Transcriptional Regulation of Endogenous Retroviruses and Their Misregulation in Human Diseases
Abstract
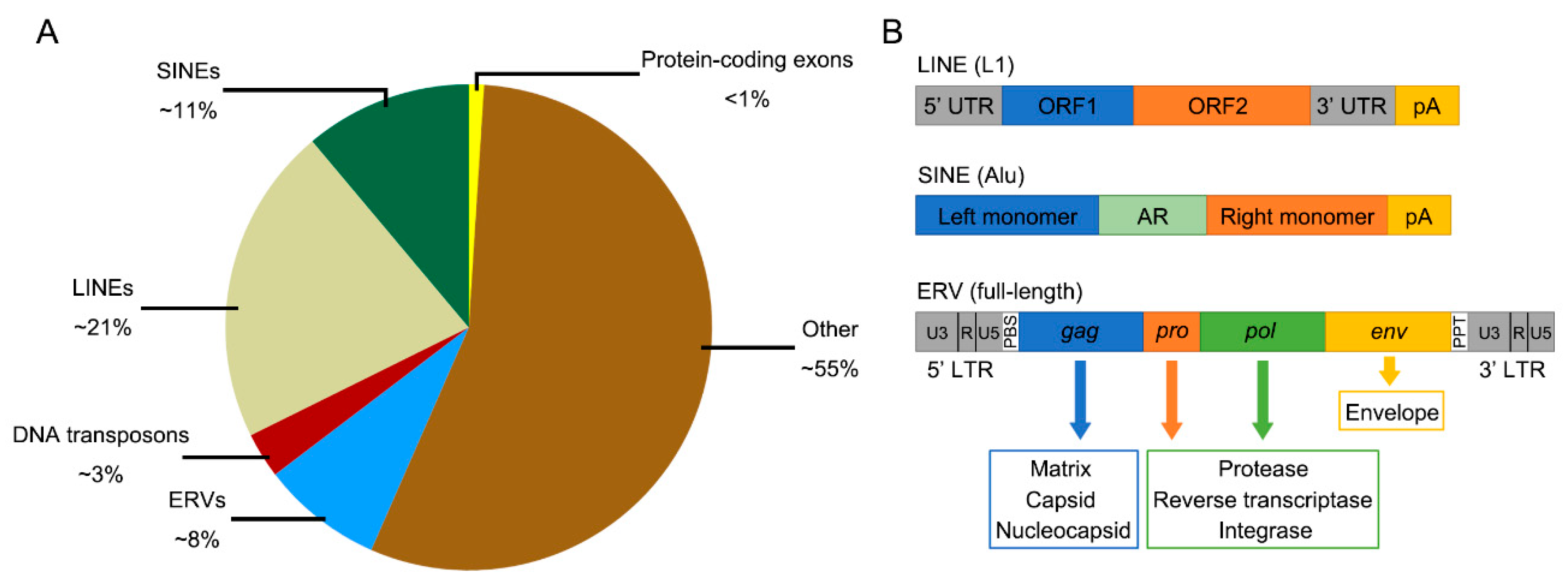
:1. Introduction
2. Silencing and Transcriptional Regulation of ERVs
2.1. KRAB-ZFPs/TRIM28 Pathway Is a Master Regulator for ERVs Silencing
2.2. Chromatin Remodeler and Histone Chaperone Maintain ERVs Silencing through KRAB-ZFPs/TRIM28 Pathway
2.3. Sumoylation of TRIM28 Contributes to ERVs Silencing
2.4. DNA Methylation in ERVs Silencing
2.5. RNA-Mediated Regulation of ERVs
2.6. Exogenous Viruses Are Associated with ERVs Activation
2.7. Additional Factors in ERVs Transcriptional Regulation
3. Transcriptional Dysregulation of ERVs in Human Diseases
3.1. HERVs in Cancer
3.2. HERVs in Aging and Neurodegenerative Diseases
4. Conclusions and Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Senft, A.D.; Macfarlan, T.S. Transposable elements shape the evolution of mammalian development. Nat. Rev. Genet. 2021, 22, 691–711. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Kazazian, H.H.; Moran, J.V. Mobile DNA in Health and Disease. N. Engl. J. Med. 2017, 377, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Kury, P.; Nath, A.; Creange, A.; Dolei, A.; Marche, P.; Gold, J.; Giovannoni, G.; Hartung, H.P.; Perron, H. Human Endogenous Retroviruses in Neurological Diseases. Trends Mol. Med. 2018, 24, 379–394. [Google Scholar] [CrossRef]
- Gorbunova, V.; Seluanov, A.; Mita, P.; McKerrow, W.; Fenyö, D.; Boeke, J.D.; Linker, S.B.; Gage, F.H.; Kreiling, J.A.; Petrashen, A.P.; et al. The role of retrotransposable elements in ageing and age-associated diseases. Nature 2021, 596, 43–53. [Google Scholar] [CrossRef]
- Geis, F.K.; Goff, S.P. Silencing and Transcriptional Regulation of Endogenous Retroviruses: An Overview. Viruses 2020, 12, 884. [Google Scholar] [CrossRef]
- Mao, J.; Zhang, Q.; Cong, Y.-S. Human endogenous retroviruses in development and disease. Comput. Struct. Biotechnol. J. 2021, 19, 5978–5986. [Google Scholar] [CrossRef]
- De Parseval, N.; Heidmann, T. Human endogenous retroviruses: From infectious elements to human genes. Cytogenet. Genome Res. 2005, 110, 318–332. [Google Scholar] [CrossRef]
- Vargiu, L.; Rodriguez-Tomé, P.; Sperber, G.O.; Cadeddu, M.; Grandi, N.; Blikstad, V.; Tramontano, E.; Blomberg, J. Classification and characterization of human endogenous retroviruses; mosaic forms are common. Retrovirology 2016, 13, 7. [Google Scholar] [CrossRef]
- Johnson, W.E. Endogenous Retroviruses in the Genomics Era. Annu. Rev. Virol. 2015, 2, 135–159. [Google Scholar] [CrossRef]
- Mager, D.L.; Stoye, J.P. Mammalian Endogenous Retroviruses. Microbiol. Spectr. 2015, 3, MDNA3-0009-2014. [Google Scholar] [CrossRef]
- Gifford, R.J.; Blomberg, J.; Coffin, J.M.; Fan, H.; Heidmann, T.; Mayer, J.; Stoye, J.; Tristem, M.; Johnson, W.E. Nomenclature for endogenous retrovirus (ERV) loci. Retrovirology 2018, 15, 59. [Google Scholar] [CrossRef]
- Teich, N.M.; Weiss, R.A.; Martin, G.R.; Lowy, D.R. Virus infection of murine teratocarcinoma stem cell lines. Cell 1977, 12, 973–982. [Google Scholar] [CrossRef]
- Niwa, O.; Yokota, Y.; Ishida, H.; Sugahara, T. Independent mechanisms involved in suppression of the Moloney leukemia virus genome during differentiation of murine teratocarcinoma cells. Cell 1983, 32, 1105–1113. [Google Scholar] [CrossRef]
- Feuer, G.; Taketo, M.; Hanecak, R.C.; Fan, H. Two blocks in Moloney murine leukemia virus expression in undifferentiated F9 embryonal carcinoma cells as determined by transient expression assays. J. Virol. 1989, 63, 2317–2324. [Google Scholar] [CrossRef]
- Wolf, D.; Goff, S.P. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 2009, 458, 1201–1204. [Google Scholar] [CrossRef]
- Wolf, G.; Yang, P.; Füchtbauer, A.C.; Füchtbauer, E.-M.; Silva, A.M.; Park, C.; Wu, W.; Nielsen, A.L.; Pedersen, F.S.; Macfarlan, T.S. The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Genes Dev. 2015, 29, 538–554. [Google Scholar] [CrossRef]
- Wang, C.; Goff, S.P. Differential control of retrovirus silencing in embryonic cells by proteasomal regulation of the ZFP809 retroviral repressor. Proc. Natl. Acad. Sci. USA 2017, 114, E922–E930. [Google Scholar] [CrossRef]
- Seah, M.K.Y.; Wang, Y.; Goy, P.-A.; Loh, H.M.; Peh, W.J.; Low, D.H.P.; Han, B.Y.; Wong, E.; Leong, E.L.; Wolf, G.; et al. The KRAB-zinc-finger protein ZFP708 mediates epigenetic repression at RMER19B retrotransposons. Development 2019, 146, dev170266. [Google Scholar] [CrossRef]
- Jacobs, F.M.J.; Greenberg, D.; Nguyen, N.; Haeussler, M.; Ewing, A.D.; Katzman, S.; Paten, B.; Salama, S.R.; Haussler, D. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 2014, 516, 242–245. [Google Scholar] [CrossRef] [Green Version]
- Tan, X.; Xu, X.; Elkenani, M.; Smorag, L.; Zechner, U.; Nolte, J.; Engel, W.; Pantakani, D.V.K. Zfp819, a novel KRAB-zinc finger protein, interacts with KAP1 and functions in genomic integrity maintenance of mouse embryonic stem cells. Stem Cell Res. 2013, 11, 1045–1059. [Google Scholar] [CrossRef] [PubMed]
- Ecco, G.; Cassano, M.; Kauzlaric, A.; Duc, J.; Coluccio, A.; Offner, S.; Imbeault, M.; Rowe, H.M.; Turelli, P.; Trono, D. Transposable Elements and Their KRAB-ZFP Controllers Regulate Gene Expression in Adult Tissues. Dev. Cell 2016, 36, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Schlesinger, S.; Lee, A.H.; Wang, G.Z.; Green, L.; Goff, S.P. Proviral silencing in embryonic cells is regulated by Yin Yang 1. Cell Rep. 2013, 4, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Rowe, H.M.; Jakobsson, J.; Mesnard, D.; Rougemont, J.; Reynard, S.; Aktas, T.; Maillard, P.V.; Layard-Liesching, H.; Verp, S.; Marquis, J.; et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 2010, 463, 237–240. [Google Scholar] [CrossRef]
- Yang, P.; Wang, Y.; Macfarlan, T.S. The Role of KRAB-ZFPs in Transposable Element Repression and Mammalian Evolution. Trends Genet. 2017, 33, 871–881. [Google Scholar] [CrossRef]
- Cammas, F.; Mark, M.; Dollé, P.; Dierich, A.; Chambon, P.; Losson, R. Mice lacking the transcriptional corepressor TIF1beta are defective in early postimplantation development. Development 2000, 127, 2955–2963. [Google Scholar] [CrossRef]
- Grassi, D.A.; Jönsson, M.E.; Brattås, P.L.; Jakobsson, J. TRIM28 and the control of transposable elements in the brain. Brain Res. 2019, 1705, 43–47. [Google Scholar] [CrossRef]
