Y. Nuclear mRNA Export and Aging. Int. J. Mol. Sci. 2022, 23, 5451. https://doi.org/10.3390/ijms23105451
Park H-S, Lee J, Lee H-S, Ahn SH, Ryu H-Y. Nuclear mRNA Export and Aging. International Journal of Molecular Sciences. 2022; 23(10):5451. https://doi.org/10.3390/ijms23105451
Chicago/Turabian StylePark, Hyun-Sun, Jongbok Lee, Hyun-Shik Lee, Seong Hoon Ahn, and Hong-Yeoul Ryu. 2022. "Nuclear mRNA Export and Aging" International Journal of Molecular Sciences 23, no. 10: 5451. https://doi.org/10.3390/ijms23105451
APA StylePark, H.-S., Lee, J., Lee, H.-S., Ahn, S. H., & Ryu, H.-Y. (2022). Nuclear mRNA Export and Aging. International Journal of Molecular Sciences, 23(10), 5451. https://doi.org/10.3390/ijms23105451