- Fukuda, K.; Shinkai, Y. SETDB1-Mediated Silencing of Retroelements. Viruses 2020, 12, 596. [Google Scholar] [CrossRef]
- Matsui, T.; Leung, D.; Miyashita, H.; Maksakova, I.A.; Miyachi, H.; Kimura, H.; Tachibana, M.; Lorincz, M.C.; Shinkai, Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 2010, 464, 927–931. [Google Scholar] [CrossRef]
- Tachibana, K.; Gotoh, E.; Kawamata, N.; Ishimoto, K.; Uchihara, Y.; Iwanari, H.; Sugiyama, A.; Kawamura, T.; Mochizuki, Y.; Tanaka, T.; et al. Analysis of the subcellular localization of the human histone methyltransferase SETDB1. Biochem. Biophys. Res. Commun. 2015, 465, 725–731. [Google Scholar] [CrossRef]
- Tsusaka, T.; Shimura, C.; Shinkai, Y. ATF7IP regulates SETDB1 nuclear localization and increases its ubiquitination. EMBO Rep. 2019, 20, e48297. [Google Scholar] [CrossRef] [PubMed]
- Bulut-Karslioglu, A.; De La Rosa-Velázquez, I.A.; Ramirez, F.; Barenboim, M.; Onishi-Seebacher, M.; Arand, J.; Galán, C.; Winter, G.E.; Engist, B.; Gerle, B.; et al. Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol. Cell 2014, 55, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Leung, D.C.; Dong, K.B.; Maksakova, I.A.; Goyal, P.; Appanah, R.; Lee, S.; Tachibana, M.; Shinkai, Y.; Lehnertz, B.; Mager, D.L.; et al. Lysine methyltransferase G9a is required for de novo DNA methylation and the establishment, but not the maintenance, of proviral silencing. Proc. Natl. Acad. Sci. USA 2011, 108, 5718–5723. [Google Scholar] [CrossRef] [PubMed]
- Maksakova, I.A.; Thompson, P.J.; Goyal, P.; Jones, S.J.; Singh, P.B.; Karimi, M.M.; Lorincz, M.C. Distinct roles of KAP1, HP1 and G9a/GLP in silencing of the two-cell-specific retrotransposon MERVL in mouse ES cells. Epigenetics Chromatin 2013, 6, 15. [Google Scholar] [CrossRef]
- Gao, T.; Chen, F.; Zhang, W.; Zhao, X.; Hu, X.; Lu, X. Nsd2 Represses Endogenous Retrovirus MERVL in Embryonic Stem Cells. Stem Cells Int. 2021, 2021, 6663960. [Google Scholar] [CrossRef] [PubMed]
- Robbez-Masson, L.; Tie, C.H.C.; Conde, L.; Tunbak, H.; Husovsky, C.; Tchasovnikarova, I.A.; Timms, R.T.; Herrero, J.; Lehner, P.J.; Rowe, H.M. The HUSH complex cooperates with TRIM28 to repress young retrotransposons and new genes. Genome Res. 2018, 28, 836–845. [Google Scholar] [CrossRef]
- Tchasovnikarova, I.A.; Timms, R.T.; Matheson, N.J.; Wals, K.; Antrobus, R.; Göttgens, B.; Dougan, G.; Dawson, M.A.; Lehner, P.J. GENE SILENCING. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science 2015, 348, 1481–1485. [Google Scholar] [CrossRef]
- Garland, W.; Müller, I.; Wu, M.; Schmid, M.; Imamura, K.; Rib, L.; Sandelin, A.; Helin, K.; Jensen, T.H. Chromatin modifier HUSH co-operates with RNA decay factor NEXT to restrict transposable element expression. Mol. Cell 2022, 82, 1691–1707.e8. [Google Scholar] [CrossRef]
- Yang, B.X.; El Farran, C.A.; Guo, H.C.; Yu, T.; Fang, H.T.; Wang, H.F.; Schlesinger, S.; Seah, Y.F.S.; Goh, G.Y.L.; Neo, S.P.; et al. Systematic identification of factors for provirus silencing in embryonic stem cells. Cell 2015, 163, 230–245. [Google Scholar] [CrossRef]
- Maksakova, I.A.; Goyal, P.; Bullwinkel, J.; Brown, J.P.; Bilenky, M.; Mager, D.L.; Singh, P.B.; Lorincz, M.C. H3K9me3-binding proteins are dispensable for SETDB1/H3K9me3-dependent retroviral silencing. Epigenetics Chromatin 2011, 4, 12. [Google Scholar] [CrossRef] [Green Version]
- Sachs, P.; Ding, D.; Bergmaier, P.; Lamp, B.; Schlagheck, C.; Finkernagel, F.; Nist, A.; Stiewe, T.; Mermoud, J.E. SMARCAD1 ATPase activity is required to silence endogenous retroviruses in embryonic stem cells. Nat. Commun. 2019, 10, 1335. [Google Scholar] [CrossRef] [PubMed]
- Mittal, P.; Roberts, C.W.M. The SWI/SNF complex in cancer—Biology, biomarkers and therapy. Nat. Rev. Clin. Oncol. 2020, 17, 435–448. [Google Scholar] [CrossRef] [PubMed]
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 2017, 18, 407–422. [Google Scholar] [CrossRef]
- Hammond, C.M.; Strømme, C.B.; Huang, H.; Patel, D.J.; Groth, A. Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell Biol. 2017, 18, 141–158. [Google Scholar] [CrossRef]
- Kaufman, P.D.; Kobayashi, R.; Kessler, N.; Stillman, B. The p150 and p60 subunits of chromatin assembly factor I: A molecular link between newly synthesized histones and DNA replication. Cell 1995, 81, 1105–1114. [Google Scholar] [CrossRef]
- Gaillard, P.-H.L.; Martini, E.M.; Kaufman, P.D.; Stillman, B.; Moustacchi, E.; Almouzni, G. Chromatin assembly coupled to DNA repair: A new role for chromatin assembly factor I. Cell 1996, 86, 887–896. [Google Scholar] [CrossRef]
- Lechner, M.S.; Begg, G.E.; Speicher, D.W.; Rauscher, F.J. Molecular determinants for targeting heterochromatin protein 1-mediated gene silencing: Direct chromoshadow domain-KAP-1 corepressor interaction is essential. Mol. Cell. Biol. 2000, 20, 6449–6465. [Google Scholar] [CrossRef]
- Thiru, A.; Nietlispach, D.; Mott, H.R.; Okuwaki, M.; Lyon, D.; Nielsen, P.R.; Hirshberg, M.; Verreault, A.; Murzina, N.V.; Laue, E.D. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. EMBO J. 2004, 23, 489–499. [Google Scholar] [CrossRef]
- Simon, B.; Lou, H.J.; Huet-Calderwood, C.; Shi, G.; Boggon, T.J.; Turk, B.E.; Calderwood, D.A. Tousled-like kinase 2 targets ASF1 histone chaperones through client mimicry. Nat. Commun. 2022, 13, 749. [Google Scholar] [CrossRef]
- Dyer, M.A.; Qadeer, Z.A.; Valle-Garcia, D.; Bernstein, E. ATRX and DAXX: Mechanisms and Mutations. Cold Spring Harb. Perspect. Med. 2017, 7, a026567. [Google Scholar] [CrossRef] [Green Version]
- Sadic, D.; Schmidt, K.; Groh, S.; Kondofersky, I.; Ellwart, J.; Fuchs, C.; Theis, F.J.; Schotta, G. Atrx promotes heterochromatin formation at retrotransposons. EMBO Rep. 2015, 16, 836–850. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Kim, H.; Huang, R.; Lu, W.; Tang, M.; Shi, F.; Yang, D.; Zhang, X.; Huang, J.; Liu, D.; et al. The Daxx/Atrx Complex Protects Tandem Repetitive Elements during DNA Hypomethylation by Promoting H3K9 Trimethylation. Cell Stem Cell 2015, 17, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Elsässer, S.J.; Noh, K.-M.; Diaz, N.; Allis, C.D.; Banaszynski, L.A. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature 2015, 522, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Trovato, M.; Patil, V.; Gehre, M.; Noh, K.M. Histone Variant H3.3 Mutations in Defining the Chromatin Function in Mammals. Cells 2020, 9, 2716. [Google Scholar] [CrossRef]
- Flotho, A.; Melchior, F. Sumoylation: A regulatory protein modification in health and disease. Annu. Rev. Biochem. 2013, 82, 357–385. [Google Scholar] [CrossRef]
- Ninova, M.; Fejes Tóth, K.; Aravin, A.A. The control of gene expression and cell identity by H3K9 trimethylation. Development 2019, 146, dev181180. [Google Scholar] [CrossRef]
- Rowe, H.M.; Friedli, M.; Offner, S.; Verp, S.; Mesnard, D.; Marquis, J.; Aktas, T.; Trono, D. De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development 2013, 140, 519–529. [Google Scholar] [CrossRef]
- Hutnick, L.K.; Huang, X.; Loo, T.-C.; Ma, Z.; Fan, G. Repression of retrotransposal elements in mouse embryonic stem cells is primarily mediated by a DNA methylation-independent mechanism. J. Biol. Chem. 2010, 285, 21082–21091. [Google Scholar] [CrossRef]
- Walsh, C.P.; Chaillet, J.R.; Bestor, T.H. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 1998, 20, 116–117. [Google Scholar] [CrossRef]
- Karimi, M.M.; Goyal, P.; Maksakova, I.A.; Bilenky, M.; Leung, D.; Tang, J.X.; Shinkai, Y.; Mager, D.L.; Jones, S.; Hirst, M.; et al. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 2011, 8, 676–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groh, S.; Schotta, G. Silencing of endogenous retroviruses by heterochromatin. Cell. Mol. Life Sci. 2017, 74, 2055–2065. [Google Scholar] [CrossRef] [PubMed]
- Fasching, L.; Kapopoulou, A.; Sachdeva, R.; Petri, R.; Jönsson, M.E.; Männe, C.; Turelli, P.; Jern, P.; Cammas, F.; Trono, D.; et al. TRIM28 represses transcription of endogenous retroviruses in neural progenitor cells. Cell Rep. 2015, 10, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.-L.; Nishi, M.; Ohtsuka, T.; Matsui, T.; Takemoto, K.; Kamio-Miura, A.; Aburatani, H.; Shinkai, Y.; Kageyama, R. Essential roles of the histone methyltransferase ESET in the epigenetic control of neural progenitor cells during development. Development 2012, 139, 3806–3816. [Google Scholar] [CrossRef]
- Collins, P.L.; Kyle, K.E.; Egawa, T.; Shinkai, Y.; Oltz, E.M. The histone methyltransferase SETDB1 represses endogenous and exogenous retroviruses in B lymphocytes. Proc. Natl. Acad. Sci. USA 2015, 112, 8367–8372. [Google Scholar] [CrossRef] [PubMed]
- Pasquarella, A.; Ebert, A.; Pereira de Almeida, G.; Hinterberger, M.; Kazerani, M.; Nuber, A.; Ellwart, J.; Klein, L.; Busslinger, M.; Schotta, G. Retrotransposon derepression leads to activation of the unfolded protein response and apoptosis in pro-B cells. Development 2016, 143, 1788–1799. [Google Scholar] [CrossRef]
- Veland, N.; Lu, Y.; Hardikar, S.; Gaddis, S.; Zeng, Y.; Liu, B.; Estecio, M.R.; Takata, Y.; Lin, K.; Tomida, M.W.; et al. DNMT3L facilitates DNA methylation partly by maintaining DNMT3A stability in mouse embryonic stem cells. Nucleic Acids Res. 2019, 47, 152–167. [Google Scholar] [CrossRef]
- Bourc’his, D.; Bestor, T.H. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 2004, 431, 96–99. [Google Scholar] [CrossRef]
- Ohtani, H.; Liu, M.; Zhou, W.; Liang, G.; Jones, P.A. Switching roles for DNA and histone methylation depend on evolutionary ages of human endogenous retroviruses. Genome Res. 2018, 28, 1147–1157. [Google Scholar] [CrossRef]
- Stein, P.; Rozhkov, N.V.; Li, F.; Cárdenas, F.L.; Davydenko, O.; Davydenk, O.; Vandivier, L.E.; Gregory, B.D.; Hannon, G.J.; Schultz, R.M. Essential Role for endogenous siRNAs during meiosis in mouse oocytes. PLoS Genet. 2015, 11, e1005013. [Google Scholar] [CrossRef]
- Li, J.; Kannan, M.; Trivett, A.L.; Liao, H.; Wu, X.; Akagi, K.; Symer, D.E. An antisense promoter in mouse L1 retrotransposon open reading frame-1 initiates expression of diverse fusion transcripts and limits retrotransposition. Nucleic Acids Res. 2014, 42, 4546–4562. [Google Scholar] [CrossRef]
- Brockdorff, N.; Bowness, J.S.; Wei, G. Progress toward understanding chromosome silencing by Xist RNA. Genes Dev. 2020, 34, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Carter, A.C.; Xu, J.; Nakamoto, M.Y.; Wei, Y.; Zarnegar, B.J.; Shi, Q.; Broughton, J.P.; Ransom, R.C.; Salhotra, A.; Nagaraja, S.D.; et al. Spen links RNA-mediated endogenous retrovirus silencing and X chromosome inactivation. eLife 2020, 9, e54508. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, Y.W.; Siomi, M.C.; Siomi, H. PIWI-Interacting RNA: Its Biogenesis and Functions. Annu. Rev. Biochem. 2015, 84, 405–433. [Google Scholar] [CrossRef] [PubMed]
- Pal-Bhadra, M.; Leibovitch, B.A.; Gandhi, S.G.; Chikka, M.R.; Rao, M.; Bhadra, U.; Birchler, J.A.; Elgin, S.C.R. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 2004, 303, 669–672. [Google Scholar] [CrossRef]
- Aravin, A.A.; Sachidanandam, R.; Girard, A.; Fejes-Toth, K.; Hannon, G.J. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 2007, 316, 744–747. [Google Scholar] [CrossRef]
- Aravin, A.A.; Sachidanandam, R.; Bourc’his, D.; Schaefer, C.; Pezic, D.; Toth, K.F.; Bestor, T.; Hannon, G.J. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 2008, 31, 785–799. [Google Scholar] [CrossRef]
- Guallar, D.; Bi, X.; Pardavila, J.A.; Huang, X.; Saenz, C.; Shi, X.; Zhou, H.; Faiola, F.; Ding, J.; Haruehanroengra, P.; et al. RNA-dependent chromatin targeting of TET2 for endogenous retrovirus control in pluripotent stem cells. Nat. Genet. 2018, 50, 443–451. [Google Scholar] [CrossRef]
- Shi, H.; Wei, J.; He, C. Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cell 2019, 74, 640–650. [Google Scholar] [CrossRef]
- Chelmicki, T.; Roger, E.; Teissandier, A.; Dura, M.; Bonneville, L.; Rucli, S.; Dossin, F.; Fouassier, C.; Lameiras, S.; Bourc’his, D. m6A RNA methylation regulates the fate of endogenous retroviruses. Nature 2021, 591, 312–316. [Google Scholar] [CrossRef]
- Chen, J.; Foroozesh, M.; Qin, Z. Transactivation of human endogenous retroviruses by tumor viruses and their functions in virus-associated malignancies. Oncogenesis 2019, 8, 6. [Google Scholar] [CrossRef] [Green Version]
- Tovo, P.-A.; Garazzino, S.; Daprà, V.; Alliaudi, C.; Silvestro, E.; Calvi, C.; Montanari, P.; Galliano, I.; Bergallo, M. Chronic HCV Infection Is Associated with Overexpression of Human Endogenous Retroviruses that Persists after Drug-Induced Viral Clearance. Int. J. Mol. Sci. 2020, 21, 3980. [Google Scholar] [CrossRef] [PubMed]
- Vincendeau, M.; Göttesdorfer, I.; Schreml, J.M.H.; Wetie, A.G.N.; Mayer, J.; Greenwood, A.D.; Helfer, M.; Kramer, S.; Seifarth, W.; Hadian, K.; et al. Modulation of human endogenous retrovirus (HERV) transcription during persistent and de novo HIV-1 infection. Retrovirology 2015, 12, 27. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, D.; Tirado-Magallanes, R.; Bhatia, S.S.; Teo, W.S.; Sian, S.; Hora, S.; Lee, K.K.; Zhang, Y.; Jadhav, S.P.; Wu, Y.; et al. TIP60 represses activation of endogenous retroviral elements. Nucleic Acids Res. 2018, 46, 9456–9470. [Google Scholar] [CrossRef]
- Macfarlan, T.S.; Gifford, W.D.; Agarwal, S.; Driscoll, S.; Lettieri, K.; Wang, J.; Andrews, S.E.; Franco, L.; Rosenfeld, M.G.; Ren, B.; et al. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev. 2011, 25, 594–607. [Google Scholar] [CrossRef] [PubMed]
- Winkler, D.D.; Luger, K. The histone chaperone FACT: Structural insights and mechanisms for nucleosome reorganization. J. Biol. Chem. 2011, 286, 18369–18374. [Google Scholar] [CrossRef]
- Chen, F.; Zhang, W.; Xie, D.; Gao, T.; Dong, Z.; Lu, X. Histone chaperone FACT represses retrotransposon MERVL and MERVL-derived cryptic promoters. Nucleic Acids Res. 2020, 48, 10211–10225. [Google Scholar] [CrossRef]
- Falco, G.; Lee, S.-L.; Stanghellini, I.; Bassey, U.C.; Hamatani, T.; Ko, M.S.H. Zscan4: A novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev. Biol. 2007, 307, 539–550. [Google Scholar] [CrossRef]
- Zhang, W.; Chen, F.; Chen, R.; Xie, D.; Yang, J.; Zhao, X.; Guo, R.; Zhang, Y.; Shen, Y.; Göke, J.; et al. Zscan4c activates endogenous retrovirus MERVL and cleavage embryo genes. Nucleic Acids Res. 2019, 47, 8485–8501. [Google Scholar] [CrossRef]
- Hendrickson, P.G.; Doráis, J.A.; Grow, E.J.; Whiddon, J.L.; Lim, J.-W.; Wike, C.L.; Weaver, B.D.; Pflueger, C.; Emery, B.R.; Wilcox, A.L.; et al. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet. 2017, 49, 925–934. [Google Scholar] [CrossRef]
- Nguyen, T.D.; Davis, J.; Eugenio, R.A.; Liu, Y. Female Sex Hormones Activate Human Endogenous Retrovirus Type K Through the OCT4 Transcription Factor in T47D Breast Cancer Cells. AIDS Res. Hum. Retrovir. 2019, 35, 348–356. [Google Scholar] [CrossRef]
- Mao, J.; Zhang, Q.; Wang, Y.; Zhuang, Y.; Xu, L.; Ma, X.; Guan, D.; Zhou, J.; Liu, J.; Wu, X.; et al. TERT activates endogenous retroviruses to promote an immunosuppressive tumour microenvironment. EMBO Rep. 2022, 23, e52984. [Google Scholar] [CrossRef]
- Mi, S.; Lee, X.; Li, X.; Veldman, G.M.; Finnerty, H.; Racie, L.; LaVallie, E.; Tang, X.Y.; Edouard, P.; Howes, S.; et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 2000, 403, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Blaise, S.; de Parseval, N.; Bénit, L.; Heidmann, T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 13013–13018. [Google Scholar] [CrossRef] [PubMed]
- Best, S.; Le Tissier, P.; Towers, G.; Stoye, J.P. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 1996, 382, 826–829. [Google Scholar] [CrossRef]
- Bénit, L.; De Parseval, N.; Casella, J.F.; Callebaut, I.; Cordonnier, A.; Heidmann, T. Cloning of a new murine endogenous retrovirus, MuERV-L, with strong similarity to the human HERV-L element and with a gag coding sequence closely related to the Fv1 restriction gene. J. Virol. 1997, 71, 5652–5657. [Google Scholar] [CrossRef] [PubMed]
- Pincus, T.; Hartley, J.W.; Rowe, W.P. A major genetic locus affecting resistance to infection with murine leukemia viruses. I. Tissue culture studies of naturally occurring viruses. J. Exp. Med. 1971, 133, 1219–1233. [Google Scholar] [CrossRef] [PubMed]
- Johnson, W.E. Origins and evolutionary consequences of ancient endogenous retroviruses. Nat. Rev. Microbiol. 2019, 17, 355–370. [Google Scholar] [CrossRef]
- Boso, G.; Buckler-White, A.; Kozak, C.A. Ancient Evolutionary Origin and Positive Selection of the Retroviral Restriction Factor in Muroid Rodents. J. Virol. 2018, 92, e00850-18. [Google Scholar] [CrossRef]
- Young, G.R.; Yap, M.W.; Michaux, J.R.; Steppan, S.J.; Stoye, J.P. Evolutionary journey of the retroviral restriction gene. Proc. Natl. Acad. Sci. USA 2018, 115, 10130–10135. [Google Scholar] [CrossRef]
- Yap, M.W.; Colbeck, E.; Ellis, S.A.; Stoye, J.P. Evolution of the retroviral restriction gene Fv1: Inhibition of non-MLV retroviruses. PLoS Pathog. 2014, 10, e1003968. [Google Scholar] [CrossRef] [Green Version]
- Mortuza, G.B.; Haire, L.F.; Stevens, A.; Smerdon, S.J.; Stoye, J.P.; Taylor, I.A. High-resolution structure of a retroviral capsid hexameric amino-terminal domain. Nature 2004, 431, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Myrum, C.; Moreno-Castilla, P.; Rapp, P.R. ‘Arc’-hitecture of normal cognitive aging. Ageing Res. Rev. 2022, 80, 101678. [Google Scholar] [CrossRef] [PubMed]
- Pastuzyn, E.D.; Day, C.E.; Kearns, R.B.; Kyrke-Smith, M.; Taibi, A.V.; McCormick, J.; Yoder, N.; Belnap, D.M.; Erlendsson, S.; Morado, D.R.; et al. The Neuronal Gene Arc Encodes a Repurposed Retrotransposon Gag Protein that Mediates Intercellular RNA Transfer. Cell 2018, 172, 275–288.e18. [Google Scholar] [CrossRef] [PubMed]
- Matteucci, C.; Balestrieri, E.; Argaw-Denboba, A.; Sinibaldi-Vallebona, P. Human endogenous retroviruses role in cancer cell stemness. Semin. Cancer Biol. 2018, 53, 17–30. [Google Scholar] [CrossRef]
- Lemaitre, C.; Tsang, J.; Bireau, C.; Heidmann, T.; Dewannieux, M. A human endogenous retrovirus-derived gene that can contribute to oncogenesis by activating the ERK pathway and inducing migration and invasion. PLoS Pathog. 2017, 13, e1006451. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Li, Z.; Wan, X.; Wang, Y.; Dong, J. Human endogenous retroviral K element encodes fusogenic activity in melanoma cells. J. Carcinog. 2013, 12, 5. [Google Scholar] [CrossRef]
- Argaw-Denboba, A.; Balestrieri, E.; Serafino, A.; Cipriani, C.; Bucci, I.; Sorrentino, R.; Sciamanna, I.; Gambacurta, A.; Sinibaldi-Vallebona, P.; Matteucci, C. HERV-K activation is strictly required to sustain CD133+ melanoma cells with stemness features. J. Exp. Clin. Cancer Res. 2017, 36, 20. [Google Scholar] [CrossRef]
- Li, M.; Radvanyi, L.; Yin, B.; Rycaj, K.; Li, J.; Chivukula, R.; Lin, K.; Lu, Y.; Shen, J.; Chang, D.Z.; et al. Downregulation of Human Endogenous Retrovirus Type K (HERV-K) Viral RNA in Pancreatic Cancer Cells Decreases Cell Proliferation and Tumor Growth. Clin. Cancer Res. 2017, 23, 5892–5911. [Google Scholar] [CrossRef]
- Kaufmann, S.; Sauter, M.; Schmitt, M.; Baumert, B.; Best, B.; Boese, A.; Roemer, K.; Mueller-Lantzsch, N. Human endogenous retrovirus protein Rec interacts with the testicular zinc-finger protein and androgen receptor. J. Gen. Virol. 2010, 91, 1494–1502. [Google Scholar] [CrossRef]
- Hanke, K.; Chudak, C.; Kurth, R.; Bannert, N. The Rec protein of HERV-K(HML-2) upregulates androgen receptor activity by binding to the human small glutamine-rich tetratricopeptide repeat protein (hSGT). Int J. Cancer 2013, 132, 556–567. [Google Scholar] [CrossRef]
- Armbruester, V.; Sauter, M.; Roemer, K.; Best, B.; Hahn, S.; Nty, A.; Schmid, A.; Philipp, S.; Mueller, A.; Mueller-Lantzsch, N. Np9 protein of human endogenous retrovirus K interacts with ligand of numb protein X. J. Virol. 2004, 78, 10310–10319. [Google Scholar] [CrossRef]
- Chen, T.; Meng, Z.; Gan, Y.; Wang, X.; Xu, F.; Gu, Y.; Xu, X.; Tang, J.; Zhou, H.; Zhang, X.; et al. The viral oncogene Np9 acts as a critical molecular switch for co-activating β-catenin, ERK, Akt and Notch1 and promoting the growth of human leukemia stem/progenitor cells. Leukemia 2013, 27, 1469–1478. [Google Scholar] [CrossRef]
- Denne, M.; Sauter, M.; Armbruester, V.; Licht, J.D.; Roemer, K.; Mueller-Lantzsch, N. Physical and functional interactions of human endogenous retrovirus proteins Np9 and rec with the promyelocytic leukemia zinc finger protein. J. Virol. 2007, 81, 5607–5616. [Google Scholar] [CrossRef]
- Wang, F.; Li, X.; Xie, X.; Zhao, L.; Chen, W. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett. 2008, 582, 1919–1927. [Google Scholar] [CrossRef]
- Hou, P.; Zhao, Y.; Li, Z.; Yao, R.; Ma, M.; Gao, Y.; Zhao, L.; Zhang, Y.; Huang, B.; Lu, J. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014, 5, e1287. [Google Scholar] [CrossRef]
- Zhan, H.X.; Wang, Y.; Li, C.; Xu, J.W.; Zhou, B.; Zhu, J.K.; Han, H.F.; Wang, L.; Wang, Y.S.; Hu, S.Y. LincRNA-ROR promotes invasion, metastasis and tumor growth in pancreatic cancer through activating ZEB1 pathway. Cancer Lett. 2016, 374, 261–271. [Google Scholar] [CrossRef]
- Takahashi, K.; Yan, I.K.; Kogure, T.; Haga, H.; Patel, T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio 2014, 4, 458–467. [Google Scholar] [CrossRef]
- Sin, H.S.; Huh, J.W.; Kim, D.S.; Kang, D.W.; Min, D.S.; Kim, T.H.; Ha, H.S.; Kim, H.H.; Lee, S.Y.; Kim, H.S. Transcriptional control of the HERV-H LTR element of the GSDML gene in human tissues and cancer cells. Arch. Virol 2006, 151, 1985–1994. [Google Scholar] [CrossRef]
- Sun, Q.; Yang, J.; Xing, G.; Sun, Q.; Zhang, L.; He, F. Expression of GSDML Associates with Tumor Progression in Uterine Cervix Cancer. Transl. Oncol. 2008, 1, 73–83. [Google Scholar] [CrossRef]
- Gosenca, D.; Gabriel, U.; Steidler, A.; Mayer, J.; Diem, O.; Erben, P.; Fabarius, A.; Leib-Mösch, C.; Hofmann, W.-K.; Seifarth, W. HERV-E-mediated modulation of PLA2G4A transcription in urothelial carcinoma. PLoS ONE 2012, 7, e49341. [Google Scholar] [CrossRef] [Green Version]
- Bjerregaard, B.; Holck, S.; Christensen, I.J.; Larsson, L.I. Syncytin is involved in breast cancer-endothelial cell fusions. Cell. Mol. Life Sci. 2006, 63, 1906–1911. [Google Scholar] [CrossRef]
- Deng, B.; Xu, W.; Wang, Z.; Liu, C.; Lin, P.; Li, B.; Huang, Q.; Yang, J.; Zhou, H.; Qu, L. An LTR retrotransposon-derived lncRNA interacts with RNF169 to promote homologous recombination. EMBO Rep. 2019, 20, e47650. [Google Scholar] [CrossRef]
- Lamprecht, B.; Walter, K.; Kreher, S.; Kumar, R.; Hummel, M.; Lenze, D.; Köchert, K.; Bouhlel, M.A.; Richter, J.; Soler, E.; et al. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat. Med. 2010, 16, 571–579. [Google Scholar] [CrossRef]
- Wu, Y.; Zhao, Y.; Huan, L.; Zhao, J.; Zhou, Y.; Xu, L.; Hu, Z.; Liu, Y.; Chen, Z.; Wang, L.; et al. An LTR Retrotransposon-Derived Long Noncoding RNA lncMER52A Promotes Hepatocellular Carcinoma Progression by Binding p120-Catenin. Cancer Res. 2020, 80, 976–987. [Google Scholar] [CrossRef]
- Gibb, E.A.; Warren, R.L.; Wilson, G.W.; Brown, S.D.; Robertson, G.A.; Morin, G.B.; Holt, R.A. Activation of an endogenous retrovirus-associated long non-coding RNA in human adenocarcinoma. Genome Med. 2015, 7, 22. [Google Scholar] [CrossRef]
- Cañadas, I.; Thummalapalli, R.; Kim, J.W.; Kitajima, S.; Jenkins, R.W.; Christensen, C.L.; Campisi, M.; Kuang, Y.; Zhang, Y.; Gjini, E.; et al. Tumor innate immunity primed by specific interferon-stimulated endogenous retroviruses. Nat. Med. 2018, 24, 1143–1150. [Google Scholar] [CrossRef]
- Lee, E.; Iskow, R.; Yang, L.; Gokcumen, O.; Haseley, P.; Luquette, L.J., III; Lohr, J.G.; Harris, C.C.; Ding, L.; Wilson, R.K.; et al. Landscape of somatic retrotransposition in human cancers. Science 2012, 337, 967–971. [Google Scholar] [CrossRef]
- Beyer, U.; Moll-Rocek, J.; Moll, U.M.; Dobbelstein, M. Endogenous retrovirus drives hitherto unknown proapoptotic p63 isoforms in the male germ line of humans and great apes. Proc. Natl. Acad. Sci. USA 2011, 108, 3624–3629. [Google Scholar] [CrossRef]
- Beyer, U.; Kronung, S.K.; Leha, A.; Walter, L.; Dobbelstein, M. Comprehensive identification of genes driven by ERV9-LTRs reveals TNFRSF10B as a re-activatable mediator of testicular cancer cell death. Cell Death Differ. 2016, 23, 64–75. [Google Scholar] [CrossRef]
- Strick, R.; Ackermann, S.; Langbein, M.; Swiatek, J.; Schubert, S.W.; Hashemolhosseini, S.; Koscheck, T.; Fasching, P.A.; Schild, R.L.; Beckmann, M.W.; et al. Proliferation and cell-cell fusion of endometrial carcinoma are induced by the human endogenous retroviral Syncytin-1 and regulated by TGF-beta. J. Mol. Med. 2007, 85, 23–38. [Google Scholar] [CrossRef]
- Zhou, F.; Li, M.; Wei, Y.; Lin, K.; Lu, Y.; Shen, J.; Johanning, G.L.; Wang-Johanning, F. Activation of HERV-K Env protein is essential for tumorigenesis and metastasis of breast cancer cells. Oncotarget 2016, 7, 84093–84117. [Google Scholar] [CrossRef]
- Grandi, N.; Tramontano, E. HERV Envelope Proteins: Physiological Role and Pathogenic Potential in Cancer and Autoimmunity. Front. Microbiol. 2018, 9, 462. [Google Scholar] [CrossRef]
- Goke, J.; Ng, H.H. CTRL+INSERT: Retrotransposons and their contribution to regulation and innovation of the transcriptome. EMBO Rep. 2016, 17, 1131–1144. [Google Scholar] [CrossRef]
- Leucci, E.; Vendramin, R.; Spinazzi, M.; Laurette, P.; Fiers, M.; Wouters, J.; Radaelli, E.; Eyckerman, S.; Leonelli, C.; Vanderheyden, K.; et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature 2016, 531, 518–522. [Google Scholar] [CrossRef]
- Flockhart, R.J.; Webster, D.E.; Qu, K.; Mascarenhas, N.; Kovalski, J.; Kretz, M.; Khavari, P.A. BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res. 2012, 22, 1006–1014. [Google Scholar] [CrossRef]
- Zhou, B.; Qi, F.; Wu, F.; Nie, H.; Song, Y.; Shao, L.; Han, J.; Wu, Z.; Saiyin, H.; Wei, G.; et al. Endogenous Retrovirus-Derived Long Noncoding RNA Enhances Innate Immune Responses via Derepressing RELA Expression. mBio 2019, 10, e00937-19. [Google Scholar] [CrossRef]
- Chiappinelli, K.B.; Strissel, P.L.; Desrichard, A.; Li, H.; Henke, C.; Akman, B.; Hein, A.; Rote, N.S.; Cope, L.M.; Snyder, A.; et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell 2015, 162, 974–986. [Google Scholar] [CrossRef] [PubMed]
- Roulois, D.; Loo Yau, H.; Singhania, R.; Wang, Y.; Danesh, A.; Shen, S.Y.; Han, H.; Liang, G.; Jones, P.A.; Pugh, T.J.; et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell 2015, 162, 961–973. [Google Scholar] [CrossRef]
- Otsuki, S.; Saito, T.; Taylor, S.; Li, D.; Moonen, J.-R.; Marciano, D.P.; Harper, R.L.; Cao, A.; Wang, L.; Ariza, M.E.; et al. Monocyte-released HERV-K dUTPase engages TLR4 and MCAM causing endothelial mesenchymal transition. JCI Insight 2021, 6, e146416. [Google Scholar] [CrossRef]
- Saito, T.; Miyagawa, K.; Chen, S.-Y.; Tamosiuniene, R.; Wang, L.; Sharpe, O.; Samayoa, E.; Harada, D.; Moonen, J.-R.A.J.; Cao, A.; et al. Upregulation of Human Endogenous Retrovirus-K Is Linked to Immunity and Inflammation in Pulmonary Arterial Hypertension. Circulation 2017, 136, 1920–1935. [Google Scholar] [CrossRef]
- Taylor, S.; Isobe, S.; Cao, A.; Contrepois, K.; Benayoun, B.A.; Jiang, L.; Wang, L.; Melemenidis, S.; Ozen, M.O.; Otsuki, S.; et al. Endogenous Retroviral Elements Generate Pathologic Neutrophils in Pulmonary Arterial Hypertension. Am. J. Respir. Crit. Care Med. 2022. [Google Scholar] [CrossRef] [PubMed]
- Ariza, M.-E.; Williams, M.V. A human endogenous retrovirus K dUTPase triggers a TH1, TH17 cytokine response: Does it have a role in psoriasis? J. Investig. Dermatol. 2011, 131, 2419–2427. [Google Scholar] [CrossRef] [PubMed]
- Fleischmann, J.; Kremmer, E.; Müller, S.; Sommer, P.; Kirchner, T.; Niedobitek, G.; Grässer, F.A. Expression of deoxyuridine triphosphatase (dUTPase) in colorectal tumours. Int. J. Cancer 1999, 84, 614–617. [Google Scholar] [CrossRef]
- Kawahara, A.; Akagi, Y.; Hattori, S.; Mizobe, T.; Shirouzu, K.; Ono, M.; Yanagawa, T.; Kuwano, M.; Kage, M. Higher expression of deoxyuridine triphosphatase (dUTPase) may predict the metastasis potential of colorectal cancer. J. Clin. Pathol. 2009, 62, 364–369. [Google Scholar] [CrossRef]
- Chen, C.-W.; Tsao, N.; Huang, L.-Y.; Yen, Y.; Liu, X.; Lehman, C.; Wang, Y.-H.; Tseng, M.-C.; Chen, Y.-J.; Ho, Y.-C.; et al. The Impact of dUTPase on Ribonucleotide Reductase-Induced Genome Instability in Cancer Cells. Cell Rep. 2016, 16, 1287–1299. [Google Scholar] [CrossRef]
- Wallace, T.A.; Downey, R.F.; Seufert, C.J.; Schetter, A.; Dorsey, T.H.; Johnson, C.A.; Goldman, R.; Loffredo, C.A.; Yan, P.; Sullivan, F.J.; et al. Elevated HERV-K mRNA expression in PBMC is associated with a prostate cancer diagnosis particularly in older men and smokers. Carcinogenesis 2014, 35, 2074–2083. [Google Scholar] [CrossRef]
- Wang-Johanning, F.; Rycaj, K.; Plummer, J.B.; Li, M.; Yin, B.; Frerich, K.; Garza, J.G.; Shen, J.; Lin, K.; Yan, P.; et al. Immunotherapeutic potential of anti-human endogenous retrovirus-K envelope protein antibodies in targeting breast tumors. J. Natl. Cancer Inst. 2012, 104, 189–210. [Google Scholar] [CrossRef]
- Krishnamurthy, J.; Rabinovich, B.A.; Mi, T.; Switzer, K.C.; Olivares, S.; Maiti, S.N.; Plummer, J.B.; Singh, H.; Kumaresan, P.R.; Huls, H.M.; et al. Genetic Engineering of T Cells to Target HERV-K, an Ancient Retrovirus on Melanoma. Clin. Cancer Res. 2015, 21, 3241–3251. [Google Scholar] [CrossRef]
- Villeponteau, B. The heterochromatin loss model of aging. Exp. Gerontol. 1997, 32, 383–394. [Google Scholar] [CrossRef]
- Pal, S.; Tyler, J.K. Epigenetics and aging. Sci. Adv. 2016, 2, e1600584. [Google Scholar] [CrossRef] [Green Version]
- Barbot, W.; Dupressoir, A.; Lazar, V.; Heidmann, T. Epigenetic regulation of an IAP retrotransposon in the aging mouse: Progressive demethylation and de-silencing of the element by its repetitive induction. Nucleic Acids Res. 2002, 30, 2365–2373. [Google Scholar] [CrossRef] [PubMed]
- De Cecco, M.; Criscione, S.W.; Peterson, A.L.; Neretti, N.; Sedivy, J.M.; Kreiling, J.A. Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging 2013, 5, 867–883. [Google Scholar] [CrossRef] [PubMed]
- Nevalainen, T.; Autio, A.; Mishra, B.H.; Marttila, S.; Jylha, M.; Hurme, M. Aging-associated patterns in the expression of human endogenous retroviruses. PLoS ONE 2018, 13, e0207407. [Google Scholar] [CrossRef]
- Balestrieri, E.; Pica, F.; Matteucci, C.; Zenobi, R.; Sorrentino, R.; Argaw-Denboba, A.; Cipriani, C.; Bucci, I.; Sinibaldi-Vallebona, P. Transcriptional activity of human endogenous retroviruses in human peripheral blood mononuclear cells. Biomed. Res. Int. 2015, 2015, 164529. [Google Scholar] [CrossRef] [PubMed]
- McGinley, M.P.; Goldschmidt, C.H.; Rae-Grant, A.D. Diagnosis and Treatment of Multiple Sclerosis: A Review. JAMA 2021, 325, 765–779. [Google Scholar] [CrossRef] [PubMed]
- Perron, H.; Geny, C.; Laurent, A.; Mouriquand, C.; Pellat, J.; Perret, J.; Seigneurin, J.M. Leptomeningeal cell line from multiple sclerosis with reverse transcriptase activity and viral particles. Res. Virol. 1989, 140, 551–561. [Google Scholar] [CrossRef]
- Perron, H.; Lalande, B.; Gratacap, B.; Laurent, A.; Genoulaz, O.; Geny, C.; Mallaret, M.; Schuller, E.; Stoebner, P.; Seigneurin, J.M. Isolation of retrovirus from patients with multiple sclerosis. Lancet 1991, 337, 862–863. [Google Scholar] [CrossRef]
- Komurian-Pradel, F.; Paranhos-Baccala, G.; Bedin, F.; Ounanian-Paraz, A.; Sodoyer, M.; Ott, C.; Rajoharison, A.; Garcia, E.; Mallet, F.; Mandrand, B.; et al. Molecular cloning and characterization of MSRV-related sequences associated with retrovirus-like particles. Virology 1999, 260, 1–9. [Google Scholar] [CrossRef]
- Blond, J.L.; Besème, F.; Duret, L.; Bouton, O.; Bedin, F.; Perron, H.; Mandrand, B.; Mallet, F. Molecular characterization and placental expression of HERV-W, a new human endogenous retrovirus family. J. Virol. 1999, 73, 1175–1185. [Google Scholar] [CrossRef]
- Rolland, A.; Jouvin-Marche, E.; Viret, C.; Faure, M.; Perron, H.; Marche, P.N. The envelope protein of a human endogenous retrovirus-W family activates innate immunity through CD14/TLR4 and promotes Th1-like responses. J. Immunol. 2006, 176, 7636–7644. [Google Scholar] [CrossRef] [Green Version]
- Kremer, D.; Schichel, T.; Förster, M.; Tzekova, N.; Bernard, C.; van der Valk, P.; van Horssen, J.; Hartung, H.-P.; Perron, H.; Küry, P. Human endogenous retrovirus type W envelope protein inhibits oligodendroglial precursor cell differentiation. Ann. Neurol. 2013, 74, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Römer, C. Viruses and Endogenous Retroviruses as Roots for Neuroinflammation and Neurodegenerative Diseases. Front. Neurosci. 2021, 15, 648629. [Google Scholar] [CrossRef]
- Kremer, D.; Gruchot, J.; Weyers, V.; Oldemeier, L.; Göttle, P.; Healy, L.; Ho Jang, J.; Kang T Xu, Y.; Volsko, C.; Dutta, R.; et al. pHERV-W envelope protein fuels microglial cell-dependent damage of myelinated axons in multiple sclerosis. Proc. Natl. Acad. Sci. USA 2019, 116, 15216–15225. [Google Scholar] [CrossRef] [PubMed]
- Kremer, D.; Förster, M.; Schichel, T.; Göttle, P.; Hartung, H.-P.; Perron, H.; Küry, P. The neutralizing antibody GNbAC1 abrogates HERV-W envelope protein-mediated oligodendroglial maturation blockade. Mult. Scler. J. 2015, 21, 1200–1203. [Google Scholar] [CrossRef] [PubMed]
- Hansen, B.; Oturai, A.B.; Harbo, H.F.; Celius, E.G.; Nissen, K.K.; Laska, M.J.; Søndergaard, H.B.; Petersen, T.; Nexø, B.A. Genetic association of multiple sclerosis with the marker rs391745 near the endogenous retroviral locus HERV-Fc1: Analysis of disease subtypes. PLoS ONE 2011, 6, e26438. [Google Scholar] [CrossRef]
- Christensen, T.; Dissing Sørensen, P.; Riemann, H.; Hansen, H.J.; Munch, M.; Haahr, S.; Møller-Larsen, A. Molecular characterization of HERV-H variants associated with multiple sclerosis. Acta Neurol. Scand. 2000, 101, 229–238. [Google Scholar] [CrossRef]
- Brudek, T.; Christensen, T.; Aagaard, L.; Petersen, T.; Hansen, H.J.; Møller-Larsen, A. B cells and monocytes from patients with active multiple sclerosis exhibit increased surface expression of both HERV-H Env and HERV-W Env, accompanied by increased seroreactivity. Retrovirology 2009, 6, 104. [Google Scholar] [CrossRef]
- Muradrasoli, S.; Forsman, A.; Hu, L.; Blikstad, V.; Blomberg, J. Development of real-time PCRs for detection and quantitation of human MMTV-like (HML) sequences HML expression in human tissues. J. Virol. Methods 2006, 136, 83–92. [Google Scholar] [CrossRef]
- Viola, M.V.; Frazier, M.; White, L.; Brody, J.; Spiegelman, S. RNA-instructed DNA polymerase activity in a cytoplasmic particulate fraction in brains from Guamanian patients. J. Exp. Med. 1975, 142, 483–494. [Google Scholar] [CrossRef]
- Andrews, W.D.; Tuke, P.W.; Al-Chalabi, A.; Gaudin, P.; Ijaz, S.; Parton, M.J.; Garson, J.A. Detection of reverse transcriptase activity in the serum of patients with motor neurone disease. J. Med. Virol. 2000, 61, 527–532. [Google Scholar] [CrossRef]
- McCormick, A.L.; Brown, R.H.; Cudkowicz, M.E.; Al-Chalabi, A.; Garson, J.A. Quantification of reverse transcriptase in ALS and elimination of a novel retroviral candidate. Neurology 2008, 70, 278–283. [Google Scholar] [CrossRef] [PubMed]
- Steele, A.J.; Al-Chalabi, A.; Ferrante, K.; Cudkowicz, M.E.; Brown, R.H.; Garson, J.A. Detection of serum reverse transcriptase activity in patients with ALS and unaffected blood relatives. Neurology 2005, 64, 454–458. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Fan, Y.; Laurie, P.; Kim, J.M.H.; Ravits, J. No evidence of HIV pol gene in spinal cord tissues in sporadic ALS by real-time RT-PCR. Amyotroph. Lateral Scler. 2010, 11, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Douville, R.; Liu, J.; Rothstein, J.; Nath, A. Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Ann. Neurol. 2011, 69, 141–151. [Google Scholar] [CrossRef]
- Li, W.; Lee, M.-H.; Henderson, L.; Tyagi, R.; Bachani, M.; Steiner, J.; Campanac, E.; Hoffman, D.A.; von Geldern, G.; Johnson, K.; et al. Human endogenous retrovirus-K contributes to motor neuron disease. Sci. Transl. Med. 2015, 7, 307ra153. [Google Scholar] [CrossRef]
- Manghera, M.; Ferguson-Parry, J.; Lin, R.; Douville, R.N. NF-κB and IRF1 Induce Endogenous Retrovirus K Expression via Interferon-Stimulated Response Elements in Its 5′ Long Terminal Repeat. J. Virol. 2016, 90, 9338–9349. [Google Scholar] [CrossRef]
- Oluwole, S.O.A.; Yao, Y.; Conradi, S.; Kristensson, K.; Karlsson, H. Elevated levels of transcripts encoding a human retroviral envelope protein (syncytin) in muscles from patients with motor neuron disease. Amyotroph. Lateral Scler. 2007, 8, 67–72. [Google Scholar] [CrossRef]
- Balestrieri, E.; Arpino, C.; Matteucci, C.; Sorrentino, R.; Pica, F.; Alessandrelli, R.; Coniglio, A.; Curatolo, P.; Rezza, G.; Macciardi, F.; et al. HERVs expression in Autism Spectrum Disorders. PLoS ONE 2012, 7, e48831. [Google Scholar] [CrossRef]
- Heidmann, O.; Béguin, A.; Paternina, J.; Berthier, R.; Deloger, M.; Bawa, O.; Heidmann, T. HEMO, an ancestral endogenous retroviral envelope protein shed in the blood of pregnant women and expressed in pluripotent stem cells and tumors. Proc. Natl. Acad. Sci. USA 2017, 114, E6642–E6651. [Google Scholar] [CrossRef]
- Balestrieri, E.; Cipriani, C.; Matteucci, C.; Benvenuto, A.; Coniglio, A.; Argaw-Denboba, A.; Toschi, N.; Bucci, I.; Miele, M.T.; Grelli, S.; et al. Children with Autism Spectrum Disorder and Their Mothers Share Abnormal Expression of Selected Endogenous Retroviruses Families and Cytokines. Front. Immunol 2019, 10, 2244. [Google Scholar] [CrossRef] [Green Version]
- Johnston, J.B.; Silva, C.; Holden, J.; Warren, K.G.; Clark, A.W.; Power, C. Monocyte activation and differentiation augment human endogenous retrovirus expression: Implications for inflammatory brain diseases. Ann. Neurol 2001, 50, 434–442. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Samimi, H.; Gamez, M.; Zare, H.; Frost, B. Pathogenic tau-induced piRNA depletion promotes neuronal death through transposable element dysregulation in neurodegenerative tauopathies. Nat. Neurosci. 2018, 21, 1038–1048. [Google Scholar] [CrossRef]
- Dembny, P.; Newman, A.G.; Singh, M.; Hinz, M.; Szczepek, M.; Krüger, C.; Adalbert, R.; Dzaye, O.; Trimbuch, T.; Wallach, T.; et al. Human endogenous retrovirus HERV-K(HML-2) RNA causes neurodegeneration through Toll-like receptors. JCI Insight 2020, 5, e131093. [Google Scholar] [CrossRef]
- Mak, M.; Samochowiec, J.; Frydecka, D.; Pełka-Wysiecka, J.; Szmida, E.; Karpiński, P.; Sąsiadek, M.M.; Piotrowski, P.; Samochowiec, A.; Misiak, B. First-episode schizophrenia is associated with a reduction of HERV-K methylation in peripheral blood. Psychiatry Res. 2019, 271, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Perron, H.; Hamdani, N.; Faucard, R.; Lajnef, M.; Jamain, S.; Daban-Huard, C.; Sarrazin, S.; LeGuen, E.; Houenou, J.; Delavest, M.; et al. Molecular characteristics of Human Endogenous Retrovirus type-W in schizophrenia and bipolar disorder. Transl. Psychiatry 2012, 2, e201. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Sabunciyan, S.; Yolken, R.H.; Lee, D.; Kim, S.; Karlsson, H. Transcription of human endogenous retroviruses in human brain by RNA-seq analysis. PLoS ONE 2019, 14, e0207353. [Google Scholar] [CrossRef]
- Huang, W.; Li, S.; Hu, Y.; Yu, H.; Luo, F.; Zhang, Q.; Zhu, F. Implication of the env gene of the human endogenous retrovirus W family in the expression of BDNF and DRD3 and development of recent-onset schizophrenia. Schizophr. Bull. 2011, 37, 988–1000. [Google Scholar] [CrossRef]
- Huang, W.-J.; Liu, Z.-C.; Wei, W.; Wang, G.-H.; Wu, J.-G.; Zhu, F. Human endogenous retroviral pol RNA and protein detected and identified in the blood of individuals with schizophrenia. Schizophr. Res. 2006, 83, 193–199. [Google Scholar] [CrossRef]
- Karlsson, H.; Bachmann, S.; Schröder, J.; McArthur, J.; Torrey, E.F.; Yolken, R.H. Retroviral RNA identified in the cerebrospinal fluids and brains of individuals with schizophrenia. Proc. Natl. Acad. Sci. USA 2001, 98, 4634–4639. [Google Scholar] [CrossRef]
- Johansson, E.M.; Bouchet, D.; Tamouza, R.; Ellul, P.; Morr, A.S.; Avignone, E.; Germi, R.; Leboyer, M.; Perron, H.; Groc, L. Human endogenous retroviral protein triggers deficit in glutamate synapse maturation and behaviors associated with psychosis. Sci. Adv. 2020, 6, eabc0708. [Google Scholar] [CrossRef]

| ERVs Classes | Exogenous Counterpart | Representative ERVs |
|---|---|---|
| Class I | Gammaretrovirus Epsilonretrovirus | FeLV, GALV, KoRV, McERV, MDEV, MuERV-C, MuRRS, MuRVY, MuLV, GLN, VL30, PERV, HERV-E, F, H, I, P, R, T, W, HERV-FRD |
| Class II | Alpharetrovirus Betaretrovirus Deltaretrovirus Lentivirus | ALV, IAP, MMTV, MPMV, MusD/ETn, MINERVa, RELIK, HERV-K (HML-1, 2, 3, 4, 5, 6, 7, 8, 9, 10) |
| Class III | Spumaretrovirus | MuERV-L, HERV-L |
| HERVs | HERVs Products/Activities | Oncogenic Mechanisms |
|---|---|---|
| HERV-K (HML-2) | ENV protein | ENV induces EMT and activates ERK pathway in breast cancer [105]. ENV mediates intercellular fusion in melanoma [106]. ENV maintains CD133+ melanoma cells with stemness features [107]. ENV promotes pancreatic cancer proliferation, tumorigenesis, and metastasis by activating RAS/MEK/ERK and JNK/c-Jun signaling pathways [108]. |
| Rec protein | Rec activates c-MYC by overcoming the transcriptional repression of testicular zinc-finger protein (TZFP) for c-MYC promoter [109]. Rec relieves the repression of androgen receptor (AR) activity by forming a trimeric complex with TZFP and AR [109], or binds to the human small glutamine-rich tetratricopeptide repeat protein (hSGT) [110]. | |
| Np9 protein | Np9 interacts with ligand of Numb protein X, affecting tumorigenesis through the LNX/Numb/Notch pathway [111]. Np9 as a critical molecular switch of multiple signaling pathways in leukemia [112]. Rec and Np9 derepressed c-MYC through the inhibition of promyelocytic leukemia zinc-finger protein (PLZF) [113]. | |
| HERV-H | lncRNA | UCA1 enhances proliferation, motility, invasion, and drug resistance of bladder cancer [114]. linc-ROR contributes to progression, metastasis, or chemoresistance in breast cancer [115], pancreatic cancer [116], and hepatocellular carcinoma [117]. |
| cis-regulatory element | LTR acts as alternative promoter for GSDML in cervical cancer [118,119]. | |
| HERV-E | cis-regulatory element | Modulating PLA2G4A transcription in urothelial cancer [120]. |
| syncytin-1/ERVW-1 | ENV protein | Syncytin-1 mediates cancer–endothelial cell fusions in breast cancer [121]. |
| ERV-9 | lncRNA | PRLH1 plays an important role in the formation of RNA–protein complex that promotes the HR-mediated DSB repair [122]. |
| MaLR | cis-regulatory element | LTR acts as alternative promoter for CSF1R in Hodgkin’s lymphoma [123]. |
| MER52A | lncRNA | lncMER52A promotes invasion and metastasis of hepatocellular carcinoma cells by stabilizing p120-catenin [124]. |
| MER48 | lncRNA/cis-regulatory element | lncRNA EVADR is associated with adenocarcinomas, and a MER48 ERV element acts as an active promoter for its specific activation [125]. |
| Multiple HERVs | dsRNA | dsRNAs derived from the bi-directional transcription of HERVs induce an immunosuppressive tumor microenvironment [91,126]. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.; Pan, J.; Cong, Y.; Mao, J. Transcriptional Regulation of Endogenous Retroviruses and Their Misregulation in Human Diseases. Int. J. Mol. Sci. 2022, 23, 10112. https://doi.org/10.3390/ijms231710112
Zhang Q, Pan J, Cong Y, Mao J. Transcriptional Regulation of Endogenous Retroviruses and Their Misregulation in Human Diseases. International Journal of Molecular Sciences. 2022; 23(17):10112. https://doi.org/10.3390/ijms231710112
Chicago/Turabian StyleZhang, Qian, Juan Pan, Yusheng Cong, and Jian Mao. 2022. "Transcriptional Regulation of Endogenous Retroviruses and Their Misregulation in Human Diseases" International Journal of Molecular Sciences 23, no. 17: 10112. https://doi.org/10.3390/ijms231710112
APA StyleZhang, Q., Pan, J., Cong, Y., & Mao, J. (2022). Transcriptional Regulation of Endogenous Retroviruses and Their Misregulation in Human Diseases. International Journal of Molecular Sciences, 23(17), 10112. https://doi.org/10.3390/ijms231710112








